1. INTRODUCTION
Globally, flowers play a substantial role in signifying numerous traditions. They also carry cultural and esthetic meanings that cross linguistic and religious boundaries, fostering social cohesion. In recent years, the phenomenal growth in floriculture has increased significantly. Market conditions of demand and supply have led to highly volatile pricing and the consideration of flowers as a market commodity [1]. Jasmine (Jasminum spp., family Oleaceae) is a flowering plant known for its fragrance [2]. Over 200 species of this genus exhibit an array of preferences and hierarchies. Some jasmine cultivars have received geographical indication (GI) tags because of their exceptional qualities from geo-specific growth conditions. Udupi jasmine [Jasminum sambac (L.) Aiton] locally known as Shankarpura mallige, received GI status on July 24, 2006 (GI No. 70; Certificate No. 36), documented at https://search.ipindia.gov.in/girpublic/application/details/70 for its fragrance and exceptional shelf life owing to its geographical origin [3].
This variety, grown in a small geographical area of 4.22 km2, has played a vibrant role not only in the cultural sphere but also in the socioeconomic condition of the jasmine growers. Most importantly, the production and marketing of the Udupi Jasmine helped alleviate the cultivators’ economic situation, impacting a better quality of life in about 6000 households [4]. There is a huge demand for these flowers on selected occasions and in the festival season, which makes them very expensive. Farmers are prone to use synthetic hormones, chemical fertilizers, and pesticides to meet soaring demand and gain profit. These practices have increased crop susceptibility to pests, diseases, and pollution. Moreover, women and youth mostly carry out cultivation, crop maintenance, harvesting, and marketing, making them vulnerable to the ill effects of chemical inputs. Increased global awareness of the adverse effects of chemical cultivation practices has shifted toward environmentally sound crop production. This has emphasized research favoring eco-friendly practices. There is a growing body of knowledge supporting plant growth-promoting rhizobacteria (PGPR) as a valuable alternative. PGPRs are microbes that inhabit the rhizosphere and improve plant health without any anomalies to the agroecosystem and human health [5].
Pseudomonas spp. are Gram-negative, rod-shaped, beneficial rhizobacteria found in the rhizosphere [6]. Although some species of rhizobacteria have been reported to cause plant diseases, most of them are known for plant growth-promoting (PGP) properties [7]. Pseudomonas spp. with PGP properties rapidly colonize by utilizing exudates secreted by plant roots [8,9]. The well-established mechanisms of plant growth promotion include the production of enzymes, metabolites, phytohormones, mineral phosphate solubilization, siderophores, and imparting resilience against biotic and abiotic stresses. Currently, other PGP microbes are not used to cultivate Udupi jasmine, mainly because of their nescience [3]. While PGP Pseudomonas are available in the local markets, they fail to bring about the desired effect as they are non-native to that location and fail to adapt to the soil conditions prevalent in jasmine-growing soils. The present study attempts to isolate native Pseudomonas strains that adapt easily to soil conditions, protecting them from the indiscriminate use of chemicals, which leads to decreased cultivation costs and soil degradation.
2. MATERIALS AND METHODS
2.1. Collection of Rhizosphere Soil Samples
Rhizosphere soil samples were collected from five well-maintained gardens (labelled as sites 1, 2, 3, 4, and 5) in Shankarpura, Udupi District, Karnataka, India. The geographic coordinates of sites 1, 2, 3, 4, and 5 are 13.259139 N, 74.771672 E; 13.256178 N, 74.774898 E; 13.261779 N, 74.767479 E; 13.261507 N, 74.767177 E, and 13.260136 N, 74.773157 E, respectively [Table 1]. Three healthy and high-yielding plants were identified from each location, and the adhering soils from the roots were aseptically drawn without damaging the plant. The collected soil samples were mixed, and representative soils were brought to the lab in sterile polyethylene bags and maintained at 4°C until analysis. An outline of the method is presented in Figure 1.
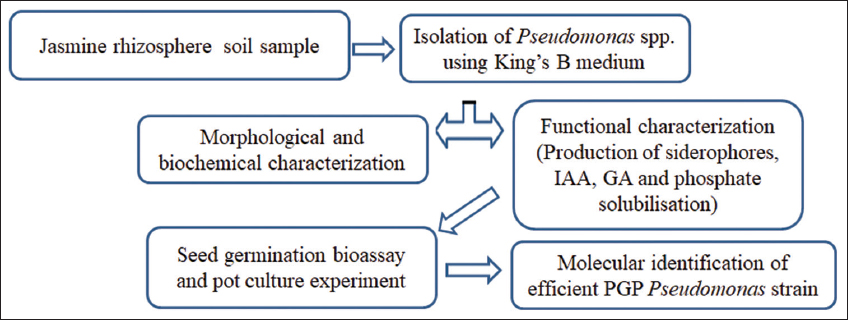 | Figure 1: Schematic representation of experimental framework. [Click here to view] |
Table 1: Details of sampling locations chosen in the present study.
| Sampling location | Latitude | Longitude |
|---|---|---|
| 1 | 13.259139 | 74.771672 |
| 2 | 13.256178 | 74.774898 |
| 3 | 13.261779 | 74.767479 |
| 4 | 13.261507 | 74.767177 |
| 5 | 13.260136 | 74.773157 |
2.2. Isolation of PGP Pseudomonas spp. from Rhizosphere Soils
Pseudomonas strains were isolated from the soil samples by dilution plating on King’s B agar medium [10]. The composition per liter of the medium used in the study was: proteose peptone 20 g; magnesium sulfate heptahydrate 1.5 g; dipotassium hydrogen phosphate 1.5 g; glycerol 15 mL; distilled water 1000 mL; pH 7.2, and agar 15 g. The plates were incubated at 28 ± 2°C for 24 h. Representative colonies were purified and maintained at 4°C [11].
