1. INTRODUCTION
The presence of drug resistance to antiretroviral (ARV) drugs poses a significant barrier to effectively treating individuals infected with human immunodeficiency virus type 1 (HIV-1). HIV-1 resistance to drugs can be acquired through the development of resistance in individuals undergoing antiretroviral therapy (ART), or transmitted when a drug-resistant virus is passed on to someone who has not been previously exposed to ARV drugs. While both acquired and transmitted drug resistance of HIV-1 are significant issues in public health, it is worth noting that transmitted resistance has the potential to more swiftly undermine the efficacy of initial ART on a population scale [1]. Individuals who have acquired drug resistance are faced with a reduced genetic threshold for resistance upon initiating ART. This leads to an increased likelihood of virological failure and a higher risk of gaining resistance to the medications in their treatment regimen, even if those treatments were initially effective [2-5]. Numerous retrospective and prospective studies have provided evidence indicating that the existence of medication resistance before initiating a treatment regimen is a distinct and influential factor in determining the efficacy of such a regimen [6,7]. Consequently, numerous expert committees have issued recommendations advocating for the utilization of HIV reverse transcriptase (RT) and protease sequencing in order to assist clinicians in the selection of appropriate ARV medicines for their patients. Additionally, genotypic resistance testing (GRT) has become an integral component of standard clinical care in recent years [8]. In developed nations, drug resistance testing (DRT) has become widespread and is widely acknowledged as a vital component of the treatment of patients with detectable plasma viremia who are receiving ART. In addition, transmission of drug-resistant viruses from one individual to another occurs in a variety of contexts, including between adults and from mother to infant [9,10]. This indicates that testing for drug resistance before initiating therapy may be advantageous, even for individuals who have never received treatment [11]. Interpretation of genotypic and phenotypic DRT continues to provide challenges, despite the substantial amount of research conducted in this area [12].
The World Health Organization (WHO) has utilized HIV DRT to inform policies regarding the distribution of ART on an individual basis in clinical practice. To provide public health recommendations about ART regimens for different groups, it is necessary to gather relevant information. This test is valuable because it can detect mutations in the viral genome that confer resistance to the patient’s regimen, allowing doctors to fine-tune the treatment they provide their patients and increase the likelihood that they will achieve virological suppression. In addition to minimizing the spread of HIV drug resistance, community-level drug resistance surveillance can improve treatment outcomes for the entire population by decreasing the use of ineffective treatments. The laboratory procedures for HIV DRT encompass several steps. These include the extraction of viral RNA from plasma or dry blood spot samples, amplification of the RNA using RT-polymerase chain reaction (PCR), subsequent nested PCR amplification, documentation of the PCR products using gel electrophoresis, purification of the nested PCR products, cycle sequencing of the purified products, purification of the cycle sequencing products, and, finally, population-based (bulk) sequencing. The utilization of multiple sequencing primers may be necessary, depending on the specific laboratory methodology, in order to achieve comprehensive and bidirectional coverage of the entirety of the HIV-1 pol region of interest during the sequencing process [13]. To combat HIV-1 drug resistance, researchers have focused on developing more effective ARV drugs in recent years. The researchers intended to combine cutting-edge technologies for in silico virtual screening/structure-based drug discovery, synthetic organic chemistry, mechanistic enzymology, and protein crystallography, as well as pharmacological assays [14].
Patients who are infected with viruses, including HIV-1, have the ability to rapidly develop mutations that make them resistant to drugs. The assessment of viral resistance plays a crucial role in determining the effectiveness of ART. Consequently, genotypic testing is conducted either at the initiation of treatment or when treatment is deemed unsuccessful. The relevant regions of the viral genome are subjected to sequencing, followed by the interpretation of the resulting amino acid sequence in order to determine the resistance to therapy [15]. The job of interpreting the outcomes of genotypic drug resistance tests for HIV-1 poses a significant challenge for doctors involved in the treatment of individuals infected with HIV-1. The observed phenomenon can be attributed to the intricate interplay of several mutations that contribute to the development of drug resistance, as well as the diverse degrees of diminished sensitivity resulting from these mutations. A constraint of DRT is their incapacity to detect subtle drug-resistant variations within a patient’s viral quasi-species, notwithstanding their potential therapeutic relevance.
The in silico method exemplifies the rapid evolution and eventual replacement of more conventional HIV-1 DRT methods in clinical diagnostics. Bioinformatics, structural biology, and the availability of three-dimensional (3D) protein structures, in particular, have played a significant role in expanding the likelihood of discovering novel medications through the application of rational methods [16]. Two distinct approaches could be employed to comprehend drug resistance: rule-based frameworks and algorithm-driven frameworks. Algorithmic systems are developed by employing statistical models that are trained on clinical or virological data using machine learning approaches. In contrast, rules-based interpretation systems rely on the knowledge and proficiency of expert panels [17]. A multitude of expert perspectives have contributed to the development of various sets of guidelines, including those from REGA [18] ANRS [19], HIVdb [20], and HIV-GRADE [21], which has arisen as a result of the comprehensive insights informing HIV-GRADE. In the same way, algorithmic techniques exhibit variations in terms of the specific machine learning algorithms employed and the datasets utilized for training the models. An illustrative instance is the utilization of geno2pheno[resistance] [22], which employs support vector regression and classification techniques.
This article examines the scientific principles that form the basis for interpreting genotypic-resistance test results. It also explores the existing web-based systems used for interpreting genotypic and phenotypic data, as well as the websites that offer clinically significant summaries of HIV-1 drug resistance mutations (DRMs).
