1. INTRODUCTION
At present, agriculture practices involve the extensive use of chemical fertilizers that lead to several impacts as groundwater pollution, eutrophication and production of greenhouse gases ultimately leading to environmental pollution and posing several health hazards. Plant growth-promoting rhizobacteria (PGPR) are a diverse collection of rhizobacteria that occur in the rhizospheric zone and interact with the plant’s root [1]. PGPRs exhibit different Plant growth promoting (PGP) traits to improve the growth and development of plants. A varied range of rhizobacteria comprising of Acinetobacter, Azotobacter, Azospirillum, Bacillus, Burkholderia, Enterobacter, Klebsiella, Pseudomonas, Serratia and Xanthomonas are known to enhance the growth of plant by various mechanisms such as biological fixation of atmospheric nitrogen, biosynthesis of phytohormones, mineral nutrient solubilization and increasing resistance of the host plant against stress factors [2].
Phosphorus is an important component required for the proper growth of plants because of its involvement in various metabolic processes. A high concentration of phosphate is found naturally in soil due to the application of chemical fertilizers [3], but this phosphate rapidly gets converted into unavailable forms by forming complexes with iron, aluminium, or calcium ions [4]. This unavailable phosphate could be transformed back into available forms by several phosphate solubilizing rhizobacteria.
Indole acetic acid (IAA) is the main phytohormone that is produced by PGPR in addition to host plants. IAA is synthesized throughout the plant body, mainly in meristematic tissues and in buds, including germinating seeds, root and shoot tips. IAA stimulates various important physiological processes such as lateral bud development, differentiation in vascular bundles and tropism responses [5].
Siderophore compounds are chelating agents having a strong affinity for ferric irons [6] and are produced by some PGPRs. This compound chelates to the unavailable form of iron and makes it available to the roots of plant in the adjacent environment [7,8]. Besides, siderophores also control the growth of plant pathogens through biological control mechanisms [9]. It has been reported that biocontrol action of siderophores is useful subsequently in many crops [10]. Rhizobacterial strains indirectly support the growth of plant by reducing the growth of phytopathogenic microbes, production of hydrolytic enzymes, hydrogen cyanide (HCN) and hydrogen sulfide (H2S) in the rhizosphere and minimize the effects of biotic stresses [11].
Potential PGPR isolates can be screened through estimations of phosphate solubilization, production of IAA, siderophore, ammonia and HCN in vitro to study their PGP traits [12,13]. PGP traits as phosphate solubilization and ammonia production can directly enhance growth of plants by increasing availability of nutrients while IAA phytohormone increase the root growth [14,15].
In the past few decades, the application of PGPR has increased extensively for sustainable agriculture. More research works show that the application of PGPR inoculants significantly increased growth and yield of crops. The present study was undertaken to screen PGPR strains having various PGP traits and the use of such PGPR strains as bioinoculant.
2. MATERIALS AND METHODS
2.1. Collection of soil sample and isolation of rhizobacteria
Nine soil samples were collected from different sites from the hilly terrain of Harda District in Madhya Pradesh. These soil samples were brought to the laboratory for further study. To isolate the rhizobacteria, 10 g of soil was added to 90 ml of sterile saline distilled water (0.85% NaCl) and placed on an orbital shaker at 120 rpm for 30 minutes. About 100 µl of soil suspension by serial dilution from 10–4 to 10–6 of each soil sample was spread on nutrient agar medium (NAM) plate and placed in an incubator at 28 ± 2°C for 2 days. Single colonies of rhizobacterial isolates were selected on the basis of faster growth and purified by repeat streaking on the NAM. After purification, these PGPR isolates were screened for PGP traits.
2.2. Morphological characteristics
After purification of the colony, each isolate was observed for their morphological dissimilar appearances of colonies such as colour, elevation, size, shape and surface, which were recorded on nutrient agar plates to minimize the number of similar isolates. Gram staining of each isolate was initially determined by using crystal violet and safranin staining, according to standard microbiological protocol.
2.3. Determination of PGP traits
2.3.1. Phosphate solubilization
The isolated rhizobacterial strains were checked for their ability to dissolve insoluble tricalcium phosphate as described by Gaur [16]. For this, 5 µl of bacterial suspension (≅3 × 108 colony forming unit ml-1) was placed on Pikovskaya’s agar medium, incubated at 28 ± 2°C and observed on 7th day after incubation. The solubilization index (SI) was determined by the ratio of total diameter (colony + halo zone) to the diameter of the colony.
