1. INTRODUCTION
Cardiovascular diseases are the primary cause for deaths worldwide. It is believed that the production, accumulation, and clotting of fibrin in blood arteries, which results in intravascular thrombosis, is the cause of cardiovascular illnesses such as ischemic heart disease, ischemic stroke, and venous thromboembolism [1]. Furthermore, a functioning hemostatic system creates a thrombus to prevent excessive blood loss. Uncontrolled hemostasis causes unsuppressed thrombosis, which can lead to several cardiovascular problems (deep vein thrombosis, vascular congestion, embolism in the pulmonary veins, acute myocardial infarction, etc.), as fibrin clots accumulate inside blood vessels and eventually obstruct blood flow [2]. Plasminogen activators, which convert inactive plasminogen (zymogen) into active plasmin, are the only fibrinolytic drugs currently available in the market. In the blood, plasmin, a serine protease, dissolves fibrin clots and prevents thrombosis [3]. Tissue-type plasminogen activators, urokinase-type plasminogen activators, and bacterial plasminogen activators are different types of plasminogen activators and have been used as thrombolytic drugs. Additionally, it has been noted that the commonly used plasminogen activators in fibrinolytic treatment have very limited fibrin specificity, have unfavorable side effects such as internal bleeding, and are neurotoxic at high doses [4]. In the past decade, various potent thrombolytic agents have been discovered and characterized from microorganisms [5-8], earthworms [9], snake venoms [10], insects [11], and leeches [12]. These enzymes are potential as both therapeutic agents and valuable tools for figuring out the fibrinolytic process. To avoid unwanted contamination and a huge cost of purification, these sources must be recommended after numerous stages of screening and feasibility studies [13]. Members of the Bacillus species are among the most studied microbes due to their innate ability to create fibrinolytic proteases without endotoxins and short fermentation cycles, with certain species being regarded as safe for medication administration [14,15]. The identification of microbial proteases that are plasmin-like and have a high level of fibrin selectivity has sparked recent interest in fibrinolytic drugs. The price of biomedical products like therapeutic enzymes depends on the quantity and kind of downstream processing processes [16]. Therefore, a simple and cost-effective purification technique is required to separate the desired bioproducts. The purification of a protein is crucial for examining its physical and biological features from the viewpoint of improving the knowledge on how enzymes work. Additionally, a substantial portion of the biochemical exams requires purifying the substances being examined because they need to be generally free of “contaminants” to be adequately characterized [17].
In this study, a novel fibrinolytic enzyme produced by Bacillus subtilis AIBL_AMSB2_M7E32 was purified by a four-step purification process. As previously reported, the strain was discovered in the soils of Coringa mangrove forests. The study also reports the in vitro activity of the fibrinolytic enzyme on blood clots, thus showing evidence of its thrombolytic potential.
2. MATERIALS AND METHODS
2.1. Chemicals
Chemicals and culture medium for this study including both thrombin and fibrinogen were purchased from Himedia Pvt. Ltd. in Mumbai. DEAE Cellulose and DEAE Sephadex-G50 were purchased from Sigma Pvt. Ltd. The agarose was purchased from Lonza Rockland, ME, USA. Trichloroacetic acid was supplied by Fisher Scientific, while Hammarsten Grade Casein was purchased from MP Biochemicals. Solvents used in this study were purchased from Merck. Fibrinogen was purchased from Sigma-Aldrich, USA. All the chemicals used for the downstream process were purchased from Sigma Pvt. Ltd. and Himedia Pvt. Ltd. in Mumbai.
2.2. Bacterial Strain
Bacillus subtilis AIBL_AMSB2_M7E32, a producer of the fibrinolytic enzyme, was isolated from Coringa Mangroves soils in the Kakinada District of Andhra Pradesh (Latitude 16°49'18.5''N, 82°17'53.2''E, Longitude 16.821803, 82.298114). By using ultraviolet mutagenesis and ethyl methyl sulfonate mutagenesis, the B. subtilis AIBL_AMSB2_M7E32 (accession number: OQ600796) strain was obtained, which has better fibrinolytic activity when compared with the wild strain. The variant was kept at –20°C as glycerol stocks and 4°C on an agar plate and was used for further studies.