2. MATERIALS AND METHODS
2.1. HIVdb
The Stanford HIV Drug Resistance Database is responsible for the maintenance of an online genotypic resistance interpretation system called HIVdb. This system is freely accessible and serves as a valuable tool for clinicians and laboratories in the interpretation of HIV-1 GRT [23]. The assays evaluate the susceptibility of protease inhibitors (PIs), integrase inhibitors, as well as nucleoside RT inhibitors (NRTIs) and non-nucleoside RT inhibitors (NNRTIs). The HIVdb genotypic resistance interpretation system offers three distinct categories of information, with a comprehensive assessment of ARV resistance mutations in a given sequence. First, it assigns penalty scores to each mutation, indicating their impact on resistance. Second, it provides estimations of reduced susceptibility to NRTIs, NNRTIs, PIs, and integrase inhibitors. Finally, it includes informative comments pertaining to each specific ARV resistance mutation. The application exhibits several notable qualities, including its user-friendly interface for sequence submission, robust quality control analysis capabilities, transparent functionality, and extensive provision for user comments. HIVdb has the ability to provide outcomes using diverse interpretation algorithms for genotypic resistance of HIV-1, following the compilation of algorithm specifications [24].
2.2. Sequence Analysis Using HIVdb
Nucleotide sequence of drug resistance HIV-1 integrases was retrieved from GenBank NCBI using accession number: BD168948.1 in FASTA format. The protein sequence of drug resistance HIV integrases contains 864 amino acids, and if only one sequence is being input, it can be entered as plain text. If multiple sequences are being input, they must be in the FASTA format as given in Figure 1.
 | Figure 1: Nucleotide sequence of drug resistance HIV-1 integrases in the FASTA format. Sequence name: WO 2002038771-A/2: Drug resistance HIV integrases. GenBank accession number: BD168948.1.
[Click here to view] |
2.3. HIV-GRADE (Genotypic Resistance-Algorithm Deutschland)
HIV-GRADE was conceived as a national strategy to standardize drug resistance interpretation in Germany and introduce standards for evaluating the impact of mutations on treatment combinations. The guidelines for HIV-GRADE are derived from a bioinformatics-driven interpretation system (geno2pheno[resistance]) and clinical follow-up data. HIV-GRADE permits users to view the rules and outcomes of alternative drug resistance algorithms for a particular sequence in a centralized location. Unique to this tool is the ability to compare side-by-side the outcomes of various drug resistance assessment techniques [25]. The HIV-GRADE program permits the analysis of multiple nucleic acid sequences in bulk. HIV-GRADE results can be contrasted with those of other systems, such as REGA [18], ANRS [19], and HIVdb [20].
2.4. Sequence Analysis Using HIV-GRADE
Nucleotide sequence of drug resistance HIV-1 reverse transcriptase was retrieved from GenBank NCBI using accession number: Z99333.1 in the FASTA format. The nucleotide sequence of drug resistance HIV-1 RT contains 777 amino acids, and if only one sequence is being input, it can be entered as plain text. If multiple sequences are being input, they must be in the FASTA format as given in Figure 2.
 | Figure 2: Nucleotide sequence of HIV-1 RT in the FASTA format. Sequence name: HIV-1 isolate C44 DNA for RT. GenBank accession number: Z99333.1.
[Click here to view] |
2.5. Geno2pheno[Resistance] System
Geno2pheno[resistance] is a data-driven method for making quantitative predictions about viral drug resistance based on a compilation of genotype–phenotype pairings using support vector regression [22]. To determine HIV-1 viral resistance, genotype–phenotype (geno2pheno) techniques [26-29] are utilized. In geno2pheno[resistance], two distinct strategies are available. In the support vector regression models that serve as the foundation for the original geno2pheno[resistance] method, a linear kernel function is utilized. To train these models, the researchers employed Sanger sequencing techniques to analyze the genetic sequences of HIV-1. Additionally, they measured drug-specific resistance factors (RFs), which are numerical values that indicate the degree of resistance to a particular medication. These factors quantify the change in inhibitory concentration required to suppress the growth of a modified sample compared with the original, non-mutated strain [26,28]. Newer methods, including g2p[drug exposure], are predicated on statistical techniques known as support vector classification models. Clinical data were used to train these models, specifically Sanger sequences labeled with whether or not they originated from a patient who had been treated with a particular medication [29].
2.6. Sequence Analysis Using Geno2pheno[Resistance]
The nucleotide sequence of drug resistance HIV-1 protease (pol) gene was retrieved from GenBank NCBI using accession number: MW110766.1 in the FASTA format. The nucleotide sequence of drug resistance HIV-1 RT contains 297 amino acids, and if only one sequence is being input, it can be entered as plain text. If multiple sequences are being input, they must be in the FASTA format as given in Figure 3.
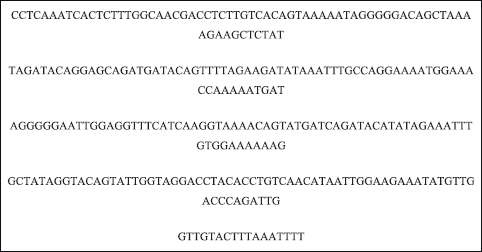 | Figure 3: Nucleotide sequence of HIV-1 proteases (pol) gene in FASTA format. Sequence name: HIV-1 isolate UVAS/PACP/011 from Pakistan protease (pol) gene, partial cds. GenBank accession number: MW110766.1.