Clear halo zone around bacterial colony indicates phosphate solubilization zone. PGPR strains were chosen for further estimation of phosphate solubilization in broth on the basis of SI. The selected PGPR was inoculated in 50 ml of Pikovskaya broth and kept in an orbital shaker at 28 ± 2°C and 120 rpm for 5 days [17]. After the requisite incubation period, rhizobacterial culture was harvested by centrifugation at 10,000 rpm for 15 minutes. The supernatant (1 ml) was mixed with 2.5 ml of Barton’s reagent and the volume made up to 50 ml with distilled water and a blank was also prepared. The absorbance of developed yellow colour was measured after 10 minutes at 430 nm in a UV-VIS spectrophotometer (Systronics-2201) and soluble phosphorus present in supernatant was quantified using a standard curve of KH2PO4.
2.3.2. Production of IAA
The production of IAA by PGPR isolates was quantified by the method of Bric et al. [18]. PGPR isolates were inoculated in 50 ml nutrient broth and incubated at 28 ± 2°C in an orbital shaker at 120 rpm for 48 hours. After the requisite incubation period, rhizobacterial culture was harvested by centrifugation at 10,000 rpm for 10 minutes and supernatant was used for quantification. Salkowski’s reagent was mixed in 2:1 proportions with supernatant and placed in the dark for 30 minutes. The developed pink supernatant showed the presence of IAA. The absorbance of this supernatant was measured at 530 nm in UV-VIS spectrophotometer and the amount of IAA was determined using the IAA standard curve.
2.3.3. Siderophore production
PGPR strains were spotted on chrome azurol S (CAS) agar plates and placed in an incubator at 28 ± 2°C for 7 days. The change of colour of medium from blue to yellow-pink around the spot indicated siderophore production.
Further, the quantitative analysis of siderophore was performed using CAS solution. PGPR strain was inoculated in sterile siderophore-inducing medium [20] and incubated on an orbital shaker at 28 ± 2°C at 120 rpm. After 7 days, rhizobacteria were harvested by centrifugation at 10,000 rpm for 5 minutes and 1 ml of supernatant was mixed with 1 ml of CAS solution and 20 μl of shuttle solution (0.2 M 5-sulfosalicyclic acid) was added. After 20 minutes of incubation at room temperature, absorbance was measured at 630 nm in UV-VIS spectrophotometer. Siderophore unit (%) was calculated by the following formula:
where Ar = reference absorbance at 630 nm
As = sample absorbance at 630 nm
2.3.4. Ammonia production
The purified isolates were grown in 10 ml of peptone broth in test tube and placed at 120 rpm for 48–72 hours at 28 ± 2°C. After incubation, 0.5 ml of Nessler’s reagent was added in the culture test tube. The change of colour from yellow to brown indicated a positive test for ammonia production [21].
2.3.5. HCN production
Lorck method [22] was performed to screen the HCN production by PGPR isolates. The modified NAM was prepared by amending it with 4.4 g L-1 glycine. PGPR strains were streaked on this medium. Pre-sterilized Whatman filter paper (no. 1) discs impregnated with filter-sterilized solution of picric acid and sodium carbonate and stuck on inside the Petri dish lid. The Petri dish was wrapped with parafilm and placed in an incubator at 28 ± 2°C for incubation of 4 days. The change of colour from yellow to the reddish-brown showed HCN production by PGPR isolates.
2.4. Biochemical characterization of selected PGPR strains
Seven PGPR isolates were selected according to their multiple PGP activity and were further characterized by standard biochemical tests. The selected PGPR strains were biochemically characterized by Gram’s staining, oxidase test, catalase test, citrate utilization, amylase assay, urea hydrolysis, methyl red (MR) test, voges proskauer (VP) test, indole test, casein hydrolysis, gelatin hydrolysis [23].