2.3. Morphological and Biochemical Characterization
The morphological properties of the isolated bacterial colonies on Luria–Bertani agar were observed after incubation for 24 h at 28 ± 2°C. After incubation, colony size, shape, elevation, and color were recorded. Isolates were characterized for Gram staining, endospore formation, and motility. The biochemical properties constituting gelatin liquefaction, starch hydrolysis, utilization of different sources of carbohydrates, oxidase test, and catalase test were determined [12,13].
2.3.1. Siderophore production
All Pseudomonas isolates were assessed for siderophore production using a standard method [14]. One milliliter of 2% FeCl3 solution was added to one mL of culture filtrate, and the appearance of an orange or red–brown color indicated siderophore production.
2.3.2. Phosphate dissolving ability
The bacteria were streaked onto Modified Sperber’s medium [15]. The inoculated plates were incubated at 28 ± 2°C for 24 h, and the formation of clear zones around the growth confirmed phosphate solubilization.
2.3.3. Indole acetic acid (IAA) producing ability
The IAA-producing ability of Pseudomonas spp. was evaluated using the Salkowski method [16]. The isolates were inoculated into King’s B broth medium supplemented with 0.5 mgL-1 of L-tryptophan and incubated at 28 ± 2°C for 3 days with continuous agitation at 120 rpm. The liquid cultures were centrifuged, and the supernatant was treated with Salkowski’s reagent. This mixture was incubated for 20 min, resulting in the development of a pink color. IAA production was estimated by measuring the intensity of the colored complex at 530 nm using a double-beam PC-based ultraviolet-visible spectrophotometer (Model 2202, Systronics India Ltd., Gujarat, India).
2.3.4. Gibberellic acid (GA) producing ability
The Pseudomonas strains isolated were screened for the production of GA [17]. The isolates were inoculated into King’s B broth and incubated for 7 days at 28 ± 2°C. Broth cultures were centrifuged at 10,000 rpm for 20 min. The supernatant was extracted using ethyl acetate 2:1 (v/v) after adjusting the pH to 2.5 using 1M HCl. The optical density of the resulting solution was measured at 254 nm using a spectrometer. Unknown concentrations were determined based on the GA standard curve.
2.4. Green Gram Seed Germination Bioassay
The PGP ability of the isolates was evaluated by conducting green gram (Vigna radiata [L.] Wilczek) seed germination bioassay [18,19]. Isolated bacteria were grown in King’s B broth in culture tubes for 24 h. The cultures were centrifuged at 10,000 rpm for 20 min, and the supernatants were collected. Seeds of uniform size were chosen, washed with 70% (v/v) ethanol for 1 min, and sterilized using 4% (v/v) sodium hypochlorite solution. The seeds were immediately rinsed thrice in sterile water. Sixty uniform-sized sterilized seeds were selected and soaked in the culture supernatant for 12 h. Twenty seeds each were placed aseptically in sterile Petri plates containing wet blotting paper (20 seeds per isolate × 3 replicates = 60 seeds). Uninoculated sterile broth-treated seeds were used as controls. Three replicates were used for each treatment. After 17 h, germination percentage and radicle length were recorded.
2.5. Pot Culture Experiment
To evaluate the PGP potential of the isolates in jasmine, a pot culture experiment was conducted. The experiment comprised seven treatments; each replicated 3 times, and was arranged in a completely randomized design. The treatments included in the study were: T1, Control (uninoculated and treated with sterilized King’s B Broth); T2, RP–3; T3, WP–10; T4, NPS–6; T5, NPS–18, and T6, NPS–16. The strains were inoculated into King’s B broth and incubated for 7 days at 28 ± 2°C. The cultures thus developed were used to inoculate the rooted jasmine plants. Stem cuttings of jasmine plants of equal length were rooted in a sand–soil mixture (SSM) of 2:1 proportion. Plastic pots of 14.2 cm diameter and 15 cm height were used for this purpose. The propagated materials were maintained in a greenhouse for 2 months. Uniformly rooted cuttings were transplanted into pots containing 200 g of sterilized SSM. The liquid culture was added to each pot. The plants were irrigated regularly with sterile water. Growth indicators, such as height, number of leaves, and internodes, were recorded 180 days after treatment.
2.6 Molecular Identification
The best-performing isolate obtained from the pot culture study was processed for 16S rRNA gene sequencing at Yaazh Xenomics, Coimbatore, Chennai, India. Genomic DNA was extracted from the isolate using an EXpure Microbial DNA Isolation Kit (Bogar Biobee Stores Pvt. Ltd., Coimbatore, India). Primers (27 F and 1492 R) were used to amplify the 16S rRNA genes. The nucleotide sequences of 27 F and 1492 R were used with 5’ AGAGTTTGATCCTGGCTCAG 3’ and 5’ TACGGTACCTTGTTACGACTT 3’, respectively.
The polymerase chain reaction products were sequenced on an ABI 3730xl sequencer (Applied Biosystems). A basic local alignment search tool (BLAST) in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare the obtained sequences. Multiple sequence alignment of sequences was performed using the program MUSCLE 3.7 (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=muscle). Divergent and poorly aligned regions were removed using the program Gblocks 0.91b (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=gblocks). Phylogenetic analysis was carried out using the PhyML 3.0 aLRT program (http://www.atgc-montpellier.fr/phyml/). Tree Dyn 198.3 (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=treedyn) was used for tree rendering [20].
2.7. Statistical Analysis
All experiments were conducted in triplicate (n = 3), and the data obtained are expressed as mean ± standard deviation (SD). Data were processed using analysis of variance (ANOVA) with a significance threshold set at P ≤ 0.05. When ANOVA was significant at P < 0.05, Tukey’s test was performed to separate group mean values. The online agricultural statistics tool, a general R-based analysis platform for experimental statistics 1.0.0, was used in the statistical analysis [21].
