1. INTRODUCTION
All recent evidence suggests that global warming-induced increase in temperature in the coming decades along with frequent droughts thus imposing adverse effect on living organisms including species ecology, geographic distribution, and phenology [1,2]. The major factors associated with climate change in the near future are an increase in the average atmospheric temperature and a change in rainfall pattern [3].
The plants have evolved to cope with high temperature stress, along with drought depending on their geographic location and the prevailing climate. Over centuries, the domestication of all major crop plants has resulted in the erosion of genetic diversity for high-temperature stress thus threatening food security. Nevertheless, thermo-tolerant plants show acquired temperature stress tolerance mainly attributed to the changes in the plant transcriptome [2,4,5].
One of the important crop plants, wheat is very sensitive to high-temperature stress. In general, wheat is a cool climate crop requiring an optimal growth temperature of 20–30°C and 15°C for reproductive growth [6-9]. With every degree of rise in temperature, a 6% decrease in wheat production is predicted [8]. Earlier reports have attributed the photosynthesis susceptibility to high temperature mainly due to the damage to thylakoid membranes, along with a decrease in the expression of genes involved in photosynthesis [5,10,11]. A major limitation for the timely approval of heat-tolerant wheat genotype for cultivation by farmers is the identification and characterization of the heat-tolerant genotypes. Until date, field-based characterization of the wheat genotypes for heat tolerance is the gold standard thus prolonging the screening and characterization of the tolerant genotypes. A number of studies had been performed in the last decade for the fast and economical screening and characterization of wheat genotypes for high-temperature tolerance [4,5,12,13]. Recently, the application of chlorophyll a fluorescence for the screening and identification of heat tolerance in wheat and its wild relative has been used extensively [4,5,10]. A combination of morphophysiological traits along with biochemical traits can be used for the identification of the heat-tolerant genotypes in wheat [5]. Transgenic approach, along with DNA modification technologies, can be employed for the production of heat stress-tolerant wheat genotypes [11,14]. Recent research has focused on targeted genome editing utilizing CRISPR/Cas9 for stable and heritable modification of wheat genome [15].
Although traditional computational methods in connivance with statistical methods have been used for studying the tolerance status of plants towards biotic as well as abiotic stress, over time, due to the complexity of the thermal stress, the use of nonlinear statistical methodology such as artificial neural network have been considered to be better in identification and characterization of the stress tolerance level in different plant species [16]. A number of recent studies have shown the robustness of using generalized regression neural network (GRNN) in diverse biological studies including prediction of miRNA, in vitro culture, analysis of plant promoters, phenomics studies, genome prediction, and transcription factors (TFs) [17,18]. In general, ANN is a type of nonlinear computational methods, which can be used for the prediction, clustering, and classification of complex systems; moreover, ANN can recognize the input and output variables and identify the inherent pattern in the datasets [19,20]. A number of interconnected neurons working in parallel aids ANN to find solution to particular problem [16], although the major shortcoming of machine learning remains the identification of the optimized solution [16,21,22].
In the present study, we have used the GRNN model in connivance with heat susceptible index for the characterization of the five Indian wheat genotypes into thermo-tolerant, moderately tolerant and thermo-susceptible. Although the characterization of these wheat genotypes for their thermo-tolerance was shown in earlier research [10], in that study, the focus of characterization was mainly on the physiology of the wheat plants in response to high-temperature stress. Whereas, in the present study, we have carried out the characterization of the wheat genotypes based on chlorophyll a fluorescence, temperature induction response technique, physiological, biochemical, and expression analysis for the identification of different Indian wheat genotypes as thermo-tolerant, moderately tolerant, and thermosusceptible. Furthermore, we have incorporated heat susceptible index in the GRNN algorithm for the screening of thermo-tolerance in non-characterized wheat genotype into thermotolerant, moderately tolerant, or thermo-susceptible genotype. The algorithm when used with already characterized genotypes for heat tolerance as controls can precisely predict the heat tolerance of un-characterized wheat genotype for heat tolerance. Hence, our characterization of wheat genotypes has suggested that the algorithm and characterization parameters can be used for wheat and other plants for the screening of plant genotypes for heat tolerance. This will reduce the time and resources for the robust characterization of plants, especially wheat for thermotolerance.
2. MATERIALS AND METHODS
2.1. Plant Material and Temperature Treatment
The five Indian bread wheat genotypes namely C306, K7903, CBW12, HD2329, and HD2428 were used in the present study and the experiments were carried out at Safa Biolabs, Delhi, India. All the wheat genotypes were raised in pots with soil prepared by mixing field soil and farmyard manure in the ratio of 3:1. A total of 10 seeds were sown in each pot and at two-leaf stage, the plants were thinned to six plants per pot. The plants were raised in a growth chamber with 13 h/10 h light and dark cycles, at 20 ± 1°C. The 60-day old wheat plants were exposed to high-temperature treatment of 34ºC/26ºC (light/dark) for the next 10 days. A set from these plants was grown at 20±1°C to maintain the control conditions in the growth chamber (SS Scientific, India). All further characterization studies were carried out at this stage for the five wheat genotypes with the top-most leaves and three biological replicates.
2.2. Temperature Induction Response Technique
The characterization of the five wheat genotypes as thermo-tolerant, moderately tolerant, and thermo-susceptible genotypes was carried out using the Temperature Induction Response Technique (TIR) [5,13]. The wheat seeds were washed with 80% ethanol followed by 3 times rinsing with sterile water. The seeds were covered with moist sterile filter paper and allowed to germinate for 24 h. The 24 h germinated seeds were exposed to a moderate temperature of 37°C/1.5 h. This was followed by increase the temperature to 54°C for 3 h in incubator (WesWox, India). After temperature treatment, the plants were kept in culture room at 20 ± 1ºC for 10-days. The recovered plants were scored for percentage survival, shoot length, and root length. The characterization of the five wheat genotypes as thermotolerant, moderately tolerant, and thermosusceptible was carried out by calculating the heat susceptibility index (HSI) and median of HSI [5].