2.5. Molecular characterization of selected isolate
Genomic DNA from selected PGPR strain was extracted as described by Araujo et al. [24] with slight modifications and the isolated genomic DNA was used as a template for the amplification of 16S rRNA using primers 27F (5′AGGCCCGGGAAGGCGTATTCAC 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′). PCR was performed in a 50 µl of the final volume of reaction mixture containing 2 µl of DNA template, 4 µl of each forward and reverse primer, 25 µl 2X PCR buffer and 15 µl Milli Q water. PCR was carried out for 30 amplification cycles in a T100™ Thermal Cycler (BioRad, USA). The initial denaturation step was set at 95°C for 3 minutes, followed by 30 amplification cycles at 95°C for 1 minute, annealing temperature at 58°C for 30 seconds and 72°C for 2 minutes and final extension at 72°C for 8 minutes. After detection of PCR product by agarose gel electrophoresis along with 1 kb DNA ladder, the PCR product was further purified for use as a sequencing template and sent for sequencing to Xcelris Labs Limited, Ahmedabad, Gujarat. BLAST program was used to identify nucleotide-related sequence similarities which were obtained from GenBank database and were determined with NCBI databases. A phylogenetic tree was constructed by neighborhood joining bootstrap method among different isolates which includes phylogeny testing using bootstrap analysis with 1,000 replicates using MEGA X Software (Version 10.1.7) [25].
2.6. Statistical analysis
All experiments were performed in triplicate. The obtained data were analyzed by analysis of variance (ANOVA) using SPSS software. The mean values were compared using Duncan’s Multiple Range Test (DMRT) at p≤0.05%.
3. RESULTS AND DISCUSSION
3.1. Isolation and primary screening of PGP traits in isolates
A total of nine rhizospheric representative soil samples were collected and used for the isolation of different PGPR isolates. Out of 56 rhizobacterial isolates, 16 isolates were found positive for different PGP traits like phosphate solubilization, production of siderophore, IAA, ammonia and H2S (Table 1).
3.2. Morphological and biochemical characterization of potential PGPR isolates
A total of seven potential PGPR isolates were selected for their morphological (Table 2) and biochemical characterization (Table 3).
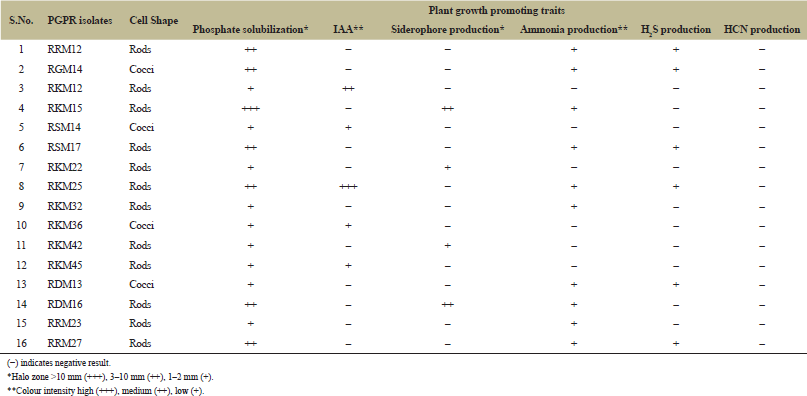 | Table 1: Screening of PGPR strains from soil for their multiple PGP traits. [Click here to view] |
3.3. Secondary screening of PGP traits in isolates
3.3.1. Phosphate solubilization
Phosphorus, an essential macronutrient, plays a key role in plant growth and development. PGPR isolates have the capability to solubilize insoluble phosphate and make it accessible for the plants [26,27]. In this study, 16 of 56 isolates were able to solubilize phosphate by making more than 3 mm clear halo zone around the colonies. On the basis of solubilizing zone, seven PGPR strains were preferred for the quantitative determination of phosphate solubilization. Maximum SI and solubilized phosphorus were shown by isolates such as RKM15 (3.4 and 339 mg L-1) and RGM14 (2.43 and 321.66 mg L-1) (Figs. 1 and 3). Minimum SI and solubilized phosphorus were observed in the case of RSM17 (1.35 and 186.67 mg L-1) (Fig. 3). Similarly, Tomer et al. [28] isolated a total of 133 isolates from soil and one isolate showed the highest solubilization efficiency of 713.11 mg L-1 which resembles with its largest SI as shown on Pikovskaya agar plate. Baliah et al. [29] observed that among 10 strains, Pseudomonas fluorescens (CTP2) released a maximum of 46.0 mg L-1 phosphorus followed by TP1 (40.6 mg L-1) in medium containing tricalcium phosphate. Liu et al. [30] reported phosphate solubilization efficiency in the range from 213.19 mg L-1 to 315.72 mg L-1. Karpagam et al. [31] reported that of 37 bacterial isolates, only six isolates showed higher SI (1.13–2.23). Phosphate-solubilizing rhizobacteria produce various types of organic acids, namely, butyric acid, fumaric acid, adipic acid, acetic acid, glyconic acid, oxalic acid, malonic acid, lactic acid, succinic acid and malic acid [32,33]. Even though organic acids may disturb the rhizosphere pH, these acids increase accessible phosphorus content in soil [34].