3. RESULTS AND DISCUSSION
3.1. Isolation of Representative Pseudomonas spp.
Six representative Pseudomonas strains were successfully obtained from the rhizosphere soils. All plants displayed PGP traits, as evidenced by the production of IAA, GA, and siderophores. NPS-18 notably exhibited phosphate solubilization ability with all other PGP traits [Figure 2], highlighting its potential as a robust bioinoculant.
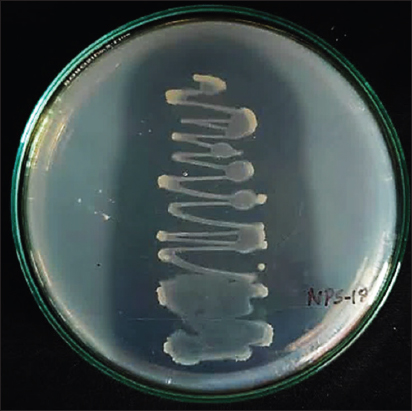 | Figure 2: Isolate NPS-18 showing clear zone when grown on modified Sperber’s medium. This is an evidence for phosphate solubilizing property of the isolate. [Click here to view] |
3.2. Morphological and Biochemical Characterization
The isolates were subjected to morphological and biochemical characterization [Table 2]. Sah et al. and Gutierrez-Albanchez et al. [22,23], reported that members of the genus Pseudomonas are Gram-negative and rod-shaped with polar flagella. Studies have also confirmed that these bacteria are typically catalase and oxidase positive [24,25], and some species of Pseudomonas also exhibit phosphate solubilization properties [26-28]. This evidence reinforces the consistency of our results with those of the previously reported studies.
Table 2: Morphological and biochemical properties of representative Pseudomonas isolates obtained from Jasminum sambac (L.) Aiton.
| Characters | Pseudomonas isolates | |||||
|---|---|---|---|---|---|---|
| RP-3 | WP-8 | WP-10 | NPS-6 | NPS-18 | NPS-16 | |
| Colony characteristics | Small, Round white, Opaque raised | Medium, irregular, yellowish green, convex | Small, round, cream, flat, opaque | Small, round, yellowish green, convex | Medium, irregular, translucent, light-green, flat | Small, round, cream, flat, opaque |
| Cell shape | Rod | Rod | Rod | Rod | Rod | Rod |
| Gram reaction | − | − | − | − | − | − |
| Catalase | + | + | + | + | + | + |
| Oxidase | + | + | + | + | + | + |
| Starch hydrolysis | − | − | − | − | − | + |
| Gelatin liquefaction | − | − | − | + | + | − |
| Siderophore production | + | + | + | + | + | + |
| Endospore formation | − | − | − | − | − | − |
| Motility | + | + | + | + | + | + |
| Phosphate solubilization | − | − | − | − | + | − |
+: Positive result, − : Negative result.
3.3. IAA and GA Biosynthesis
One of the key mechanisms by which PGPR enhances plant growth is the synthesis of IAA and GA [29-31]. All the strains produced IAA and GA [Table 3]. Among the strains, maximum IAA production was observed in WP-10 (38.06 ± 1.47 µg/mL), followed by NPS −18 (37.87 ± 0.56 µg/mL), while RP-3 recorded the lowest (24.62 ± 0.18 µg/mL). Notably, many species of Pseudomonas have been reported to produce higher levels of IAA than other PGPR [32]. In the current study, the highest GA production was observed in strain RP-3 (278.63 ± 2.89 µg/mL), followed by WP-8 (278 ± 3.96 µg/mL), WP-10 (276.80 ± 8.79 µg/mL), and NPS-18 (245.60 ± 1.55 µg/mL) [Table 3]. Sharma et al., [33] observed a range of GA production in Pseudomonas from 419.2 to 485.8 µg/mL. As evidenced by several studies, IAA and GA are well known for their roles in promoting overall plant growth [34,35]. Seed germination bioassays and pot culture experiments were conducted to evaluate the influence of these phytohormones on early plant development.
Table 3: Phytohormone producing efficiency of Pseudomonas spp.
| Isolate name | IAA (µg/mL) | GA (µg/mL) |
|---|---|---|
| NPS-16 | 30.21±0.98b | 225.70±1.64c |
| NPS-6 | 30.30±1.28b | 243.10±2.25b |
| NPS-18 | 37.87±0.56a | 245.60±1.55b |
| RP-3 | 24.62±0.18c | 278.63±2.89a |
| WP-10 | 38.06±1.47a | 276.80±8.79a |
| WP-8 | 24.73±1.11c | 278.00±3.96a |
3.4. Seed Germination Bioassay
Pseudomonas strains obtained in the current investigation significantly improved seed germination and radicle elongation in V. radiata (L.) R. Wilczek when compared to the uninoculated control [Figure 3]. Among the various treatments, NPS-18 and NPS-6 demonstrated remarkable performance, exhibiting 100% germination after 17 h of incubation. Similarly, seed inoculation with NPS-16 was 93.33% within the same duration and was statistically at par with NPS-18 and NPS-6. In contrast, the uninoculated control exhibited the lowest germination percentage (6.66 ± 0.28%), indicating the positive influence of bacterial inoculation on early seedling vigor. The enhanced germination observed in the inoculated treatments is likely associated with the increased activity of hydrolytic enzymes, such as α-amylases, which are crucial for breaking down endospermic starch into soluble sugars that fuel embryonic growth. The production of such enzymes is often regulated by the microbial production of phytohormones [36], particularly IAA and GA, which are known to stimulate enzyme activity, cell division, and tissue differentiation. Several studies have documented the role of these hormones in promoting seed germination, root initiation, elongation, and overall plant development [37-39].