2.3. Chlorophyll a Fluorescence
All the chlorophyll a fluorescence parameters were measured using Junior PAM (Walz, Germany) accordingly [10]. The 30-min dark-adapted leaves were given a 0.8 s saturating pulse of 6,000 μmol photon m−2s−1 for the determination of Fv/Fm. The maximum fluorescence (Fm’) under light-adapted state was determined by exposing the leaf to another actinic light of 6000 μmol photon m−2s−1. For the determination of fluorescence under light-adapted conditions, the actinic light was turned off and the leaf sample was provided with a 3 s of far red light. All the remaining parameters were then calculated including ETR, θPSII, qL, qP, and NPQ [Table 1]. The peak and the inflection temperature were determined by keeping the dark-adapted leaf in the water bath and taking the readings at every temperature between 25°C and 60°C.
Table 1: The description of abbreviations and their formula.
| Fluorescence abbreviation | Fluorescence explanation |
|---|---|
| Fo’ | Minimum fluorescence of leaf sample under light conditions |
| Fm’ | Maximum fluorescence of leaf sample under light conditions |
| PPFD | Photosynthetic photon flux density |
| A | The default leaf absorbance |
| Fm | Maximum fluorescence of 30 min dark-adapted leaf sample |
| Fs | Fluorescence with a steady state |
| Fo | Minimum fluorescence of 30 min dark-adapted leaf sample |
| Fv/Fm=Fm-Fo/Fm | The light energy used in photochemistry |
| NPQ=Fm/Fm’-1 | Light energy changed to heat for dissipation |
| θPSII=Fm’-Fs/Fm’ | The stored light energy in photochemistry |
| ETR=PSIIXPPFDXAX0.5 | PSII utilized electron transport rate |
| qP=qP×Fo’/Fs | Puddle model used for the calculation of QA |
| qL=Fm’-Fs/Fm’-Fo’ | Lake model used for the calculation of QA |
Fo: Minimum fluorescence, Fv/Fm: Photosynthetic efficiency, qP: Quantum efficiency through Puddle model, qL: Quantum efficiency through lake model, NPQ: Nonphotochemical quenching, ETR: Electron transport rate, θPSII: Photosynthetic yield.
2.4. Determination of the Total Chlorophyll Content
The fresh leaf (1 g) of the control and high-temperature treated plants was taken in falcon tubes, along with 20 mL of DMSO (dimethyl sulphoxide). The falcon tubes were closed and kept at 65°C for 4 h. The final absorbance was checked at 645 and 663 nm in a UV-Vis spectrophotometer and the total chlorophyll content was measured accordingly [23].
2.5. Radical Scavenging Activity by DPPH
Fresh plant leaves (0.1 g) were crushed in a mortar and pestle. The leaves were extracted with 1 mL of 60% (v/v), acidic methanol (methanol and HCl in a ratio of 99:1), and incubated for 16 h at 10°C. This was followed by centrifuging the sample at 15,000 rpm for 15 min. The supernatant was used for checking the ROS scavenging activity using DPPH (1,1- diphenyl-2-picryl-hydrazyl). The ascorbic acid was used for preparing the calibration curve using the serial dilution method. The leaf extract (900 μl) was mixed with DPPH solution (100 μl) followed by incubation at room temperature for 30 min. The absorbance of the sample was taken at 525 nm against a methanol blank accordingly [4].
2.6. Real-Time Expression Analysis
The mRNA was isolated from the leaf samples using trizol reagent and the gDNA was removed by on column treatment with DNase I accordingly [11]. The cDNA was synthesized from 1.5 μg of RNA and SYBR Green PCR Master Mix was used for real-time PCR analysis with three biological replicates per sample. The gene actin was used for internal control for normalization.
2.7. Calculation of Heat Susceptible Index
The Heat Susceptible Index was calculated using the equation SI = (1–Xh/X)/(1–Yh/Y) accordingly [24]. Where X is the trait for the parameter of a single genotype under control condition and Xh is the trait for the parameter of a single genotype under the heat treatment condition. Similarly, Y is the means of parameter of all the genotypes under control conditions and Yh is the means of parameter of all the genotypes under heat stress conditions.
2.8. Statistical Analysis
The data collected were the mean of values from three replications. The standard error was calculated from the mean of three replicates. The data were analyzed using SPSS software (version 20.0). The Tukey honestly significant difference (HSD) test at p<0.05 was used for post-hoc comparisons. The significance of the differences between treatments (for each species), between species (for each treatment), and their interaction were evaluated using a two-factorial analysis of variance (ANOVA).
2.9. Data and Model Development
The model is based on the characterization of five Indian bread wheat genotypes in response to high-temperature treatment as thermo-tolerant (best performance), moderately tolerant (moderate performance), and thermo-susceptible (poor performance) using Generalized Regression Neural Network (GRNN). The different chlorophyll a fluorescence parameters, TIR, physiological parameters, and biochemical parameters were used for the calculation of HSI using the GRNN model.
The input layer, pattern layer, summation layer, and output layer are four layers of GRNN. All the layers are connected with the corresponding layer. Each neuron of the pattern layer is linked with the stress condition parameter of a single genotype and non-stress condition parameter for a single genotype, along with sum of stress condition parameter of all genotype and sum of non-stress condition parameter of all genotype. S-summation and D-summation neuron, measure the performance that is based on different parameters as outputs of the pattern neuron, respectively. The connection between stress condition and non-stress condition is summation neurons and neuron pattern layer is equal to the target output. While connection between sum of stress reading of all genotypes and sum of non-stress reading of all genotypes for D-summation is unity. The output layer obtain the unknown output value corresponding to the input vector, only through dividing by stress condition and non-stress condition of genotype by sum of the stress condition of all genotype and sum of non-stress condition of all genotype. The indexing of the calculation is carried out by applying following equation.