2.3. Purification of Fibrinolytic Protease
After 12 h fermentation of B. subtilis AIBL_AMSB2_M7E32 in the optimized skim milk broth consisting of 3% sucrose, 3% yeast extract, 0.3% MgSO4, 2% inoculum, and 1% skim milk with a pH of 7.2, the cell-free supernatant was obtained by centrifuging the whole broth at 10,000 rpm for 20 min at 4ºC. Ammonium sulfate (AS) was added to the cell-free supernatant until 60% saturation was attained and the mixture was kept in equilibrium at 4ºC for 16 h. The precipitate was obtained by further centrifugation of the mixture at 10,000 rpm for 15 min at 4ºC. The precipitate was dissolved in a 10 mM Tris–HCl buffer of pH 7.2 and dialyzed overnight at 4ºC against the same buffer using a 30 kDa MWCO membrane. After the completion of dialysis, the dialyzed enzyme was further purified by anion exchange chromatography on DEAE Cellulose (diethylaminoethyl) (Sigma Chemical Co.). The dialyzed fraction was passed through 10 ml of Sephadex column (13 cm × 30 cm), equilibrated against 25 mM Tris–HCl and 25 mM NaCl, and washed with the same buffer. The desired protein was eluted at an elution rate of 1 ml/min with different elution buffers of 25, 50, 75, 100, 125, and 150 mM of Tris–HCl. Fractions showing higher protein absorbance at 280 nm were pooled, concentrated, and further loaded in Sephadex-G 50 column and eluted against 0.1 M phosphate buffer (pH 7.2) at an elution rate of 1 ml per 2 min. All fractions were collected separately and the absorbance of each fraction was measured at 280 nm with a UV spectrophotometer (UV-1900 Shimadzu). All the purification experiments were carried out at 4ºC.
2.4. Enzyme Activity Assay
Enzyme activity was assessed by measuring the total protein content (TPC) using Bradford’s method for protein estimation. This was followed by conducting protease and fibrin assays. For the determination of the TPC, 2.5 ml of Bradford’s reagent was added to the 0.5 ml of the enzyme-purified fractions, the reaction mixture was incubated in the dark for 10 min, and the absorbance was read at 595 nm. The total protein was calculated from the standard plot made by using different concentrations of standard protein bovine serum albumin [18]. For protease assay, 0.25 ml of the purified fractions of the enzyme was mixed with 1.25 ml of 0.6% casein in 50 mM potassium phosphate buffer having pH 7.5 and 0.25 ml and was incubated at 37°C for 15 min. The reaction was then stopped by the addition of 1.25 ml of 110 mM trichloroacetic acid and incubated at room temperature for 30 min. The reaction mixtures were centrifuged at 10,000 rpm for 15 min, and the absorbance of the supernatant was read at 280 nm. A tyrosine standard curve was used as a reference for protease activity. A unit of enzyme activity is defined as the amount of tyrosine (mg) released per minute per milliliter at 37°C [19].
For the fibrin assay, 0.1 ml of enzyme supernatant and 2.5 ml of Tris–HCl buffer (0.1 M, pH 7.8) containing calcium chloride (0.01 M) were added. To this mixture, 2.5 ml of fibrin (1%) was added and incubated at 37°C for 30 min. The reaction was then terminated by adding 5.0 ml of trichloroacetic acid (0.11 M) containing sodium acetate (0.22 M) and acetic acid (0.33 M). Furthermore, the reaction mixture was centrifuged at 10,000 rpm for 10 min, and the supernatant was collected to read absorbance at 275 nm. One unit of fibrinolytic activity was referred to in terms of the amount of enzyme per 1 mg of L-tyrosine per minute under standard assay conditions [20].
2.5. SDS and Zymography
To determine the molecular weight of the purified enzyme, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 12% polyacrylamide separating gel with 5% stacking gel. Following electrophoresis, the gel was incubated in a fixing solution consisting of methanol: glacial acetic acid: distilled water solution (5:1:4). To the gel, silver staining was performed [21]. A wide-range molecular marker, pre-stained protein ladder (Himedia) with a range of (11–100 kDa) was used for calibration. Fibrin zymography was carried out to determine the fibrinolytic activity of the samples on SDS-PAGE. SDS-PAGE of 12% was polymerized with 0.2% fibrinogen and 50 µl (10 NIH Units). The samples to be loaded were unheated, and the loading buffer was devoid of the reducing agent. Fibrin zymography was carried out at 4ºC for 1.5 h at 100 V. After zymography, the gel was soaked in Triton X-100 containing 50 mM Tris (pH 7.4) for 30 min and further washed with distilled water. After this, the gel was incubated in 30 mM Tris buffer (pH 7.4) containing 200 mM NaCl, 10 mM CaCl2, and 0.02% NaN3 (0.1 M, pH 7.2) at 37ºC for 16 h. The following day, the gel was stained with Coomassie Blue-R-250 for 1 h and de-stained [22].