[Click here to view] |
3. RESULTS AND DISCUSSION
The HIVdb GRT interpretation system is a rules-based approach that assesses NRTI, NNRTI, PI, and/or integrase strand transfer inhibitor (INSTI) susceptibility using the ARV penalty score for DRMs in an HIV-1 protease, RT, or integrase sequence (Table 1). DRM penalty scores (or ARV penalty scores) have been developed for both singular DRMs and sets of DRMs. Each ARV is classified as potentially low-level resistant, susceptible, low-level resistant, intermediate-level resistant, or highly resistant, indicating varying degrees of drug resistance (also referred to as reduced susceptibility). The Knowledgebase appendix entitled “DRM penalty scores” describes the relationship between DRM penalty scores and the five reduced susceptibility levels.
Table 1: Drug resistance HIV integrases: Sequence summary (HIVdb 9.4.1 software).
| Subtype (HIVdb 9.4.1 software) | B (1.74%)
KJ704787: United States (1983); B (1.74%); best match
L31963: France (1983); B (2.55%)
HQ026550: Korea, Republic of (1992); B (2.66%) D10112: United Kingdom (1983); B (2.78%)
FJ647145: South Africa (1985); B (2.78%)
KT427710: Brazil (2010); B (2.78%)
AF042100: Australia (1986); B (2.89%)
AY173951: Thailand (1990); B (2.89%)
U34603: The Netherlands (1986); B (2.89%)
EF514709: Denmark (2001); B (3.01%) |
| IN SDRMs (HIVdb 9.4.1 software) | None |
The categorization process relies on the HIVdb GRT interpretation system. Viruses are categorized as “susceptible” when they do not display any signs of reduced susceptibility in comparison with wild-type viruses. The attribution of “potential low-level resistance” to a virus is contingent upon the presence of DRMs that are indicative of prior exposure to ARV drugs or are linked with resistance. However, this attribution is only applicable when these DRMs occur in conjunction with other DRMs. When a virus exhibits DRMs that are linked to reduced sensitivity to ARV drugs in laboratory settings or has a suboptimal virological response to ARV therapy, it is categorized as having “low-level resistance.” The phrase “intermediate resistance” refers to a scenario wherein the efficacy of an ARV drug is predicted to be reduced in the presence of DRMs in a virus. However, it is anticipated that the ARV will still exhibit substantial antiviral activity against the virus. A virus is categorized as “high-level resistant” when it possesses DRMs that are anticipated to provide a resistance level comparable with viruses demonstrating the most significant reductions in susceptibility to ARV treatment in laboratory settings or viruses that display limited or no virological response to ARV therapy.
There are two objectives associated with DRM penalty scoring. First, they serve an informative function by demonstrating the degree to which a DRM affects the clinical activity of an ARV. In addition, the scores are calibrated so that the total DRM penalty scores for a given ARV yield an estimate of reduced susceptibility for that ARV that is consistent with available research and expert opinion. As part of the HIVdb GRT interpretation system, which also includes DRM penalty scores and projected levels of diminished ARV susceptibilities, users can find the mutation comments equally helpful and informative. The HIVdb GRT interpretation as a whole includes DRM comments.
3.1. Sequence Quality Assessment
According to the results given in Figure 4, there are no known sequence quality issues.
 | Figure 4: Sequence quality assessment of HIV-1 integrase. Sequence name: WO 2002038771-A/2: Drug resistance HIV integrases. GenBank accession number: BD168948.1. Drug resistance interpretation: IN HIVdb 9.4.1. INSTI major mutations: None. INSTI accessory mutations: None. In other mutations: I72V · I113V · V151I.
[Click here to view] |
3.2. Integrase (IN)
Table 2 interprets drug resistance HIV-1 integrases susceptible to the above-mentioned INSTI drugs. V151I is an accessory INSTI-selected mutation that occurs in 1–3% of viruses from ART-naive persons depending on subtype. Alone, it appears to have less or no effect on INSTI susceptibility. No DRMs were found for INSTI [23].
Table 2: Integrase strand transfer inhibitors (INSTIs).
| Bictegravir (BIC) | Susceptible |
| Cabotegravir (CAB) | Susceptible |
| Dolutegravir (DTG) | Susceptible |
| Elvitegravir (EVG) | Susceptible |
| Raltegravir (RAL) | Susceptible |
The HIV-GRADE interpretation system refers to four different levels of drug resistance. Generally, drugs with lower levels of resistance should be preferred as long as clinically appropriate combinations are possible.

3.3. Results for HIV-GRADE
Table 3 shows that the user can give the desired sequence name to the entered nucleotide sequences and can also select the different algorithms to compare the results.
Table 3: Title of the sequence and chosen algorithms.
| Sequence Name | HIV reverse Transcriptase_1 sample_one |
| Algorithms | GRADE, ANRS, HIVdb, Rega |
The length of sequences that incorporated the same sequence as of RT and protease can be compared with the length of the entered nucleotide sequence of HIV-1 Integrase [Table 4].
Table 4: The total length of incorporated sequences.
| Sequence Includes | Codons: Subtype |
|---|
| RT | 1–258 B (95.1%) |
| PR | 1–98 B (93.5%) |
The results given in Table 5 reveal the differences between the genes from consensus B strains and DRM strains.