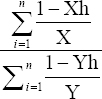 |
Xh – High temperature treatment condition/genotype
X – Control condition/genotype
Yh –Sum of high-temperature treatment condition for all genotypes
Y – Sum of control conditions for all genotypes
In each iteration, a model was developed using the training data and used to predict the outcome of the validation set (that is attributed to find out thermotolerant, moderately tolerant, and thermo-susceptible wheat genotypes on the basis of the calculated HSI and its median).
3. RESULTS AND DISCUSSION
The ability of the plant to cope with extremely high temperature is known as thermo-tolerance. All plants show basal thermo-tolerance, that is, the inherent ability to survive high temperature, and acquired thermo-tolerance, that is, induced by the gradual increase in temperature reaching lethal levels as is experienced in the natural environment [4,5,10]. The present study was carried out with five Indian bread wheat genotypes namely, C306, K7903, CBW12, HD2329, and HD2428 to check their thermo-tolerance in response to high-temperature treatment and to develop a GRNN algorithm for the speedy identification of the thermotolerance in wheat genotypes.
3.1. Characterization of the Wheat Genotypes
3.1.1. Temperature induction response technique
The 24 h old germinating seeds of the five Indian wheat genotypes taken in this study including C306, K7903, CBW12, HD2428, and HD2329 were exposed to lethal temperature treatment of 54°C/3 h with prior treatment at 37°C for 1.5 h. The scoring of the plants was carried out on the tenth day of recovery. All five genotypes showed significant differences in the percentage survival, shoot length, and root length (P < 0.001). The minimum seedlings’ survivability was observed in HD2329 and HD2428, whereas maximum survivability was observed in C306, followed by K7903 and CBW12 [Figure 1]. A similar profile was observed for shoot length and root length. Here again, HD2329 and HD2428 showed maximum decrease in the shoot and root length in response to lethal temperature treatment. In contrast, C306 and K7903 showed a minimum decrease in the shoot and root length [Figure 1].
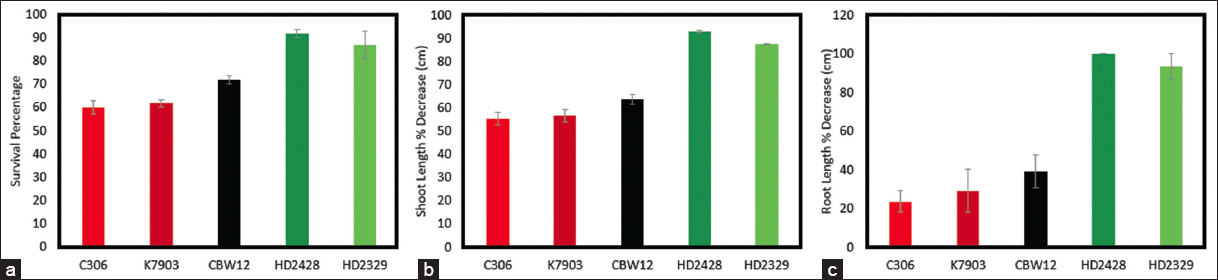 | Figure 1. Temperature Induction Response of the five Indian bread wheat genotypes. The 24 h germinating seeds were exposed to 37°C for 1.5 h followed by 54°C for 3 h. The scoring of the seedlings was carried out on the tenth day from the date of lethal temperature treatment. a. The percentage decrease in survival in response to lethal temperature treatment. b. The percentage decrease in shoot in response to lethal temperature treatment. c. The percentage decrease in root in response to lethal temperature treatment. [Click here to view] |
The previous studies have used temperature induction response techniques for the identification of thermotolerance in different plant species [5,13,25-27]. The underlying principle of the temperature induction response technique is the innate ability of the plant for acquired thermo-tolerance, that is, induced by the gradual increase in the surrounding temperature as is observed in the natural environment [4-5,10]. A number of reports have suggested the successful screening and identification of high-temperature tolerant plants including groundnut, capsicum, sunflower, and wheat using acquired thermo-tolerance [5,13,25-27]. Earlier reports have suggested the increased expression of the stress-associated genes in response to high temperature treatment in thermo-tolerant Ethiopian wheat genotypes [13].
The screening and characterization of the five wheat genotypes taken in this study were also carried out at mature plant stage. The 60-day-old potted plants were given heat treatment at 34°C/26°C (D/N) for 10 consecutive days. All the chlorophyll a fluorescence parameters along with the physiological and biochemical parameters were scored on the tenth day of heat treatment [Table 2].
Table 2: Two-way analysis of variance of genotypes, treatment, and their interactions, for the indicated parameters.
| Parameters | Sample | Column | Interaction |
|---|---|---|---|
| Fo | 934*** | 1132*** | 1418*** |
| Fv/Fm | 0.105*** | 0.272*** | 0.112*** |
| qL | 0.135*** | 0.231*** | 0.124*** |
| qP | 0.085*** | 0.800*** | 0.172*** |
| NPQ | 0.080*** | 0.800*** | 0.172*** |
| Fv’/Fm’ | 0.218*** | 0.463*** | 0.216*** |
| ETR | 1361.79*** | 2787*** | 1564*** |
| TIR survivability | 49.53*** | 165*** | 49.53*** |
| TIR root length | 213*** | 360*** | 3.45* |
| TIR shoot length | 63.53*** | 50.96*** | 9.21*** |
| Total chlorophyll | 1.038*** | 3.181*** | 1.275*** |
| DPPH activity | 16.54*** | 3.011*** | 19.26*** |
*** , **, and
* indicate significance at P < 0.001, P < 0.01, and P < 0.05, respectively, whereas NS indicates non-significant. Fo: Minimum fluorescence, Fv/Fm: Photosynthetic efficiency, qP: Quantum efficiency through Puddle model, qL: Quantum efficiency through lake model, NPQ: Nonphotochemical quenching, ETR: Electron transport rate, TIR: Temperature induction response, DPPH: 1,1- diphenyl-2-picryl-hydrazyl.