2.6. Assessment of Fibrinolytic Activity of the Enzyme In Vitro
To evaluate the in vitro activity of the purified fibrinolytic enzyme, a modified protocol by D Souza et al. (2020) was followed, wherein 0.5 ml of blood was transferred in a pre-weighed sterile 1.5-ml centrifuge tube and incubated at 37ºC for 45 min for the formation of a clot [23]. After clot formation, the serum was removed without disturbing the clot formed. Each 1.5-ml centrifuge tube with the clot was weighed and labeled W1. To determine the clot weight, the following formula was used: (weight of clot containing the 1.5-ml centrifuge tube – the weight of the 1.5-ml centrifuge tube alone). Furthermore, different volumes of 5, 10, 50, and 100 µl of the purified enzyme samples were added to the 1.5-ml centrifuge tubes. To the control tube, 100 µl of saline was added. All the 1.5-ml centrifuge tubes were incubated at 37ºC for 24 and 48 h and observed for clot lysis. After incubation, the dissolved blood was removed and the 1.5-ml centrifuge tube was weighed again to observe the difference in clot lysis. The difference in the weight taken before and after lysis is given as the percentage of clot lysis. The experiment was carried out in triplicates. The formula of clot lysis is given as follows:
Lysis percentage = 100 – {(W3 – W1) / (W2 – W1)} × 100
where W1 is the weight of the empty 1.5-ml centrifuge tube, W2 is the weight of the 1.5-ml centrifuge tube with the blood clot, and W3 is the weight of the 1.5-ml centrifuge tube after clot lysis.
2.7. Statistical Analysis
IBM SPSS Statistics Version 24.0, one-way analysis of variance (ANOVA) was combined with Duncan multiple range test. With a statistical difference of p < 0.05, the results were presented as means SE (variable).
3. RESULTS
3.1. Purification of Fibrinolytic Protease
The fibrinolytic enzyme was extracted from the supernatant of B. subtilis AIBL_AMSB2_M7E32 using AS precipitation, and dialysis was followed by DEAE-Cellulose and Sephadex G-50 chromatography. The TPC (mg/ml) of the purified enzymes revealed that the final purified TPC of the enzyme sample after gel filtration (GF1) was found to be 0.05 ± 0e mg/ml as illustrated in Figure 1. Subsequently, the enzyme activity in each purification step was determined, and it was found that the final fraction after gel filtration (GF1) had an enzyme activity (U) of 0.42 ± 0.008e as shown in Figure 2. The purified sample exhibited fibrinolytic activity (U/ml) at the rates of 564.28 ± 0.04a, 453.18 ± 12.87b, 362.65 ± 0.00c, 332.48 ± 0.00d, and 320.82 ± 0.00d after AS, Dialyzed, DEAE-Cellulose (IE), and Sephadex G 50 (GF1), respectively, as shown in Figure 3. The effectiveness of filtration at each stage has had a major effect. Ion exchange chromatography and gel filtration chromatography achieved a higher degree of purification compared with the previous stage. The final purification degree was 43.33-fold with a specific activity of 6368.20 (U/mg). The purification steps are summarized in Table 1. The active fraction was evaluated at 280 nm and the majority of contaminants were removed by Sephadex G 50 yielding the desired protein using SDS-PAGE.
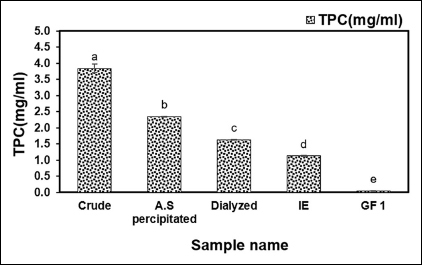 | Figure 1: TPC (mg/ml) after four-step purification of B. subtilis AIBL_AMSB2_M7 isolated from Coringa mangroves soil samples. The values indicate the mean ± SE of three replicates. Duncan’s multiple range test ANOVA revealed that values without a matching alphabet were substantially different (p < 0.05). [Click here to view] |
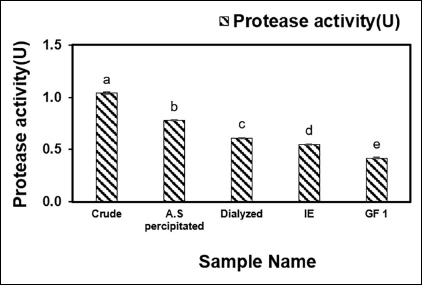 | Figure 2: Protease activity (U) after four-step purification of B. subtilis AIBL_AMSB2_M7 isolated from Coringa mangroves soil samples. The values indicate the mean ± SE of three replicates. Duncan’s multiple range test ANOVA revealed that values without a matching alphabet were substantially different (p < 0.05). [Click here to view] |
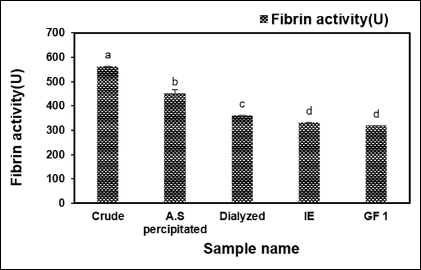 | Figure 3: Fibrin activity (U/ml) after four-step purification of B. subtilis AIBL_AMSB2_M7 isolated from Coringa mangroves soil samples. The values indicate the mean ± SE of three replicates. Duncan’s multiple range test ANOVA revealed that values without a matching alphabet were substantially different (p < 0.05). [Click here to view] |
Table 1: A stepwise purification of fibrinolytic enzyme from B. subtilis AIBL_AMSB2_M7E32.