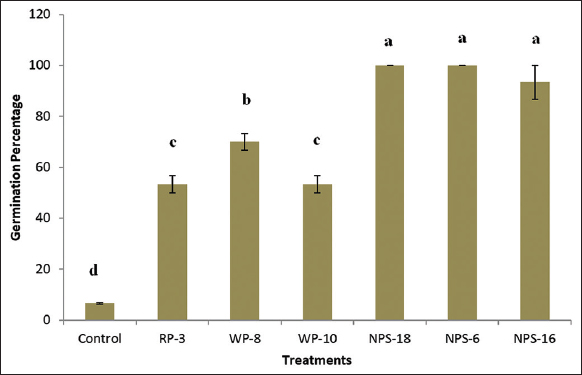 | Figure 3: Effect of Pseudomonas spp. on green gram seed germination. Letters designates significant differences among values (P ≤ 0.05). Treatment with the same letters is not significantly different. Values are means ± SD (n = 3). [Click here to view] |
Furthermore, radicle growth from the germinated seeds (after 17 h of incubation) was significantly influenced by the bacterial treatments [Figure 4]. Seeds inoculated with NPS-6 exhibited the highest radicle length (63.63 ± 2.31 mm), which was significantly superior to all other treatments. The strains NPS-16 and NPS-18 resulted in the radicle lengths of 45.30 ± 0.60 mm and 43.30 ± 1.57 mm, respectively, and were statistically at par with each other, ranking next best to NPS-6. The uninoculated control recorded the minimum radicle length (3.60 ± 0.26 mm) compared to all the other treatments. The increased radicle length in these treatments may be attributed to the synthesis and release of phytohormones by Pseudomonas strains, which enhance root elongation through cell expansion and division [40]. The results of this study align with those of several reports emphasizing the beneficial effects of PGPR on seed germination and root development [41,42].
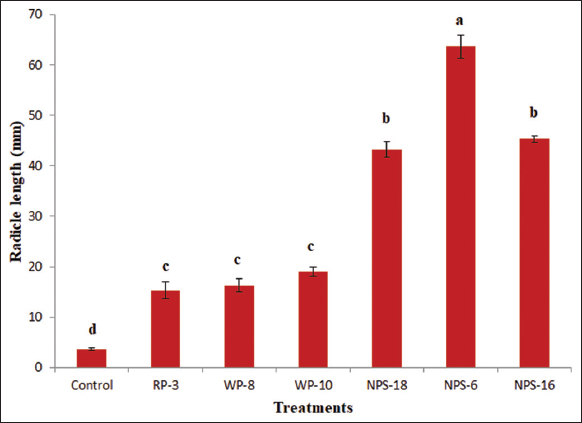 | Figure 4: Effect of Pseudomonas spp. on green gram radicle growth. Letters designates significant differences among values (P ≤ 0.05). Treatment with the same letters is not significantly different. Values are means ± SD (n = 3). [Click here to view] |
3.5. Pot Culture Experiment
The effect of Pseudomonas isolates on jasmine growth was evaluated using a pot culture experiment. Compared with the uninoculated control, treatment with Pseudomonas significantly enhanced plant growth [Table 4 and Figure 5]. NPS-18 demonstrated outstanding improvement in plant growth, recording a significantly greater plant height (61.33 ± 4.16 cm) and number of internodes (28.00 ± 2.64) compared to all the other treatments, including control. This accounted for an increase of 95.8% and 13.94% compared to plant height and number of internodes, respectively, relative to the control [Table 4]. Treatments with NPS-6 and NPS-18 recorded the highest number of leaves of 24.66 ± 3.78 and 24.33, respectively, which are significantly higher than the uninoculated control (20.00 ± 2.00).
Table 4: Effect of inoculation of Pseudomonas spp, on plant height, number of leaves and number of internodes of Jasminum sambac (L.) Aiton after 180 days after inoculation.
| Treatment | Plant height (cm) | Number of leaves | Number of internodes |
|---|---|---|---|
| Uninoculated control | 31.33±2.51e | 20.00±2.00b | 24.66±0.57b |
| NPS-16 | 47.66±3.21c | 20.33±2.30b | 20.33±1.15c |
| NPS-6 | 55.33±2.08b | 24.66±3.78a | 25.66±0.57ab |
| NPS-18 | 61.33±4.16a | 24.33±1.15a | 28.00±2.64a |
| RP-3 | 33.00±3.46de | 20.00±1.00b | 21.00±1.00c |
| WP-10 | 34.33±0.57de | 18.66±2.30b | 20.66±1.52c |
| WP-8 | 36.66±0.57d | 20.33±1.52b | 21.66±1.15c |
 | Figure 5: Effect of Pseudomonas spp. on growth of J. sambac (L.) Aiton. [Click here to view] |
Previous studies suggested that the PGP effect mediated by Pseudomonas is due to the secretion of phytohormones [43,44], production of siderophores, and increased bioavailability of nutrients [45]. In the current study, the production of phytohormones (IAA and GA) and siderophores in all isolates was observed. Moreover, NPS-18 demonstrated phosphate-solubilizing activity, thereby strengthening its role in plant development. Our previous studies documented the importance of phytohormone secretion and phosphate-solubilizing ability of PGP organisms in stimulating plant growth [46-50]. Therefore, the enhanced growth observed in plants treated with Pseudomonas is likely due to synergistic effects from its multiple beneficial traits.