3.1.2. High temperature affects PSII
The consistent heat treatment had a significant effect on all the chlorophyll a fluorescence parameters taken in the present study. The chlorophyll a fluorescence parameters, Fv/Fm and Fo were significantly affected in response to the heat treatment in all the five bread wheat genotypes taken in this study (P < 0.001) [Table 2]. The highest increase in Fo value was observed in HD2428 (47%) and HD2329 (45%) relative to CBW12 (35%), K7903 (27%) and C306 (26%). A similar trend of negative effect of heat treatment on all the five bread wheat genotypes was observed for Fv/Fm (P < 0.001). Here again, HD2329 and HD2428 were maximally affected by heat treatment. Maximum decrease in Fv/Fm was observed in HD2428 (41%) and HD2329 (40%) relative to CBW12 (16%), K7903 (4%), and C306 (2%).
Earlier studies have shown the high level of heat sensitivity of PSII to heat stress [4,10,28]. An increase in Fo suggests the decrease in the efficiency of the light reaction of photosynthesis [4,13]. Whereas, an increase in Fv/Fm indicates damage to PSII complex. Both Fv/Fm and Fo have been shown to be reliable parameters for the identification of photoinhibition [4,11].
A similar profile was observed for ?PSII, ETR, and NPQ. A significantly higher negative effect in all these parameters was observed in HD2329 and HD2428 relative to the rest of the genotypes including CBW12, K7903, and C306 (P < 0.001) [Table 2]. By the 10th day of heat treatment, a steep decrease was observed in ?PSII and ETR, whereas a corresponding increase in NPQ was observed in all five genotypes. The genotype HD2428 showed a decrease of 65% in ?PSII, followed by HD2329 that showed a decrease of 49%. The relatively lower decrease in ?PSII in response to heat treatment was observed in CBW12, K7903, and C306, with a percentage decrease of 28%, 9.7%, and 9.2%, respectively. A similar trend was observed for ETR, here again, maximum decrease was observed in HD2428 with a percentage decrease of 62%, this was followed by HD2329 (53%). The genotypes CBW12, K7903, and C306 in response to heat treatment showed a decrease in ETR percentage by only 19%, 11%, and 7%, respectively. In response to heat treatment, a significantly high increase in NPQ was observed for all the five wheat genotypes under study (P < 0.001). The maximum increase in NPQ was observed in HD2428, followed by HD2329, CBW12, K7903, and C306 [Table 3]. A higher decrease in the ETR and θPSII, along with a corresponding increase in the NPQ in HD2329 and HD2428 suggests that lesser level of light energy is utilized in photochemistry and more amount is lost through xanthophyll-dependent energy transfer.
Table 3: The mean±standard error values of any parameter are shown (n=5).
| Parameters | C306 | K7903 | CBW12 | HD2329 | HD2428 |
|---|---|---|---|---|---|
| Fo | 89.33±2.906a,b,c,d | 92.33±2.906a,b,c,d | 104.33±2.906a,b,c,d | 112.33±1.763a,b,c,d | 122.33±1.763a,b,d |
| Fv/Fm | 0.791±0.006a,b,d | 0.784±0.006a,c,d | 0.671±0.006b,c | 0.535±0.035a,b,d | 0.505±0.035a,d |
| qP | 0.817±0.011a,d | 0.814±0.011a,d | 0.712±0.011b,d | 0.508±0.04a,b,d | 0.496±0.04a,b,d |
| qL | 0.681±0.011b,d | 0.679±0.011b,d | 0.592±0.007b,d | 0.517±0.06c,d | 0.325±0.06b,d |
| NPQ | 0.253±0.027b,d | 0.273±0.027c,d | 0.303±0.027c,d | 0.49±0.08d | 0.574±0.08b,c,d |
| qPSII | 0.693±0.016a,d | 0.693±0.016a,d | 0.547±0.016b,d | 0.383±0.025a,b,d | 0.263±0.025a,b,d |
| ETR | 57.62±1.723a,d | 55.62±1.723a,d | 49.72±1.723a,d | 29.1±2.309a,d | 24±2.309a,d |
| TPS | 60±2.88a,b,d | 61.66±1.66a,b,d | 71.66±1.66b,c,d | 91.66±1.66a,b,d | 86.66±6.009b,c,d |
| TRL | 23.64±5.37ab | 29.16±10.98a,b,d | 39.13±8.69a,d | 100±0a,d | 93.33±6.66a,d |
| TSL | 55.30±2.59a,b | 56.56±2.64a,b,c | 63.54±2.25a,d | 92.66±0.45a,d | 87.34±0.35a,d |
| TC | 2.13±0.01a,d | 2.34±0.008a,d | 1.77±0.02a | 1.47±0.02d | 1.31±0.05d |
| DS | 5.9±0.09a | 5.25±0.24a | 3.7±0.09a | 2.17±0.15a,c | 2.14±0.14a,c |
| IT | 45.33±0.33a,d | 43.66±0.33a,c,d | 41.66±0.33a,b,c | 39±0.57a,b,d | 39±0.33a,b,d |
| PT | 53.33±0.33a,c | 51.33±0.33a,b,c | 49±0.57a,b | 46.66±0.33a,b,d | 46.33±0.33a,b,d |
Same letters between the wheat genotypes shows the homogeneous groups between treatments for each genotype, according to the Tukey HSD test (p<0.05). Fo: Minimum fluorescence, Fv/Fm: Photosynthetic efficiency, qP: Quantum efficiency through Puddle model, qL: Quantum efficiency through lake model, NPQ: Nonphotochemical quenching, θPSII: Photosynthetic yield, ETR: Electron transport rate, TPS: TIR percentage survival, TSL: TIR shoot length, TRL: TIR root length, TC: Total chlorophyll, DS: DPPH ROS scavenging, IT: Infection temperature, PT: Peak temperature.