Table 5: Gene differences from consensus B/DRMs.
| RT | I2M, V35T, M41L, V60I, K122E, D123E, I135T, Q207E, R211K, T215Y, V245K, M41L, T215Y |
| PR | L10V, I13V, F53L, L63P, I64V, A71V, T74A, N83S, I84V, L90M, I93M, T96S, N98D, L10V, F53L, A71V, T74A, N83S, I84V, L90M |
HIV-GRADE results were compared with those of other systems, such as REGA, ANRS, HIVdb, and the geno2pheno[resistance] system results simultaneously as given in Tables 6 and 7.
Table 6: Comparison of HIV-GRADE results with ANRS, HIVdb, and REGA and mutations scored for NNRTI drug class.
| NNRTI | GRADE 01/2023
Mutation List Rating | SIR | ANRS 33_10/2022
Mutation List Rating | SIR | HIVdb 9.4
Mutation List Rating | SIR | Rega 10.0.0
Mutation List
Rating | SIR |
|---|
| DOR | Susceptible |  | Susceptible |  | Susceptible |  | |
| EFV | Susceptible |  | Susceptible |  | Susceptible |  | Susceptible GSS 1 |  |
| ETR | Susceptible |  | Susceptible |  | Susceptible |  | Susceptible GSS 1 |  |
| NVP | Susceptible |  | Susceptible |  | Susceptible |  | Susceptible GSS 1 |  |
| RPV | Susceptible |  | Susceptible |  | Susceptible |  | Susceptible GSS 1 |  |
Table 7: Scored mutations for drug class NRTI: M41L, T215Y.
| NRTI | GRADE 01/2023
Mutation List Rating | SIR | ANRS 33_10/2022
Mutation List Rating | SIR | Mutation List | HIVdb 9.4
Rating | SIR | Mutation List | Rega 10.0.0
Rating | SIR |
| 3TC | | Susceptible |  | | Susceptible |  | T215Y, M41L | Susceptible (Score: 5) |  | | Susceptible GSS 1 |  |
| ABC | M41L, T215Y | Flagged mutations |  | M41L, T215Y | Possible resistance |  | M41L, T215Y | Low-level resistance (Score: 25) |  | | Susceptible GSS 1 |  |
| AZT | M41L, T215Y | Intermediate |  | T215Y | Resistance |  | M41L, T215Y | High-level resistance (Score: 85) |  | M41L, T215Y | Intermediate Resistant GSS 0.5 |  |
| AZT_SP | M41L, T215Y | Intermediate |  | | | |
| D4T | M41L, T215Y | Intermediate |  | | M41L, T215Y | High-level resistance (Score: 65) |  | M41L, T215Y | Intermediate Resistant GSS 0.5 |  |
| D4T_SP | M41L, T215Y | Intermediate |  | | | |
| ddI | | | M41L, T215Y | Intermediate resistance (Score: 35) |  | M41L, T215Y | Intermediate Resistant GSS 0.5 |  |
| FTC | | Susceptible |  | | Susceptible |  | T215Y, M41L | Susceptible (Score: 5) |  | | Susceptible GSS 1 |  |
| ISL | | | Susceptible |  | | |
| TDF/TAF | M41L, T215Y | Intermediate |  | | Susceptible |  | M41L, T215Y | Low-level resistance (Score: 25) |  | | Susceptible GSS 1 |  |
| TDF/TAF_SP | M41L, T215Y | Intermediate |  | | | |
| APV/FPV_RTV | I84V, L90M | Intermediate |  | | F53L, I84V, L90M | High-level resistance (Score: 90) |  | L10V, I84V, L90M | Intermediate Resistant GSS 0.75
(Score: 2) |  |
| ATV | I84V, L90M, F53L, A71V | Resistance |  | | | |
| ATV_RTV | F53L, A71V, I84V, L90M | Resistance |  | L10V, A71V, I84V, L90M | Resistance |  | F53L, I84V, L90M | High-level resistance (Score: 105) |  | L10V, A71V, T74A, I84V, L90M | Intermediate Resistant GSS 0.75
(Score: 2.75) |  |
| ATV_SP | F53L, A71V, I84V, L90M | Resistance |  | | | |
| DRV | I84V | Flagged mutations |  | | Susceptible |  | I84V | Low-level resistance (Score: 15) |  | I84V | Susceptible GSS 1.5
(Score: 1.5) |  |
| DRV_QD | | | Susceptible |  | | |
| IDV_RTV | | | F53L, I84V, L90M | High-level resistance (Score: 100) |  | L10V, A71V, T74A, I84V, L90M | Resistant GSS 0
(Score: 3.75) |  |
| LPV | F53L, A71V, I84V, L90M | Intermediate |  | L10V, F53L, L63P, A71V, I84V, L90M | Resistance |  | I84V, L90M | Intermediate resistance (Score: 45) |  | L10V, F53L, I64V, A71V, I84V, L90M | Intermediate Resistant GSS 0.75
(Score: 2.25) |  |
| NFV | | | | | F53L, I84V, | High-level resistance |  | L10V, I64V, A71V, | Resistant GSS 0 |  |
| | | | L90M | (Score: 140) | | T74A, I84V, L90M, | (Score: 4) | |
| | | | | | | I93M | | |
| SQV_RTV | I84V | Resistance |  | F53L, I84V, | High-level resistance |  | L10V, F53L, A71V, | Resistant GSS 0 |  |
| | | | L90M | (Score: 130) | | T74A, I84V, L90M | (Score: 5.5) | |
| SQV_SP | I84V | Resistance |  | | | | | | |
| TPV | I84V | Flagged mutations |  | | I84V | Intermediate resistance (Score: 30) |  | L90M, I84V | Susceptible GSS 1.5 (Score: 1.25) |  |
3.4. Comments on PIs
According to the actual version of the label, dosage adaptions for DRV should be considered.