3.6. Molecular Characterization of Efficient PGPR
Based on all PGP traits, the most efficient strain identified in the present study was NPS-18. Therefore, this strain was subjected to molecular characterization by 16S rRNA sequencing [Figures 6 and 7]. Sequence analysis of NPS-18 revealed 100% homology with Pseudomonas aeruginosa as depicted in the phylogenetic tree [Figure 8]. The 16S rRNA gene sequence of isolate NPS-18 was submitted to the NCBI GenBank (accession number PV862270). This result is consistent with a study by Chopra et al. [51], who isolated P. aeruginosa from the tea rhizosphere and demonstrated its attributes, including phytohormone synthesis, phosphate solubilization, and siderophore production. Several independent studies have documented using this bacterium as a potential bioinoculant to promote plant growth and control plant diseases [52-55]. In addition, the Directorate of Groundnut Research (ICAR), Junagadh, developed a microbial consortium, NutBoost, that includes P. aeruginosa BM 6 (PGPR4) as one of its components and is recommended for groundnut cultivation [56]. However, recent studies have underscored the inherently pathogenic nature of certain P. aeruginosa strains, which possess a sophisticated arsenal of virulence determinants, such as quorum-sensing circuits, biofilm formation, secretion systems, and antibiotic resistance that enable them to colonize immunocompromised hosts [57-60]. Furthermore, Vasco et al. [61] highlighted the need to distinguish naturally occurring environmental isolates from clinical isolates, observing that clinical isolates often show traits such as greater motility, higher stress tolerance, and longer persistence in host-associated environments that are usually absent in strains from natural environments. The isolate P. aeruginosa (NPS-18), obtained in this study, was from the rhizosphere soil of jasmine with no clinical background and therefore, may be used as a bioinoculant. Nevertheless, we also acknowledge the importance of conducting additional risk assessments, such as genome-based analysis of virulence and resistance genes, before proceeding with field-level deployment.
 | Figure 6: Agarose gel electrophoresis of polymerase chain reaction amplified 16S rRNA gene region. Lane 108: 1kb DNA ladder used as molecular weight marker, Lane 121: Amplified 16s rRNA gene product (approximately 1000 bp) from isolate NPS-18. [Click here to view] |
 | Figure 7: 16S rRNA partial gene sequence of NPS-18. [Click here to view] |
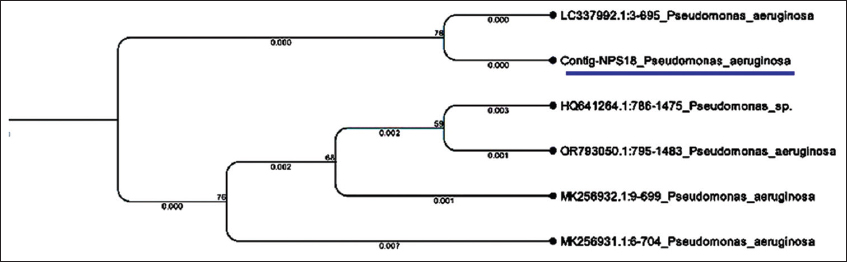 | Figure 8: Phylogenetic tree based on 16S rRNA sequence. The analysis indicates that isolate NPS-18 clusters closely with other strains of Pseudomonas aeruginosa. [Click here to view] |
Finally, we hypothesized that [Figure 9] the Pseudomonas strain NPS-18 isolated in this study enhances plant growth by producing siderophores and phytohormones (IAA and GA). IAA produced by the isolate is transported to the root hair cells via ATP-binding cassette transporters, resulting in elevated intracellular IAA levels. This may lead to the ubiquitination and subsequent degradation of the Aux/IAA repressor protein [62]. This process activates auxin-responsive genes, prompting growth and development of the plant. In addition, GA produced by the isolates binds to the gibberellin insensitive dwarf (GID 1) receptors located on the root hair cell membrane, forming the GA-GID 1 complex. This complex interacts with the TVHYNP motif of the DELLA protein. As a result, the DELLA protein undergoes ubiquitylation and is consequently degraded via the 26S proteasome pathway, activating PIF transcription factors. Furthermore, the process induces GA-responsive genes [63], thereby promoting tissue growth and development. Similarly, siderophores (BSP) produced by bacterial strains chelate Fe3+ ions present in the soil. Successively, they are transported to plant root hair cells via ABCG37 transporters [64]. The enhanced intracellular availability of iron boosts the activity of iron-dependent enzymes involved in phytohormone biosynthesis, DNA replication, photosynthesis, and respiration, culminating in plant growth.
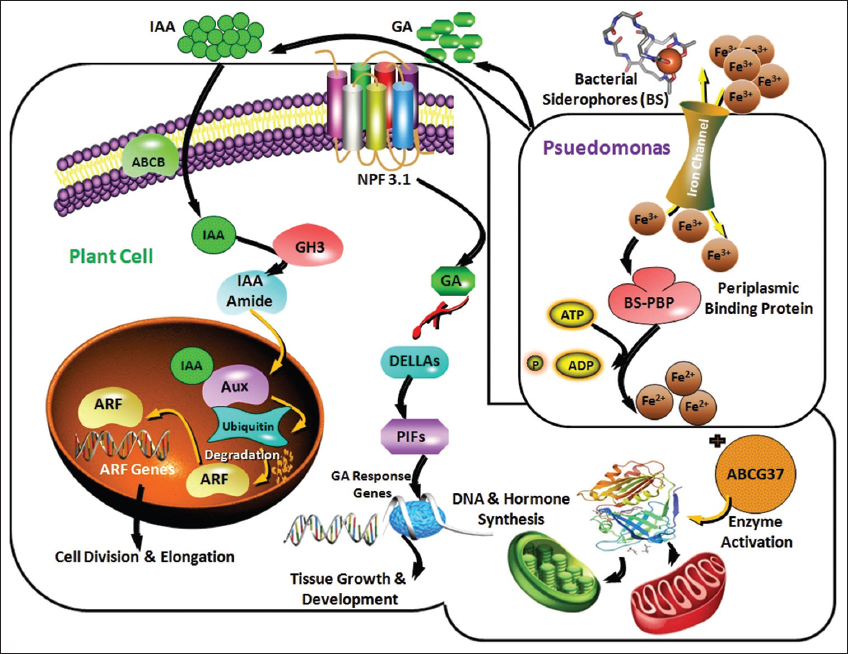 | Figure 9: The probable mode of action of Pseudomonas spp. obtained in the current study. [Click here to view] |
The findings of the study hold significant promise for green floriculture, particularly for jasmine cultivation. The identified efficient bacterial strains are promising bioinoculants for enhancing flower production in an eco-friendly manner. With this background, the findings of the current study are protected under Indian patent rights with application No. 202541007306 dated February 07, 2025. They are being made available to needy farmers on a non-profit basis, in alignment with the Nation’s vision for digital and smart agriculture.