3.1.3. High-temperature effect on light reaction connectivity
The connectivity parameters depicting electron channeling were calculated using two different models, namely “Lake model” (qL) and “Puddle model” (qP). The Lake model assumes that all the PSII units remain connected [29] were as the “Puddle model” assumes that all the PSII units are separate [30]. Although under control conditions, no significant difference was observed in any genotype for qL and qP, heat treatment significantly affected qL and qP for all the five wheat genotypes (P < 0.001). The maximum decrease in qL and qP was observed in HD2428, which showed a decrease in qL by 30% and qP by 40%. A similar decreasing trend was observed for the rest of the four genotypes. The minimum decrease in qL and qP in response to heat treatment was observed for C306 and K7903. With qL showing a decrease of 6% and 7% in C306 and K7903, whereas qP showed a decrease of 11% and 12% in C306 and K7903 in response to heat treatment [Table 3]. The reduction of QA involved in the light reaction of photosynthesis chiefly depends on the connectivity between different components of the light reaction of photosynthesis. In general, both the qL and qP indicates the proportion of reaction centers that are open. Earlier studies have used these models for the identification of the photoinhibition in plants and for the identification of the photoprotective quenching in response to decreased efficiency in utilizing fluorescence [4].
3.1.4. Inflection temperature and peak temperature
Inflection temperature and peak temperature suggest the membrane stability at high temperature. A significant difference in inflection temperature and peak temperature was observed for all the five wheat genotypes taken in this study (P < 0.001). The minimum inflection temperature and peak temperature were observed in HD2428 and HD2329 [Figure 2]. Whereas, the maximum inflection temperature and peak temperature were observed in C306 and K7903 [Figure 2]. Both inflection temperature and peak temperature are affected by membrane composition. Earlier reports have attributed the higher inflection temperature and peak temperature to increased thermo-tolerance [4].
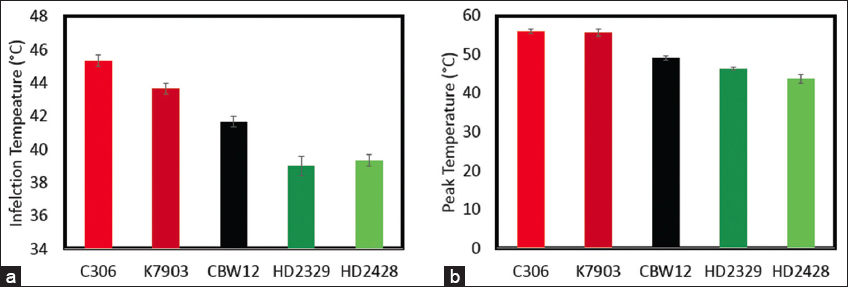 | Figure 2. The effect of lethal temperature on chlorophyll a fluorescence. a. Inflection temperature; b. Peak Temperature. [Click here to view] |
3.1.5. Distribution of absorbed light and ROS-induced damage
High-temperature treatment significantly decreased the total chlorophyll content in all five genotypes (P < 0.001). A decrease of 73% and 95% was observed by day 10 of the heat treatment in HD2329 and HD2428 [Figure 3]. Whereas, the minimum decrease in total chlorophyll was observed in C306, which showed only 6% decrease observed on day-10 of the heat treatment [Figure 3]. A higher decrease in the total chlorophyll content can be due to the ROS induced damage in HD2329 and HD2428. Furthermore, a low chlorophyll content in the leaf decrease the energy absorption thus decreasing the chances of further damage to the thylakoid membrane by ROS generation. A similar trend was observed for DPPH ROS scavenging activity. A higher reduction of DPPH was observed in C306, K7903, and CBW12 as compared to HD2329 and HD2428 in response to heat treatment, thus suggesting better ROS scavenging in C306, K7903, CBW12 relative to HD2329 and HD2428. This can be due to the higher expression of the ROS scavenging genes in C306, K7903, and CBW12 relative to HD2329 and HD2428 [Figure 3]. Earlier studies have attributed the higher ROS scavenging to higher expression of genes involved in ROS scavenging [4].
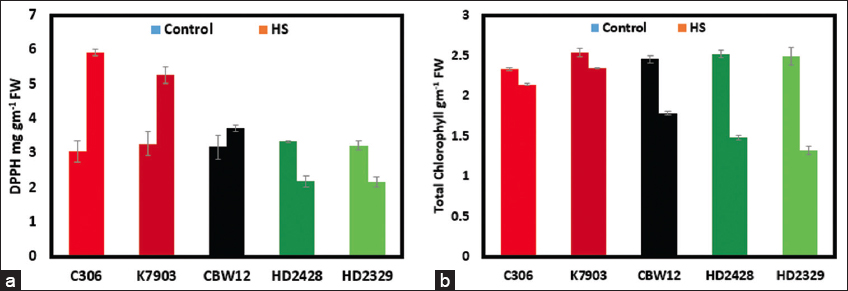 | Figure 3: The effect of high temperature on the five wheat genotypes. The first bar indicates the control, whereas the second bar indicates the high-temperature treated sample. a. The plant DPPH reduction ability. b. The decrease in total chlorophyll in response to the heat treatment. [Click here to view] |
Heat treatment primarily affects the membrane mobility thus imparting damage to the biological membranes. Heat treatment increases the mobility of the membrane lipids thus changes the membrane fluidity resulting in the damages to the biological membranes [31]. The damage to the biological membranes affects the photosynthesis since the photosynthetic apparatus especially the light reaction of photosynthesis is membrane-embedded. This resulting damage to the thylakoid membrane results in an imbalance between the light energy utilized and the light energy absorbed by the photosynthetic apparatus. The excess energy not utilized in the light reaction of photosynthesis is channelized toward the generation of the ROS (reactive oxygen species) [4,10,32,33].