A is a mutation at the resistance-associated codon 74 that is not scored by GRADE, ANRS, and HIVdb.
S is a mutation at the resistance-associated codon 83 that is not scored by GRADE, ANRS, and HIVdb.
V is a mutation at the resistance-associated codon 10 that is not scored by HIVdb.
V is a mutation at the resistance-associated codon 71 that is not scored by HIVdb.
3.5. GRADE Interpretation
According to the actual version of the label, dosage adaptions for DRV should be considered.
3.6. HIVdb Interpretation
The polymorphic mutations A71V/T are accessory mutations that have been selected by PIs and have the ability to enhance the reproduction of viruses carrying other PI-resistance mutations.
The F53L mutation is an accessory mutation that exhibits nonpolymorphic characteristics. It is largely selected by ARV drugs such as saquinavir (SQV), indinavir (IDV), atazanavir (ATV), and lopinavir (LPV). When combined with other mutations, it has been observed to be linked to a decrease in susceptibility to ATV and maybe LPV. The F53Y mutation is a relatively rare nonpolymorphic accessory mutation that has not been extensively investigated in scientific research.
I84V is a substrate-cleft mutation that has been picked by each of the principal investigators. This mutation is nonpolymorphic in nature. The I84V mutation confers decreased resistance to ARV drugs such as LPV, ATV, and DRV. The L10I/V mutations are known to be polymorphic and have been selected as accessory alterations that enhance the replication of viruses carrying other mutations associated with resistance to PIs. The L90M mutation is a non-polymorphic mutation that has been shown to decrease susceptibility to ATV and, to a lesser degree, LPV.
The M41L mutation is commonly observed in conjunction with the T215Y mutation in the context of ART. The combination of M41L and T215Y mutations results in a moderate to high level of resistance to azidothymidine (AZT) and stavudine (d4T) while also contributing to decreased sensitivity to didanosine (ddI), abacavir (ABC), and tenofovir disoproxil fumarate (TDF). The T215Y/F mutations are known as thymidine analog mutations and are associated with the development of intermediate to high-level resistance to AZT, as well as the possibility for low-level resistance to ABC and TDF.
3.7. Geno2pheno[Resistance] Results
The protease substitutions involving the insertion of 36I between positions 33 and 41 do not demonstrate any evidence of being specifically targeted by PIs or leading to a decrease in PI susceptibility, as observed in Table 8.
Table 8: Different protease substitutions.
| Aligned Amino Acids | 99 |
| Matches in Reference Sequence | 89.9 % |
| Matches in Alignment | 89.9 % |
| Substitutions | V 3 I, I 13 V, E 35 D, M 36 I, S 37 N, R 41 K, R 57 K, L 63 H, H 69 K, L 89 M |
3.8. Phenotype Prediction
The drugs are represented by three-letter codes. These codes correspond to specific classes of drugs, including nucleoside inhibitors of the RT, NNRTIs, and PIs. Examples of nucleoside inhibitors of the RT include zidovudine (ZDV), zalcitabine (ddC), ddI, d4T, lamivudine (3TC), ABC, and TDF. NNRTIs include nevirapine (NVP), delavirdine, and efavirenz (EFV). PIs encompass SQV, IDV, ritonavir (RTV), nelfinavir (NFV), amprenavir (APV), LPV, and ATV.
In Table 9, (**) positions are ordered according to their impact on the phenotype prediction. Differences with respect to HXB2 strain are underlined. Positions shown in red and green contribute to an increase or decrease in resistance, respectively. At most 15 positions are shown for each drug. In addition, (***) resistance predictions and scored mutations for ETR and RPV were performed with rules-based drug resistance interpretation models by HIV-GRADE.
Table 9: Prediction of phenotype based on resistance predictions and scored mutation subtype prediction.
| Drug | RF (*) | z-Score | Scored Positions (**) |
|---|
| ZDV | 0 | 0 | |
| ddI | 0 | 0 | |
| d4T | 0 | 0 | |
| 3TC | 0 | 0 | |
| ABC | 0 | 0 | |
| TDF | 0 | 0 | |
| NVP | 0 | 0 | |
| EFV | 0 | 0 | |
| ETR (***) | Susceptible | |
| RPV (***) | Susceptible | |
| SQV | 0.731 | –1.059 | 48G 73G 90L 84I 54I 11V 74T 88N 53F 95C 26T 1P 71A 80T 34E |
| IDV | 0.8 | –1.139 | 54I 82V 88N 46M 29D 1P 73G 21E 65E 84I 11V 71A 85I 30D 90L |
| NFV | 0.887 | –0.848 | 88N 54I 30D 46M 82V 97L 20K 73G 68G 90L 71A 31T 75V 74T 84I |
| APV | 1.085 | –0.094 | 54I 76L 50I 84I 46M 32V 85I 22A 1P 47I 82V 89M 97L 45K 21E |
| LPV | 0.626 | –1.23 | 54I 82V 46M 84I 50I 76L 10L 22A 71A 7Q 24L 20K 25D 47I 92Q |
| TPV | 0.578 | –1.212 | 48G 84I 33L 54I 47I 89M 71A 72I 15I 91T 20K 69K 90L 82V 74T |
| DRV | 0.832 | –0.692 | 47I 84I 54I 33L 76L 74T 43K 73G 46M 71A 65E 89M 48G 93I 10L |
| ATV | 0.899 | –0.968 | 54I 48G 73G 84I 82V 4T 71A 88N 90L 7Q 46M 20K 24L 76L 45K |
Table 10 shows that the sequence is predicted (fit 97%) to be of HIV subtype A1.