| Sample Name | Total Protein (mg/ml) | Total Fibrinolytic Activity (U)/ml | Specific Activity (U/mg) | Purification Fold (× fold) |
|---|---|---|---|---|
| Crude | 3.84 ± 0.15a | 564.28 ± 0.04a | 146.94 | 1 |
| AS precipitation | 2.35 ± 0.00b | 453.18 ± 12.87b | 192.51 | 1.31 |
| Dialyzed | 1.63 ± 0.01c | 362.65 ± 0.00c | 222.48 | 1.51 |
| DEAE Cellulose 52 | 1.13 ± 0.01d | 332.48 ± 0.00d | 293.87 | 2.00 |
| DEAE G-50 | 0.05 ± 0e | 320.82 ± 0.00d | 6368.20 | 43.33 |
The data are presented as mean ± SD (n = 3). ANOVA revealed that values without a matching alphabet in the same column were substantially different (p < 0.05).
3.2. SDS and Zymography
To verify the molecular weight and purity of the purified enzyme, SDS-PAGE and zymography were carried out. The molecular weight of the enzyme when estimated by SDS-PAGE was approximately 28 kDa when compared with a standard protein marker (Himedia). Fibrin zymography results indicated that purified protein from the culture B. subtilis AIBL_AMSB2_M7E32 showed fibrinolytic activity by the formation of a thin clear band of fibrinolysis on the separating gel (non-denaturing) [Figure 4]. The result of zymography correlated with the molecular weight of the purified protein in SDS-PAGE.
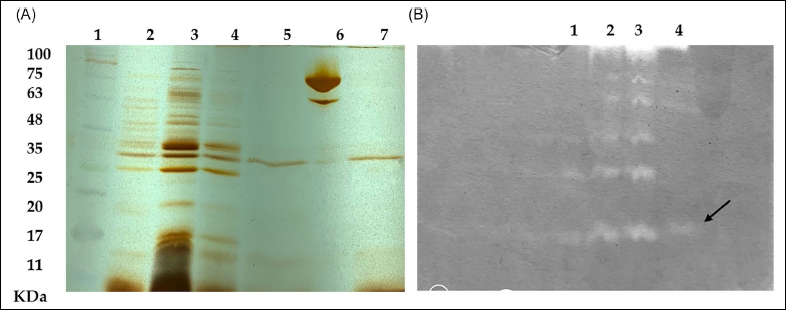 | Figure 4: (A) SDS-PAGE of the purified fibrinolytic enzyme from B. subtilis AIBL_AMSB2_M7E32. Lane 1: molecular weight markers; Lane 2: crude enzyme; Lane 3: enzyme dialysate; Lane 4: DEAE cellulose anion exchange active fraction; Lane 5: purified enzyme after DEAE Sephadex-G50 Gel filtration; Lane 6: streptokinase marker; Lane 7: purified enzyme after DEAE Sephadex-G50 Gel filtration corresponding to 28 kDa. (B) Fibrin zymography (12%) of the purified fibrinolytic enzyme from B. subtilis AIBL_AMSB2_M7E32 as indicated by the zone of clearance (as indicated by the arrow). [Click here to view] |
3.3. Assessment of Fibrinolytic Activity of the Enzyme In Vitro
The purified fibrinolytic enzyme isolated from B. subtilis AIBL_AMSB2_M7E32 was evaluated for lysis of the blood clot after 24 and 48 h, respectively. Different volumes of purified enzyme samples 5, 10, 50, and 100 µl were used. The percentage of clot lysis after the treatment of the enzyme is shown in Figure 5. Clot digested with 100 µl of the enzyme showed a maximum lysis percentage of 72.96 ± 0.16a after incubation at 37ºC for 48 h and 28.982 ± 0.33a after incubation at 37ºC for 24 h. It is well acknowledged that examining the thrombolytic activity of fibrinolytic enzymes using in vitro blood clot lysis is an accurate approach [24].