3.3.2. IAA production
IAA is the most common phytohormone that is synthesized by bacteria. Out of 56 isolates, only five PGPR isolates were found positive for the production of IAA without the addition of L- tryptophan in nutrient broth and were selected for further quantitative analysis. The data in Figure 4 shows maximum IAA production in RKM25 (35.56 µg ml-1), whereas minimum IAA production was observed in RKM36 (7.01 µg ml-1). Comparable results of IAA by PGPR isolates were reported by other researchers as well [35,36]. Pandey and Gupta [37] observed IAA production in a range from 10.96 to 37.78 µg ml-1 by PGPR isolates in the presence of L-tryptophan. Several studies showed that some rhizobacteria are able to synthesize IAA without L-tryptophan precursor. IAA is mainly synthesized by tryptophan (Trp) precursor by two pathways (dependent and independent) in both plants and bacteria. Bacteria have been found to be able to produce IAA through more than two pathways [38,39].
3.3.3. Siderophore production
PGPR isolates are also known to release iron-chelating compounds that increase the availability of iron to plants in iron-limiting soils [40]. Four isolates were found positive, that produced siderophore. Among them, RKM15 showed a large yellow halo zone on CAS agar plate (Fig. 2).
CAS agar assay confirmed the production of siderophore. The siderophore chelated to the iron of medium resulted in change of colour from blue to yellow-pink [41]. Four selected PGPR isolates were evaluated quantitatively of which, RKM15 (70.54%) and RDM16 (64.25%) showed the highest production of siderophore (Fig. 5). Similarly, Gupta and Gopal [42] reported siderophore production in the range from 21% to 70% by six isolates, from which P. fluorescens showed maximum siderophore production. Pahari and Mishra [43] screened 31 isolates, of which only two BGBA-1 and BRBA-1 produced maximum siderophore units. Ghavami et al. [44] reported that of 50 isolates, 10 bacterial isolates were able to produce siderophore.
 | Table 2: Morphological features of selected PGPR strains. [Click here to view] |
 | Table 3: Biochemical characterization of selected PGPR strains. [Click here to view] |
 | Figure 1: Solubilization of tricalcium phosphate by rhizobacterial isolate RKM15 on Pikovskaya agar medium. [Click here to view] |
 | Figure 2: Production of siderophore by rhizobacterial isolate RKM15 on CAS agar medium. [Click here to view] |
 | Figure 3: Phosphate solubilization by selected rhizobacterial isolates. Bars sharing the same letter do not differ significantly (p ≤ 0.05) and error bars (?) show SE. [Click here to view] |
3.4. Molecular characterization of potential PGPR isolate
RKM15 isolate was found to be the most prominent among all PGPRs. Hence, this isolate was selected for further characterization through 16S rRNA gene sequencing. According to the sequencing result, RKM15 demonstrated sequence homology with Pantoea dispersa, as presented in phylogenetic tree (Fig. 6). This isolate showed maximum similarity (96.25%) with P. dispersa. Accession number MN629239 of RKM15 isolates was obtained by submitting the sequences to GenBank database of NCBI.