Table 10: Prediction of HIV-1 subtype.
| Subtype | Probability |
|---|
| A1 (1) | 0.97 |
3.9. Drug Exposure Prediction
In Table 11, the drug-exposure score (DES) is an estimated number that relates to the extent of drug exposure based on this method. Because RFs and DESs vary widely between medications, geno2pheno[resistance] converts them to z-scores, where z represents the number of standard deviations above or below the mean of therapy-naive patients. Ultimately, each z-score is translated into one of three clinically motivated levels of resistance [15]: susceptible, intermediate, or resistant as shown in Figure 5.
Table 11: Prediction of drug exposure based on drug-exposure score and z-score.
| Drug | DES | z-Score | Drug Exposure | Resistance | Scored Positions |
|---|
| SQV | –1.240 | –0.957 | Unexposed | Susceptible | 84I 48G 24L 74T 54I 90L 7Q 69K 91T 53F 38L 39P 96T 8R |
| IDV | –1.250 | –1.192 | Unexposed | Susceptible | 82V 4T 84I 88N 5L 90L 46M 54I 66I 24L 48G 60D 73G 23L 93I |
| NFV | –1.084 | –0.526 | Unexposed | Susceptible | 88N 30D 54I 84I 90L 48G 22A 5L 6W 83N |
| APV | –1.256 | –0.717 | Unexposed | Susceptible | 50I 84I 54I 30D 25D 24L 47I 83N 92Q 73G 74T 33L 76L 21E 90L |
| LPV | –1.091 | –0.830 | Unexposed | Susceptible | 54I 47I 84I 48G 73G 76L 32V 46M 10L 30D 82V 36I 50I 60D |
| TPV | –1.226 | –0.277 | Unexposed | Susceptible | 84I 47I 50I 24L 33L 43K 83N 54I 48G 30D 76L 66I 90L |
| DRV | –1.035 | –0.471 | Unexposed | Susceptible | 43K 84I 11V 50I 30D 87R 42W 33L 55K 57K 6W 10L 90L 76L 34E |
| ATV | –1.122 | –0.995 | Unexposed | Susceptible | 84I 88N 48G 76L 54I 74T 73G 23L 50I 89M 24L 32V 10L 71A 11V |
 | Figure 5: (*) Number of standard deviations above the mean of drug-naive patients. Negative z-scores may indicate hypersusceptibility.
[Click here to view] |
4. DISCUSSION
HIV/AIDS, which is a prevalent global public health issue, is ascribed to HIV. The implementation of ART has substantially improved the prognosis and quality of life for HIV-positive individuals. Despite this, the development of drug resistance in HIV is a significant barrier to the continued efficacy of treatment protocols. To customize therapeutic strategies for maximum efficacy, it is crucial to accurately forecast drug resistance [30].
Highly active ART, which involves the use of a combination of ARV drugs, is currently considered the established protocol for preventing HIV-1 infection and the development of resistance. The HIV/AIDS epidemic is expected to persist for an extended duration, emphasizing the imperative to pursue the development of innovative and enhanced therapeutic approaches. Several factors that should be taken into account for the development of enhanced anti-HIV-1 drugs encompass reduced long-term toxicity, the capacity to combat the establishment of drug-resistant variations, and the creation of a long-acting treatment that necessitates less frequent administration [14]. The integration of GRT has become a standard component in the diagnostic process for managing patients with HIV infection. Nevertheless, the clinical efficacy of this treatment is constrained in practical settings due to the complex association between genotypic changes and phenotypic resistance observed in vitro, as well as the corresponding treatment response observed in vivo [26].
The ability to accurately anticipate the virological response to a novel ARV drug treatment regimen is contingent upon the presence of pre-existing HIV-1 drug resistance before treatment begins. Numerous studies have demonstrated that the implementation of GRT before initiating a new treatment regimen enhances the probability of achieving a virological response to such a regimen. However, the process of interpreting the results obtained from HIV-1 medication resistance tests presents significant difficulties. It is essential to first recognize the presence of several mutations linked to drug resistance, often known as DRMs. Moreover, the presence of DRMs leads to varying levels of reduced susceptibility to various ARV medications. In addition, traditional GRTs lack the capability to detect DRMs that may be present in a patient’s viral population at minimal rates [30].
In recent years, the field of HIV drug resistance prediction has undergone a significant transformation due to the emergence of computational tools and algorithms that enable the use of in silico techniques. The novel methodology of this study provides numerous advantages, including expedited examination of large genetic datasets, efficient resource utilization, and the ability to predict patterns of resistance across a broad spectrum of ARV medications. The utilization of in silico methodologies serves as a prime example of the swift progression and eventual substitution of traditional HIV-1 DRT techniques in the realm of clinical diagnostics.