3.1.6. High temperature influence on selected gene expression
The selected genes involved in photosynthesis and ROS scavenging showed higher expression in C306 and K7903 relative to other three genotypes namely CBW12, HD2329, and HD2428. Higher expression of ascorbate peroxidase and glutathione reductase was observed in C306, K7903, and CBW12, whereas least expression was observed in HD2329 and HD2428 [Figure 4]. Similarly, the rubisco large subunit and rubisco small subunit showed higher expression in C306, K7903 and CBW12 in response to heat treatment relative to HD2329 and HD2428 [Figure 4]. The expression analysis studies for selected genes involved in photosynthesis and ROS scavenging revealed higher expression of all the genes in C306, K7903, and CBW12 relative to HD2329 and HD2428. The higher expression of stress-induced genes in response to heat treatment was reported earlier in C306 and K7903 [5,10]. The reason for the difference in the genotype tolerance to lethal temperature is due to the induction of different protective molecular pathways in response to high-temperature treatment. A number of transcriptomic, proteomic, and metabolomic studies in recent times have highlighted the differential induction of specific genes/products in response to heat treatment. Earlier reports have suggested the differential expression of stress-induced genes in response to heat treatment in thermo-tolerant and thermo-susceptible wheat genotypes [5]. The proteome analysis between in two Indian bread wheat genotypes, Raj4014 (heat tolerant susceptible) and WH730 (heat tolerant) suggested the induction of stress-induced genes in response to high-temperature treatment the heat tolerant genotypes relative to the heat sensitive genotypes [34]. Another report also suggested the differential regulation of proteins in response to heat treatment in thermo-tolerant genotypes “810” and thermo-susceptible genotypes “1039” [35]. These studies have highlighted the differential regulation of stress-induced genes between the thermo-tolerant and thermo-susceptible genotypes in response to heat stress in wheat.
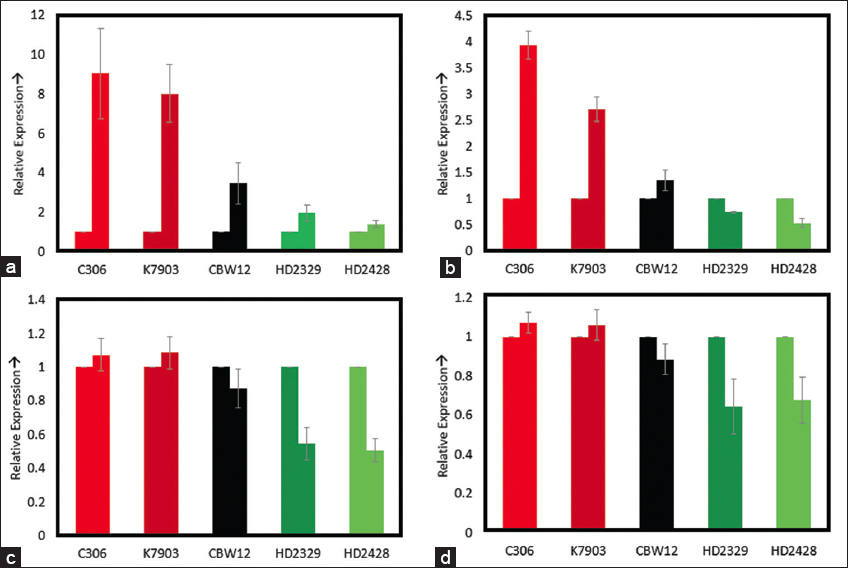 | Figure 4: Expression analysis of selected genes involved in photosynthesis and ROS scavenging. The first bar indicates the control, whereas the second bar indicates the high temperature treated sample. a. Glutathione Reductase; b. Ascorbate Peroxidase; c. Rubisco Small Subunit; d. Rubisco Large Subunit. [Click here to view] |
3.1.7. Heat Susceptibility Index
The calculation of heat susceptibility index was carried out on the basis of different chlorophyll a fluorescence parameters along with the different physiological and biochemical parameters [Figure 5]. Heat susceptibility index takes into consideration all the control scores along with the treatment scores. In the present study, we have used HSI for the characterization of the five Indian bread wheat genotypes into thermo-tolerant, moderately tolerant, and thermosusceptible genotypes. The genotype C306 showed the least HSI, followed by K7903 and CBW12, whereas the genotypes HD2329 and HD2428 showed the highest HSI. Earlier, HSI was used for the identification of thermotolerant and thermosusceptible wheat genotypes [5,10,13].