 | Figure 4: IAA production by selected rhizobacterial isolates. Bars sharing the same letter do not differ significantly ( [Click here to view] |
 | Figure 5: Siderophore production by selected rhizobacterial isolates. Bars sharing the same letter do not differ significantly (p ≤ 0.05) and error bars (?) show SE. [Click here to view] |
 | Figure 6: Phylogenetic analysis of the RKM15 isolates using MEGA X version 10.0.5 by neighbor-joining method at bootstrap replicate value of n = 1000. Phylogenetic tree was constructed with branch lengths in the same units that inferred evolutionary distances. The PGPR isolates identified in the present study have been highlighted, and 16S rRNA gene accession numbers are given within brackets. [Click here to view] |
4. CONCLUSION
Finally, the present study emphasized on the capacity of the rhizobacteria isolated from soils of hilly regions of Harda District in Madhya Pradesh, having PGP traits such as phosphate solubilization, production of siderophores and ammonia. The RKM15 strain may be concluded as a very efficient phosphate solubilizer along with other PGP traits such as siderophore and ammonia production and showed maximum similarity with P. dispersa. RKM15 strain may be used as a bioinoculant to enhance plant growth by increasing nutrient availability through diverse mechanisms in soil.
ACKNOWLEDGMENT
Authors are thankful to Council of Scientific and Industrial Research, New Delhi and Department of Science and Technology, Government of India, for providing J.R.F. and S.R.F. The authors also acknowledge the Director, Dayalbagh Educational Institute, for the financial support to carry out this work.
CONFLICTS OF INTEREST
Authors declare that they do not have any conflicts of interest.
FINANCIAL SUPPORT
None.
REFERENCES
1.Ahmad F, Ahmad I, Khan MS. Screening of free living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 2008; 263:173–81. CrossRef
2.Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci 2014; 26(1):1–20. CrossRef
3.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003; 255:571–86. CrossRef
4.Havlin JL, Beaton JD, Tisdale SL, Nelson WL. Soil fertility and fertilizers: an introduction to nutrient management (No. 631.422/H388). Pearson Prentice Hall, Upper Saddle River, NJ, 2005.
5.Son JS, Sumayo M, Hwang YJ, Kim BS, Ghim SY. Screening of plant growth-promoting rhizobacteria as elicitor of systemic resistance against gray leaf spot disease in pepper. Appl Soil Ecol 2014; 73:1–8. CrossRef
6.Lankford CE. Bacterial assimilation of iron. Crit Rev Microbiol 1973; 2:273–331. CrossRef
7.Loper JE, Buyer JS. Siderophores in microbial interactions on plant surfaces. Mol Plant Microbe In 1991; 4(1):5–13. CrossRef
8.Gyaneshwar P, Naresh KG, Parekh LJ. Effect of buffering on the phosphate solubilizing ability of microorganisms. World J Microbiol Biotechnol 1998; 14:669–73. CrossRef
9.Schippers B, Bakker AW Bakker PAHM. Interactions of deleterious and beneficial rhizosphere micro-organisms and the effect of cropping practices. Ann Rev Phytopathol 1987; 25:339–58. CrossRef
10.Manwar AV, Khandelwal SR, Chaudhari, BL, Meyer JM, Chincholkar SB. Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl Biochem Biotech 2004; 118(1–3):243–51. CrossRef
11.Ribeiro CM, Cardoso EJBN. Isolation, selection and characterization of root associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia). Microbiol Res 2012; 167(2):69–78. CrossRef
12.Park M, Kim C, Yang J, Lee H, Shin W, Kim S, et al. Isolation and characterization of diazotrophic growth promoting bacteria from rhizosphere of agricultural crops of Korea. Microbiol Res 2005; 160(2):127–33. CrossRef
13.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 2009; 63:541–56. CrossRef
14.Bhattacharyya PK, Jha DK. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 2012; 28:1327–50. CrossRef
15.Goswami D, Dhandhukia P, Patel P, Thakker JN. Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol Res 2014; 169(1): 66–75. CrossRef
16.Gaur AC. Physiological functions of phosphate solubilizing micro-organisms. In: Gaur AC (ed.). Phosphate solubilizing microorganisms as biofertilizers. Omega Scientific Publishers, New Delhi, India, pp 16–72, 1990.
17.King EJ. The colorimetric determination of phosphorus. Biochem J 1932: 26(2):292. CrossRef
18.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indole acetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol 1991; 57:535–38. CrossRef
19.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 1987; 160(1):47–56. CrossRef
20.Alexander DB, Zuberer DA. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils 1991; 12:39–45. CrossRef
21.Cappuccino JG, Sherman N. Biochemical activities of microorganisms. In: Microbiology, A laboratory manual. The Benjamin/Cummings Publishing Co., San Francisco, CA, 1992.