Computational models and simulations used in in silico approaches may not fully capture the complexities of biological systems. It is possible that in silico models do not account for all interactions and factors influencing drug resistance. In vitro experiments can reveal previously unknown interactions, side effects, and other elements that may not be fully represented by computer predictions alone. HIV is well known for its rapid evolution and high mutation rate. In vitro studies can shed light on the dynamic nature of viral evolution by observing and comprehending the viral evolutionary processes. In silico models may struggle to keep up with the virus’s evolving nature in the absence of real-time experimental data.
The fields of bioinformatics and structural biology, with a specific focus on the accessibility of 3D protein structures, have greatly contributed to the increased potential for the discovery of new pharmaceuticals by employing logical approaches. This article explores the scientific foundations that underlie the interpretation of genotypic-resistance test outcomes. In addition, this study examines the currently employed web-based platforms for the interpretation of genotypic and phenotypic data, along with the websites that provide concise and clinically relevant summaries of DRMs in HIV-1.
The HIVdb program evaluates the potential efficacy of an ARV against a specific mutant virus in relation to its effectiveness against a wild-type virus. The integration of a comprehensive comprehension of the principles of ART with the analysis and accompanying remarks aids healthcare professionals in gaining a deeper knowledge of the outcomes derived from HIV-1 GRTs [14]. HIVdb is an advanced computational system designed to analyze HIV-1 sequences provided by users. It employs sophisticated algorithms to determine the potential resistance levels of these sequences to a comprehensive range of 24 FDA-approved ARV drugs. The pharmacological medications under consideration encompass a collective sum of eight PIs, seven NRTIs, five NNRTIs, and four INSTIs. The HIV-GRADE platform provides users with access to a centralized repository where they may access information regarding the rules and results of various alternative drug resistance algorithms for a specific sequence. One distinguishing feature of this tool is its capability to conduct a comparative analysis of different drug resistance assessment approaches, allowing for a side-by-side evaluation of their respective outcomes [25]. The HIV-GRADE program enables the examination of many nucleic acid sequences collectively. The data obtained via HIV-GRADE can be compared with those obtained from other systems, including REGA [18], ANRS [19], and HIVdb [20]. The Geno2pheno system was created with the purpose of aiding in the analysis and interpretation of sequence data derived from GRTs. The Geno2pheno[resistance] tool uses regression models to effectively predict the degree of change in drug susceptibility by analyzing an individual’s genotype. These models enable the transformation of complex mutational patterns into a unified measure of drug resistance for each specific medication. The Geno2pheno[resistance] approach utilizes a data-centric methodology to generate quantitative predictions on the development of drug resistance in viruses. This is achieved by using a comprehensive collection of genotype–phenotype associations through support vector regression. Genotype–phenotype approaches (namely, geno2pheno) are employed for the purpose of assessing HIV-1 viral resistance. Hence, HIVdb, HIV-GRADE, and geno2pheno[resistance] are web servers that are freely available to the public. These servers are designed to provide a quick analysis of viral drug resistance based on genotypic information. This in silico approach is founded on the utilization of these three significant resistance prediction tools, namely, HIVdb, HIV-GRADE, and Geno2pheno. The aforementioned tools have been meticulously developed and improved through extensive research, making them indispensable in the ongoing HIV-related fight against drug resistance. These methodologies collectively provide a comprehensive framework for evaluating genotypic data, predicting resistance mutations, and optimizing treatment approaches.
5. CONCLUSION
In silico HIV drug resistance prediction can help physicians and healthcare researchers choose the right ARV drugs to treat drug-resistant HIV patients. The Stanford HIVdb, HIV-GRATE, and Geno2pheno[resistance] databases help us forecast how genetic alterations may affect ARV medication efficacy. These platforms combine massive databases, computer models, and biological insights to inform personalized treatment methods for doctors and researchers. These databases and technologies speed up in silico DRM discovery, guiding treatment decisions and improving HIV management. However, in silico predictions must be considered with clinical expertise and experimental validation. Computational biologists, doctors, and virologists will collaborate to improve these prediction systems for use in real-world medical settings. In silico HIV medication resistance prediction provides personalized therapy options that improve patient outcomes and aid the global fight against HIV/AIDS.
The distinctive characteristic of this methodology resides in its ability to utilize the computational capabilities of in silico analysis, enabling efficient, economical, and thorough evaluations of genetic information. The integration of HIVdb, HIV-GRADE, and Geno2pheno within this framework provides medical professionals and researchers with a comprehensive perspective on resistance mutations and the complexities of HIV drug resistance. This study has demonstrated the significant potential of the in silico method for accurately predicting HIV drug resistance. In conclusion, this approach presents a flexible and adaptable strategy for confronting the ever-changing landscape of HIV/AIDS therapy by integrating cutting-edge tools and methods. The ongoing development of this methodology has the potential to improve the precision and efficacy of HIV treatment, ultimately benefiting those affected by this global health issue.
6. ACKNOWLEDGMENTS
This is part of my Ph.D. work and I would like to thank my guide Dr. S. Pushkala and the department faculties for their constant support.
7. AUTHOR CONTRIBUTIONS
The first author TB confirms sole responsibility for the study conception and design, data collection, analysis and interpretation of results, and manuscript preparation. The second author reviewed the results and approved the final version of the manuscript.
8. FINANCIAL SUPPORT AND SPONSORSHIP
There is no funding to report.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
13. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Shafer RW, Rhee S-Y, Pillay D, Miller V, Sandstrom P, Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS [Internet]. 2007;21(2):215–23. [CrossRef]
2. Little SJ, Holte S, Routy J-P, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med [Internet]. 2002;347(6):385–94. [CrossRef]
3. Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–8.