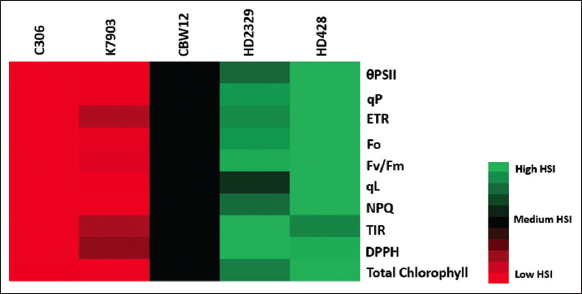 | Figure 5: Heat Susceptibility Index: Heat map showing the response of the five Indian wheat genotypes at seedling stage (TIR) and mature plant stage. The seedling were given heat treatment of 37°C/1.5 h followed by 54°C/3 h, whereas the mature plants were given heat treatment of 34°C/26°C (Light/Dark) for 10 days. The heat susceptibility index was calculated based on different chlorophyll a fluorescence parameters, physiological parameters, biochemical parameters, and temperature induction response techniques and depicted in the form of heat map. [Click here to view] |
3.1.8. Optimization of GRNN model
On the basis of the chlorophyll a fluorescence parameters, the GRNN model was applied for the prediction of thermo-tolerant, moderately tolerant, and thermo-susceptible wheat genotypes by calculating HSI [Figure 6]. The HSI calculations were incorporated with the GRNN algorithm for the speedy screening and characterization of the five wheat genotypes. Our model was able to correctly predict the wheat genotypes, C306 and K7903 as thermo-tolerant, whereas CBW12 was predicted as moderate and HD2329 and HD2428 were predicted as thermo-susceptible. Our prediction corroborates earlier reports of the high-temperature stress tolerance of the genotypes used in the present study [5]. Recent biological studies have used GRNN in In-vitro plant tissue culture, genome prediction, miRNA prediction, and phenomic studies [17,18]. A number of interconnected neurons working in parallel aids the efficient working of ANN in finding relevant solution [16]. Hence, the GRNN model can be successfully applied for the speedy identification of the high-temperature stress tolerance of the plant species when already characterized abiotic stress tolerant and susceptible control plants were used.
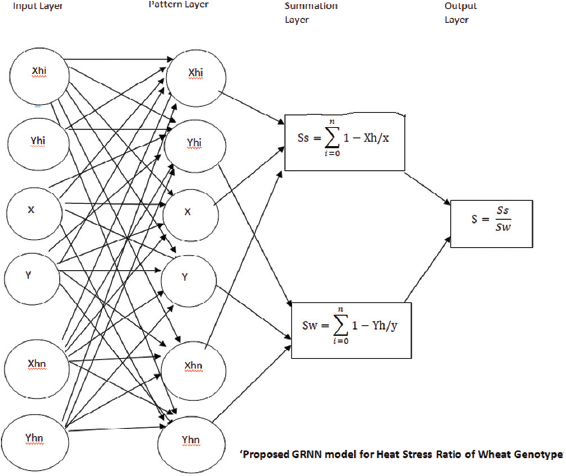 | Figure 6: Proposed GRNN Model for Heat Stress Ratio of Wheat Genotype The thermo-tolerance of the different chlorophyll a fluorescence parameters, TIR, physiological parameters, and biochemical parameters for the five wheat genotypes, namely, C306, K7903, CBW12, HD2329, and HD2428 was established according to GRNN. [Click here to view] |
Furthermore, the GRNN model suggests that C306 and K7903 show considerably higher level of heat tolerance by the increased ability to scavenge ROS generated under high-temperature treatment along with increased stability of the photosynthetic apparatus relative to HD2329 and HD2428. Hence, C306 and K7903 can be used in wheat breeding programs for imparting high temperature stress tolerance to wheat.
4. CONCLUSION
This study has highlighted the effective and consistent use of the TIR technique in combination with chlorophyll a fluorescence in the identification of thermo-tolerant, moderately tolerant, and thermo-susceptible Indian bread wheat genotypes. The fast, cheap, and repeatability of TIR technique along with chlorophyll a fluorescence give it a competitive edge over other screening methods for the screening of thermo-tolerance in plants, especially in wheat. In the present study, the genotypes, C306 and K7903 were found to be having a minimum heat susceptibility index, the genotype CBW12 showed a moderate heat susceptibility index, whereas the genotype HD2329 and HD2428 showed the maximum heat susceptibility index. Correspondingly, the genotypes C306, K7903, and CBW12 showed a relatively higher expression of selected genes involved in photosynthesis and ROS scavenging as compared to HD2329 and HD2428. On the basis of HSI, the GRNN model used in this study was able to clearly identify the thermo-tolerant, moderately tolerant, and thermosusceptible wheat genotypes taken in this study. Hence, the GRNN model can be used for the identification of the thermo-tolerance nature of the plant species when used with known tolerant and susceptible genotypes as controls thus saving time and resources.
5. ACKNOWLEDGMENT
All the authors are grateful to Safa Biolabs and Dr Akhilesh Das Gupta Institute of Professional Studies (Formerly ADGITM), Delhi, India, for providing expertise, facilities and technical assistance in successfully carrying out the present research.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst 2006;37:637-69. [CrossRef]
2. Agisho HA, Hairat S. Understanding drought stress in plants for facing challenges and management in wheat breeding:A review. Plant Cell Biotechnol Mol Biol 2021;140-56.
3. Masson-Delmotte V, Zhai P, Pörtner HO, Roberts D, Skea J, Shukla PR, et al. Intergovernmental Panel on Climate Change. Press;2019. 9. Available from:https://www.ipcc.ch/site/assets/uploads/sites/2/2019/06/SR15_full_report_high_res (Last accessed on 2020 Feb 28].