4. CONCLUSION
The study provides insights into PGP Pseudomonas spp. associated with rhizosphere soils of GI tagged Udupi jasmine. NPS-18 demonstrated the highest efficiency among the six isolates, exhibiting multifunctional traits that enhance plant growth. Based on 16S rRNA gene sequencing, the best-performing isolate was identified as P. aeruginosa. These experiments were performed under regulated greenhouse conditions, which may not accurately reflect the complexity of the natural field conditions. Further, expanding the scope of sampling to diverse geographic regions could help identify more effective PGPR strains. Despite these limitations, this study is the first report on the isolation and functional characterization of indigenous Pseudomonas spp. from the rhizosphere of Udupi jasmine, laying the groundwork for future applications in sustainable floriculture.
5. ACKNOWLEDGMENT
The authors would like to thank Alva’s Education Foundation, Moodubidire, Karnataka, India, for providing laboratory facilities to conduct this research. We are also grateful to Dr. Farhan Zameer, Chief Scientific Officer, ATMA Research Center, Alva’s Ayurveda Medical College, Moodubidire, and Dr. Vishnumoorthy Prabhu, Associate Professor, Department of English, GFGC, Thenkanidiyoor, Udupi, for proofreading the manuscript. Special thanks are extended to Dr. Sudharshan Kini, Associate Professor, NUCSER, Nitte University, Derlakatte, Mangalore, and Dr. Chithaya, Assistant Professor, Zonal Agricultural and Horticultural Research Station, Brahmavar, for valuable research inputs.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declare that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Anumala NV, Kumar R. Floriculture sector in India:Current status and export potential. J Hortic Sci Biotechnol. 2021;96(5):673-80.[CrossRef]
2. Lu Z, Wang X, Lin X, Mostafa S, Bao H, Ren S, et al. Genome-wide identification and characterization of long non-coding RNAs associated with floral scent formation in Jasmine (Jasminum sambac). Biomolecules. 2023;14(1):45.[CrossRef]
3. Chaitanya HS, Nataraja S, Vikram HC, Jayalakshmi NH. Review on production techniques of GI Crop, Udupi Mallige (Jasminum sambac (L.) Aiton). J Pharmacogn Phytochem. 2018;3:50-2.
4. Handy F, Cnaan RA, Bhat G, Meijs LC. Jasmine growers of coastal Karnataka:Grassroots sustainable community-based enterprise in India. Entrep Reg Dev. 2011;23(5-6):405-17.[CrossRef]
5. Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, et al. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci 2013;4:356.[CrossRef]
6. Zboralski A, Filion M. Pseudomonas spp. can help plants face climate change. Front Microbiol. 2023;14:1198131.[CrossRef]
7. Beneduzi A, Ambrosini A, Passaglia LMP. Plant growth-promoting rhizobacteria (PGPR):Their potential as antagonists and biocontrol agents. Genet Mol Biol 2012;35(4 Suppl):1044-51.[CrossRef]
8. Zhang H, Zheng D, Hu C, Cheng W, Lei P, Xu H, et al. Certain tomato root exudates induced by Pseudomonas stutzeri NRCB010 enhance its rhizosphere colonization capability. Metabolites. 2023;13(5):664.[CrossRef]
9. Singh P, Singh RK, Zhou Y, Wang J, Jiang Y, Shen N, et al. Unlocking the strength of plant growth promoting Pseudomonas in improving crop productivity in normal and challenging environments:A review. J Plant Interact. 2022;17(1):220-38.[CrossRef]
10. King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. Transl Res. 1954;44(2):301-7.
11. DeBritto ST, Gajbar D, Satapute P, Sundaram L, Lakshmikantha RY, Jogaiah S, et al. Isolation and characterization of nutrient dependent pyocyanin from Pseudomonas aeruginosa and its dye and agrochemical properties. Sci Rep. 2020;10(1):1-12.[CrossRef]
12. Aneja KR. Experiments in Microbiology, Plant Pathology and Biotechnology. 6th ed. New Delhi:New Age International;2022.
13. Ramya Rai M, Rao RB. Isolation of phosphate solubilizing microorganisms from geographical indication tagged Shankarpura jasmine [Jasminum sambac (L.) Aiton] and their plausible role in plant growth promotion. Bioscene. 2024;21(3):856-74.
14. Kotasthane AS, Agrawal T, Zaidi NW, Singh US. Identification of siderophore producing and cyanogenic fluorescent Pseudomonas and a simple confrontation assay to identify potential biocontrol agent for collar rot of chickpea. 3 Biotech. 2017;7(2):137.[CrossRef]
15. Krishnaraj PU. Genetic Characterization of Mineral Phosphate Solubilization in Pseudomonas sp. [Ph.D. Thesis]. New Delhi:Indian Institute of Agricultural Sciences;1996.