22.Lorck H. Production of hydrocyanic acid by bacteria. Physiol Plant 1948; 1:142–46. CrossRef
23.Cappuccino JG, Sherman N. Microbiology. A Laboratory manual.7th Ed. Pearson Education, Inc., San Francisco, CA, 2005.
24.Araújo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JW, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 2002; 68(10): 4906–14. CrossRef
25.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35:1547–49. CrossRef
26.Duangpaenga A, Phetcharata P, Chanthaphoa S, Okuda N. Screening of endophyte bacteria for phosphate solubilization from organic rice. In: Proceeding—Science and Engineering, pp 61–66, 2013.
27.Zheng BX, Ibrahim M, Zhang DP, Bi QF, Li HZ, Zhou GW, et al. Identification and characterization of inorganic-phosphate-solubilizing bacteria from agricultural fields with a rapid isolation method. AMB Express 2018; 8(1):47. CrossRef
28.Tomer S, Suyal DC, Shukla A, Rajwar J, Yadav A, Shouche Y, et al. Isolation and characterization of phosphate solubilizing bacteria from Western Indian Himalayan soils. Biotech 2017; 7(2): 95. CrossRef
29.Baliah NT, Pandiarajan G, Kumar B M. Isolation, identification and characterization of phosphate solubilizing bacteria from different crop soils of SrivilliputturTaluk, Virudhunagar District, Tamil Nadu. Trop Ecol 2016; 57(3):465–74.
30.Liu C, Mou L, Yi J, Wang J, Liu A, Yu J. The Eno Gene of Burkholderia cenocepacia Strain 71-2 is involved in phosphate solubilization. Curr Microbiol 2019; 76(4): 495–502. CrossRef
31.Karpagam T, Nagalakshmi PK. Isolation and characterization of phosphate solubilizing microbes from agricultural soil. IJCMAS 2014; 3(3): 601–14.
32.Rodriguez H, Reynaldo F. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 1999; 17:319–39. CrossRef
33.Chen YP, Rekha PD, Arun AB, Shen FT, Lai WA, Young CC. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl Soil Ecol 2006; 34:33–41. CrossRef
34.Gaur AC, Madan M, Ostwal KP. Solubilization of phosphatic compounds by native microflora of rock phosphate. Ind J ExpBiol 1973; 11:427–9.
35.Zarrin F, Saleemi M, Zia M, Sultan T, Aslam M, Rehman RU, Chaudhary FM. Antifungal activity of plant growth promoting rhizobacteria isolates against Rhizoctonia solani in wheat. Afr J Biotechnol 2009; 8(2):219–25.
36.Rana A, Saharan B, Joshi M, Prasanna R, Kumar K, Nain L. Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Ann Microbiol 2011; 61(4): 893–900. CrossRef
37.Pandey S, Gupta S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front Microbiol 2019; 10:1506. CrossRef
38.Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 1996; 42(3): 207–20. CrossRef
39.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signalling. FEMS Microbiol Rev 2007; 31(4):425–48. CrossRef
40.Szilagyi-Zecchin VJ, Ikeda AC, Hungria M, Adamoski D, Kava-Cordeiro V, Glienke C, Galli-Terasawa LV. Identification and characterization of endophytic bacteria from corn (Zea mays L.) roots with biotechnological potential in agriculture. AMB Express 2014; 4(1):26. CrossRef
41.Patel PR, Shaikh SS, Sayyed RZ. Modified chrome azurol S method for detection and estimation of siderophores having an affinity for metal ions other than iron. Environ Sustain 2018; 1:81–7. CrossRef
42.Gupta A, Gopal M. Siderophore production by plant growth promoting rhizobacteria. Indian J Agric Res 2008; 42(2): 153–6.
43.Pahari A, Mishra BB. Characterization of siderophore producing rhizobacteria and its effect on growth performance of different vegetables. Int J Curr Microbiol App Sci 2017; 6(5): 1398–405. CrossRef
44.Ghavami N, Alikhani HA, Pourbabaei AA, Besharati H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J Plant Nutr 2017; 40(5): 736–46. CrossRef