4. Markowitz M, Mohri H, Mehandru S, Shet A, Berry L, Kalyanaraman R, et al. Infection with multidrug resistant, dual-tropic HIV-1 and rapid progression to AIDS: a case report. Lancet [Internet]. 2005;365(9464):1031–8. [CrossRef]
5. Daar ES, Richman DD. Confronting the emergence of drug-resistant HIV type 1: impact of antiretroviral therapy on individual and population resistance. AIDS Res Hum Retroviruses. 2005;21:343–57.
6. Rhee S-Y. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res [Internet]. 2003;31(1):298–303. [CrossRef]
7. Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev [Internet]. 2002;15(2):247–77. [CrossRef]
8. Hirsch MS, Brun-Vézinet F, D’Aquila RT, Hammer SM, Johnson VA, Kuritzkes DR, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an international AIDS society-USA panel. JAMA [Internet]. 2000;283(18):2417. [CrossRef]
9. Booth CL, Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. J Antimicrob Chemother [Internet]. 2007;59(6):1047–56. [CrossRef]
10. Arrivé E, Newell M-L, Ekouevi DK, Chaix M-L, Thiebaut R, Masquelier B, et al. Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol [Internet]. 2007;36(5):1009–21. [CrossRef]
11. Hirsch MS, Günthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, Hammer SM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an international AIDS society–USA panel. Clin Infect Dis [Internet]. 2008;47(2):266–85. [CrossRef]
12. Johnson VA, Brun-Vézinet F, Clotet B, Günthard HF, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med. 2007;15(4):119–25.
13. Kingwara L, Karanja M, Ngugi C, Kangogo G, Bera K, Kimani M, et al. From sequence data to patient result: a solution for HIV drug resistance genotyping with exatype, end to end software for pol-HIV-1 Sanger based sequence analysis and patient HIV drug resistance result generation. J Int Assoc Provid AIDS Care [Internet]. 2020;19:232595822096268. [CrossRef]
14. Kudalkar SN, Beloor J, Quijano E, Spasov KA, Lee WG, Cisneros JA, et al. From in silico hit to long-acting late-stage preclinical candidate to combat HIV-1 infection. Proc Natl Acad Sci USA [Internet]. 2018;115:E802–11. [CrossRef]
15. Vercauteren J, Vandamme AM. Algorithms for the interpretation of HIV-1 genotypic drug resistance information. Antiviral Res [Internet]. 2006;71(2-3):335–42. [CrossRef]
16. Nayak C, Chandra I, Singh SK. An in silico pharmacological approach toward the discovery of potent inhibitors to combat drug resistance HIV-1 protease variants. J Cell Biochem [Internet]. 2019;120(6):9063–81. [CrossRef]
17. Döring M, Büch J, Friedrich G, Pironti A, Kalaghatgi P, Knops E, et al. Geno2pheno[ngs-freq]: a genotypic interpretation system for identifying viral drug resistance using next-generation sequencing data. Nucleic Acids Res [Internet]. 2018;46:W271–7. [CrossRef]
18. Kuleuven.be [Internet]. Rega algorithm. 2023 [cited 2023 Aug 21]. Available from: https://rega.kuleuven.be/cev/avd/software/rega-algorithm
19. Hivfrenchresistance.org [Internet]. HIV-1 genotypic drug resistance interpretation’s algorithms. 2023 [cited 2023 Aug 21]. Available from: https://hivfrenchresistance.org/
20. Stanford.edu [Internet]. HIV drug resistance database. 2023 [cited 2023 Aug 21]. Available from: https://hivdb.stanford.edu/
21. Hiv-Grade [Internet]. Hiv-grade.de. 2023 [cited 2023 Aug 8]. Available from: https://www.hiv-grade.de/grade_new/
22. Geno2pheno.org [Internet]. Geno2pheno resistance. 2023 [cited 2023 Aug 21]. Available from: https://www.geno2pheno.org/
23. Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis [Internet]. 2006;42(11):1608–18. [CrossRef]
24. Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology [Internet]. 2012;55(2):98–101. [CrossRef]
25. Obermeier M, Pironti A, Berg T, Braun P, Däumer M, Eberle J, et al. HIV-GRADE: a publicly available, rules-based drug resistance interpretation algorithm integrating bioinformatic knowledge. Intervirology [Internet]. 2012;55(2):102–7. [CrossRef]
26. Beerenwinkel N. Geno2pheno: estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res [Internet]. 2003;31(13):3850–5. [CrossRef]
27. Pironti A, Walter H, Pfeifer N, Knops E, Lübke N, Büch J, et al. Determination of phenotypic resistance cutoffs from routine clinical data. J Acquir Immune Defic Syndr [Internet]. 2017;74(5):e129–37. [CrossRef]
28. Lengauer T, Sing T. Bioinformatics-assisted anti-HIV therapy. Nat Rev Microbiol [Internet]. 2006;4(10):790–7. [CrossRef]
29. Pironti A, Pfeifer N, Walter H, Jensen BEO, Zazzi M, Gomes P, et al. Using drug exposure for predicting drug resistance – a data-driven genotypic interpretation tool. PLoS One [Internet]. 2017;12(4):e0174992. [CrossRef]
30. Günthard HF, Saag MS, Benson CA, del Rio C, Eron JJ, Gallant JE, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2016 Recommendations of the international antiviral society-USA panel. JAMA [Internet]. 2016;316(2):191. [CrossRef]