16. Gang S, Sharma S, Saraf M, Buck J, Schumacher J. Production in Klebsiella by LC-MS/MS and the Salkowski method. Bio Protoc. 2019;9(9):3230.[CrossRef]
17. Berríos J, Illanes A, Aroca G. Spectrophotometric method for determining gibberellic acid in fermentation broths. Biotechnol Lett. 2004;26:67-70. [CrossRef]
18. Nathan Vinod Kumar K, Rajam S, Rani ME. Plant growth promotion efficacy of indole acetic acid (IAA) produced by a mangrove associated fungi Trichoderma viride VKF3. Int J Curr Microbiol Appl Sci. 2017;6(11):2692-701. [CrossRef]
19. Bhagobaty RK, Joshi SR. Promotion of seed germination of green gram and chickpea by Penicillium verruculosum RS7PF, a root endophytic fungus of Potentilla fulgens L. Adv Biotech. 2009;8:16-8.
20. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr:Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:gkn180.[CrossRef]
21. Gopinath PP, Prasad R, Joseph B, Adarsh VS. Collection of shiny apps for data analysis in agriculture grapesAgri1. J Open Source Softw. 2021;6(63):3437. https://doi.org/10.21105/joss.03437 [CrossRef]
22. Sah S, Krishnani S, Singh R. Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr Res Microb Sci. 2021;2:100084.[CrossRef]
23. Gutierrez-Albanchez E, García-Villaraco AJ, Lucas I, Horche B, Ramos-Solano E, Gutierrez-Mañero FJ. Pseudomonas palmensis sp. nov., a novel bacterium isolated from Nicotiana glauca microbiome:Draft genome analysis and biological potential for agriculture. Front Microbiol 2021;12:672751.[CrossRef]
24. Domínguez-Bello MG, Reyes N, Teppa-Garrán A, Romero R. Interference of Pseudomonas strains in the identification of Helicobacter pylori. J Clin Microbiol. 2000;38(2):937.[CrossRef]
25. Dodd CER. Pseudomonas - introduction. In:Batt CA, Tortorello ML, editors. Encyclopedia of Food Microbiology. 2nd ed. New York:Elsevier;2014. 244-7.
26. Ou K, He X, Cai K, Zhao W, Jiang X, Ai W, et al. Phosphate-solubilizing Pseudomonas sp. strain WS32 rhizosphere colonization-induced expression changes in wheat roots. Front Microbiol. 2022;13:927889.[CrossRef]
27. Yadav AN. Phosphate-solubilizing microorganisms for agricultural sustainability. J Appl Biol Biotechnol. 2022;10(3):1-6.[CrossRef]
28. Bakki M, Banane B, Marhane O, Esmaeel Q, Hatimi A, Barka EA, et al. Phosphate solubilizing Pseudomonas and Bacillus combined with rock phosphates promoting tomato growth and reducing bacterial canker disease. Front Microbiol. 2024;15:1289466.[CrossRef]
29. Orozco-Mosqueda MD, Santoyo G, Glick BR. Recent advances in the bacterial phytohormone modulation of plant growth. Plants. 2023;12(3):606.[CrossRef]
30. Zhang T, Jian Q, Yao X, Guan L, Li L, Liu F, et al. Plant growth-promoting rhizobacteria (PGPR) improve the growth and quality of several crops. Heliyon. 2024;10(10):e31553. [CrossRef]
31. Khoso MA, Wagan S, Alam I, Hussain A, Ali Q, Saha S, et al. Impact of plant growth-promoting rhizobacteria (PGPR) on plant nutrition and root characteristics:Current perspective. Plant Stress. 2024;11:100341.[CrossRef]
32. Singh TB, Sahai V, Ali A, Prasad M, Yadav A, Shrivastav P, et al. Screening and evaluation of PGPR strains having multiple PGP traits from the hilly terrain. J Appl Biol Biotech. 2020;8(4):38-44.[CrossRef]
33. Sharma S, Sharma A, Kaur M. Extraction and evaluation of gibberellic acid from Pseudomonas sp.:plant growth promoting rhizobacteria. J Pharmacogn Phytochem. 2018;7(1):2790-5.
34. Lata DL, Abdie O, Rezene Y. IAA-producing bacteria from the rhizosphere of chickpea (Cicer arietinum L.):Isolation, characterization, and their effects on plant growth performance. Heliyon. 2024;10(21):e39702.[CrossRef]
35. Hasan A, Tabassum B, Hashim M, Khan N. Role of plant growth promoting rhizobacteria (PGPR) as a plant growth enhancer for sustainable agriculture:A review. Bacteria. 2024;3(2):59-75.[CrossRef]
36. Alybayev S, Smekenov I, Kuanbay A, Sarbassov D, Bissenbaev A. Gibberellic-acid-dependent expression of a-amylase in wheat aleurone cells is mediated by target of rapamycin (TOR) signaling. Curr Plant Biol. 2024;37:100312.[CrossRef]
37. Kaneko M, Itoh H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. The alpha-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002;128(4):1264-70.[CrossRef]
38. Meliani A, Bensoltane A, Benidire L, Oufdou K. Plant growth promotion and IAA secretion with Pseudomonas fluorescens and Pseudomonas putida. Res Rev J Bot Sci. 2017;6(2):16-24.