4. Hairat S, Khurana P. Evaluation of Aegilops tauschii and Aegilops speltoides for acquired thermotolerance:Implications in wheat breeding programmes. Plant Physiol Biochem 2015;95:65-74. [CrossRef]
5. Hairat S, Khurana P. Photosynthetic efficiency, temperature induction response, carbon isotope discrimination correlate with expression profiling in Indian wheat cultivars. Plant Signal Behav 2016;11:e1179416. [CrossRef]
6. Farooq M, Bramley H, Palta JA, Siddique KH. Heat stress in wheat during reproductive and grain-filling phases. CRC Crit Rev Plant Sci 2011;30:491-507. [CrossRef]
7. Zampieri M, Ceglar A, Dentener F, Toreti A. Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ Res Lett 2017;12:064008. [CrossRef]
8. Comastri A, Janni M, Simmonds J, Uauy C, Pignone D, Nguyen HT, et al. Heat in wheat:Exploit reverse genetic techniques to discover new alleles within the Triticum durum sHsp26 family. Front Plant Sci 2018;9:1337. [CrossRef]
9. Hairat S, Agisho HA, Zaki M. Challenges and management of wheat under global climate change. Plant Cell Biotechnol Mol Biol 2021;22:136-51.
10. Hairat S, Khurana P. Improving photosynthetic responses during recovery from heat treatments with brassinosteroid and calcium chloride in Indian bread wheat cultivars. Am J Plant Sci 2015b;6:1827. [CrossRef]
11. Hairat S, Baranwal VK, Khurana P. Identification of Triticum aestivum nsLTPs and functional validation of two members in development and stress mitigation roles. Plant Physiol Biochem 2018;130:418-30. [CrossRef]
12. Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M, Siddique KH. Physiological responses to drought stress in wild relatives of wheat:implications for wheat improvement. Acta Physiol Plant 2017;39:106. [CrossRef]
13. Hairat S, Zaki M. Screening and identification of high temperature tolerant ethiopian bread wheat cultivars based on temperature induction response technique. Plant Cell Biotechnol Mol Biol 2021;22:257-64.
14. Zang X, Geng X, He K, Wang F, Tian X, Xin M, et al. Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and Arabidopsis. Front Plant Sci 2018;9:1710. [CrossRef]
15. Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 2017;89:1251-62. [CrossRef]
16. Silva JC, Teixeira RM, Silva FF, Brommonschenkel SH, Fontes EP. Machine learning approaches and their current application in plant molecular biology:A systematic review. Plant Sci 2019;284:37-47. [CrossRef]
17. Hesami M, Alizadeh M, Naderi R, Tohidfar M. Forecasting and optimizing Agrobacterium-mediated genetic transformation via ensemble model-fruit fly optimization algorithm:A data mining approach using chrysanthemum databases. PLoS One 2020;15:e0239901. [CrossRef]
18. Hesami M, Jones AM. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol 2020;104:9449-85. [CrossRef]
19. Niazian M, Shariatpanahi ME, Abdipour M, Oroojloo M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019;256:1317-32. [CrossRef]
20. Hesami M, Naderi R, Yoosefzadeh-Najafabadi M, Rahmati M. Data-driven modeling in plant tissue culture. J Appl Environ Biol Sci 2017;7:37-44.
21. Moravej M, Amani P, Hosseini-Moghari SM. Groundwater level simulation and forecasting using interior search algorithm-least square support vector regression (ISA-LSSVR). Groundw Sustain Dev 2020;11:100447. [CrossRef]
22. Araghinejad S, Fayaz N, Hosseini-Moghari SM. Development of a hybrid data driven model for hydrological estimation. Water Resour Manage 2018;32:3737-50. [CrossRef]
23. Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Canad J Bot 1979;57:1332-4. [CrossRef]
24. Fischer RA, Maurer R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust J Agric Res 1978;29:897-912. [CrossRef]
25. Mishra S, Laxman RH, Reddy KM, Venugopalan R. TIR approach and stress tolerance indices to identify donor for high-temperature stress tolerance in pepper (Capsicum annuum L.). Plant Genet Resour 2020;18:19-27. [CrossRef]
26. Selvaraj MG, Burow G, Burke JJ, Belamkar V, Puppala N, Burow MD. Heat stress screening of peanut (Arachis hypogaea L.) seedlings for acquired thermotolerance. Plant Growth Regul 2011;65:83-91. [CrossRef]
27. Senthil-Kumar M, Srikanthbabu V, Mohan Raju B, Ganeshkumar, Shivaprakash N, Udayakumar M. Screening of inbred lines to develop a thermotolerant sunflower hybrid using the temperature induction response (TIR) technique:A novel approach by exploiting residual variability. J Exp Bot 2003;54:2569-78. [CrossRef]
28. Berry J, Bjorkman O. Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol 1980;31:491-543. [CrossRef]
29. Kramer DM, Johnson G, Kiirats O, Edwards GE. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 2004;79:209-18. [CrossRef]
30. Schreiber U, Schliwa U, Bilger W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 1986;10:51-62. [CrossRef]
31. Quartacci MF, Pinzino C, Sgherri CL, Dalla Vecchia F, Navari-Izzo F. Growth in excess copper induces changes in the lipid composition and fluidity of PSII-enriched membranes in wheat. Physiol Plant 2000;108:87-93. [CrossRef]
32. Osmond CB. What is photoinhibition?Some insights from comparisons of shade and sun plants. In:Baker NR, Bowyer JR, editors. Photoinhibition of Photosynthesis. From Molecular Mechanisms to the Field. Oxford:BIOS Scientific Publishers;1994. 1-24.
33. Sharkey TD. Effects of moderate heat stress on photosynthesis:Importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ 2005;28:269-77. [CrossRef]
34. Gupta OP, Mishra V, Singh NK, Tiwari R, Sharma P, Gupta RK, et al. Deciphering the dynamics of changing proteins of tolerant and intolerant wheat seedlings subjected to heat stress. Mol Biol Rep 2015;42:43-51. [CrossRef]
35. Wang X, Dinler BS, Vignjevic M, Jacobsen S, Wollenweber B. Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Sci 2015;230:33-50. [CrossRef]