39. Tsukanova KA, Chebotar VK, Meyer JJ, Bibikova TN. Effect of plant growth-promoting rhizobacteria on plant hormone homeostasis. S Afr J Bot. 2017;113:91-102.[CrossRef]
40. Fatima T, Arora NK. Pseudomonas entomophila PE3 and its exopolysaccharides as biostimulants for enhancing growth, yield and tolerance responses of sunflower under saline conditions. Microbiol Res. 2021;244:126671.[CrossRef]
41. Xie H, Pasternak JJ, Glick BR. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr Microbiol. 1996;32:67-71.[CrossRef]
42. Gupta R, Chakrabarty SK. Gibberellic acid in plant:Still a mystery unresolved. Plant Signal Behav. 2013;8(9):e25504.[CrossRef]
43. Qessaoui R, Bouharroud R, Furze JN, El Aalaoui M, Akroud H, Amarraque A, et al. Applications of new rhizobacteria Pseudomonas isolates in agroecology via fundamental processes complementing plant growth. Sci Rep. 2019;9:12832.[CrossRef]
44. Jakubowska Z, Gradowski M, Dobrzynski J. Role of plant growth-promoting bacteria (PGPB) in enhancing phenolic compounds biosynthesis and its relevance to abiotic stress tolerance in plants:A review. Antonie van Leeuwenhoek. 2025;118(9):123.[CrossRef]
45. Zhang S, Deng Z, Borham A, Ma Y, Wang Y, Hu J, et al. Significance of soil siderophore-producing bacteria in evaluation and elevation of crop yield. Horticulturae. 2023;9(3):370.[CrossRef]
46. Bhavyashree P, Rao RB, Bhat P. A study on plant growth promotional and antagonistic properties of lactic acid bacteria. J Adv Sci Res. 2021;2020(CSTSS):1-10. Available from: https://sciensage.info/index.php/JASR/article/view/1637
47. Ashritha, Rao RB, Rai MR, Nagaraj P, Visweswara P. Characterization of phosphate solubilising bacteria isolated from rhizosphere soils of Piper nigrum L. Biotechnol. 2021;20(1):15-21.[CrossRef]
48. Ramya Rai M, Rao RB. Identification of native Trichoderma for augmentation of plant growth in geographical indication tagged fragrant Shankarpura jasmine [Jasminum sambac (L.) Aiton]. J Biol Control. 2024;38(4):389-97. [CrossRef]
49. Zameer F, Meghashri S, Gopal S, Rao BR. Chemical and microbial dynamics during composting of herbal pharmaceutical industrial waste. J Chem. 2010;7(1):143-8.[CrossRef]
50. Kavya H, Akki AJ, Rao RB, Goveas SW, Sridhar KR. Plant growth-promotion and antipathogenic fungal activity of four rhizosphere isolates of Trichoderma from southwest India. Plant Fungal Res 2025;8(1):6.[CrossRef]
51. Chopra A, Bobate S, Rahi P, Banpurkar A, Mazumder PB, Satpute S. Pseudomonas aeruginosa RTE4:A tea rhizobacterium with potential for plant growth promotion and biosurfactant production. Front Bioeng Biotechnol. 2020;8:861.[CrossRef]
52. Ghadamgahi F, Tarighi S, Taheri P, Saripella GV, Anzalone A, Kalyandurg PB, et al. Plant growth-promoting activity of Pseudomonas aeruginosa FG106 and its ability to act as a biocontrol agent against potato, tomato and taro pathogens. Biology. 2022;11(1):140.[CrossRef]
53. Krishnan Kutty S, Skandasamy N, Padma Devi R, Djearamane S, Tey LH, Wong LS, et al. Field trial to correlate mineral solubilization activity of Pseudomonas aeruginosa and biochemical content of groundnut plants. Open Life Sci. 2025;20:20221008.[CrossRef]
54. Katiyar P, Pandey N, Varghese B, Sahu KK. Biopriming of Pseudomonas aeruginosa abates fluoride toxicity in Oryza sativa L. by restricting fluoride accumulation, enhancing antioxidative system, and boosting activities of rhizospheric enzymes. Plants. 2025;14(8):1223.[CrossRef]
55. Thakker JN, Rathod K, Patel K, Pandya J, Badrakia J, Dhandhukia P. Blue-green partnership:Unravelling potential of marine Pseudomonas aeruginosa OG as a biocontrol agent against Fusarium oxysporum f. sp. vasinfectum. Discov Oceans. 2025;2(1):13.[CrossRef]
56. ICAR-National Bureau of Agriculturally Important Microorganisms (NBAIM). Technologies for Sustainable Agriculture:Compilation of Technologies Developed by ICAR-NBAIM. Mau:ICAR-NBAIM;2022. Available from: https://icar.org.in/sites/default/files/2022-06/technologies-compilation_nbaim.pdf
57. Zameer F, Rukmangada MS, Chauhan JB, Khanum SA, Kumar P, Devi AT, et al. Evaluation of adhesive and anti-adhesive properties of Pseudomonas aeruginosa biofilms and their inhibition by herbal plants. Iran J Microbiol. 2016;8(2):108-19.
58. Avinash MG, Aishwarya S, Zameer F, Gopal S. Pseudomonas aeruginosa biofilm and their molecular escape strategies. J App Biol Biotech. 2023;11(3):28-37. [CrossRef]
59. Morganal, Diggle SP, Whiteley M. Pseudomonas aeruginosa:Ecology, evolution, pathogenesis and antimicrobial susceptibility. Nat Rev Microbiol. 2025;Advance online publication.[CrossRef]
60. Vidal-Cortés P, Campos-Fernández S, Cuenca-Fito E, Del Río-Carbajo L, Fernández-Ugidos P, López-Ciudad VJ, et al. Difficult-to-Treat Pseudomonas aeruginosa infections in critically Ill patients:A comprehensive review and treatment proposal. Antibiotics. 2025;14(2):178.[CrossRef]
61. Vasco G, Martínez R, Noboa D, Vasco K, Trueba G. Physiological adaptations of clinical vs. indoor environmental strains of Pseudomonas aeruginosa in a hospital setting. FEMS Microbiol Lett 2025;372:fnaf027.[CrossRef]
62. Gao J, Zhuang S, Zhang W. Advances in plant auxin biology:Synthesis, metabolism, signaling, interaction with other hormones, and roles under abiotic stress. Plants. 2024;13(17):2523.[CrossRef]
63. Chebotar GO, Chebotar SV. Gibberellin-signaling pathways in plants. Cytol Genet. 2011;45(4):259-68.CrossRef]
64. Singh P, Chauhan PK, Upadhyay SK, Singh RK, Dwivedi P, Wang J, et al. Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded land. Front Microbiol. 2022;13:898979.[CrossRef]