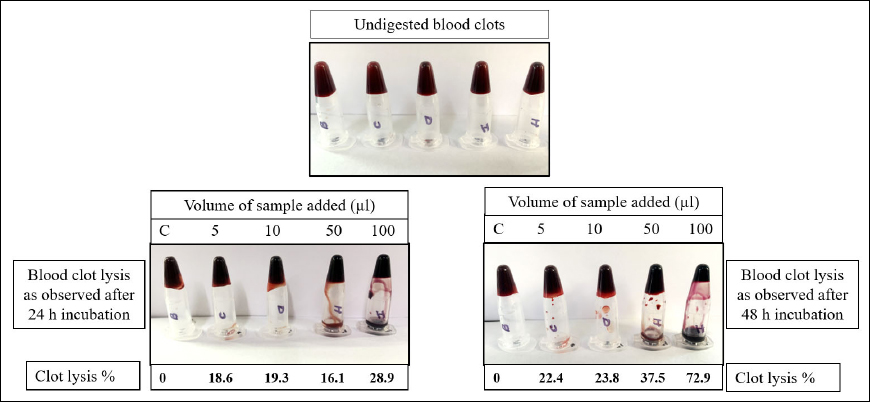 | Figure 5: Blood clot lysis experiment with purified fibrinolytic enzyme from B. subtilis AIBL_AMSB2_M7E32 for a total of 48 h of incubation. [Click here to view] |
4. DISCUSSION
In recent decades, several thrombolytic drugs have been discovered and characterized in the past few decades from a variety of sources, of which the most prominent ones are fermented foods [25]. Therefore, increasing attention is being paid to fibrinolytic enzymes from non-food sources. However, the majority of findings demonstrated that a range of extracellular and intracellular fibrinolytic enzymes are produced by Bacillus species. The fibrinolytic enzyme produced by B. subtilis was reportedly employed as a fibrinolytic treatment as early as the 1990s [16]. The use of an enzyme ultimately determines the degree of purity it achieves. Numerous methods for purifying enzymes are designed to provide better purity and strong catalytic activity [26]. This study reported that a Coringa mangrove isolate, namely, B. subtilis AIBL_AMSB2_M7E32, showed fibrinolytic activity, with a 43.33-fold purification and a molecular weight of 28 kDa. To the best of our knowledge, there are no reports of a purified fibrinolytic enzyme of 28 kDa produced by B. subtilis AIBL_AMSB2_M7E32 from Coringa mangrove soils. Furthermore, the fibrinolytic enzyme showed a specific activity of 6368.20 (U/mg), which was higher than the previously reported studies where a study reported that a purified fibrinolytic enzyme from Bacillus sp. like Bacillus tequilensis showed an activity of 2373.59 ± 54.81a with a purification fold of 329.76 ± 12.34a [27]. Another fibrinolytic enzyme from Bacillus velezensis Z01 showed a specific activity of 76.46 U/µg with a purification of 7.77-fold [28]. Also, the purification of BSF 1 Protease from B. subtilis A26 resulted in a specific activity of 3211 U/mg with a purification of 4.97-fold [29]. A five-step purification of fibrinolytic enzyme-producing B. subtilis DC33 resulted in a 34.6-fold purification with a specific activity of 15495.9 U/mg [30]. This molecular weight was similar to the previously reported nattokinase of approximately 27.7 kDa [31]. Some studies have reported a subtilisin DFE of 28 kDa that was isolated from Bacillus amyloliquefaciens DC-4 [32]. However, the molecular weight of the purified enzyme of our study was lower than the commonly used thrombolytic agents such as streptokinase (47 kDa) and urokinase (54 kDa), thus showing lesser chances for antigenicity and being a safer thrombolytic drug [33]. Some fibrinolytic enzymes so far reported by Bacillus sp and from fermented food sources are a 27.7 kDa enzyme from B. subtilis DC 33 isolated from Douchi a fermented food [30], and 27 or 29.60 kDa enzyme from Bacillus. sp. JS2 isolated from Jeotgal [34], a 31 kDa enzyme BK17 from Bacillus BK-17 [35], and a 45 kDa enzyme from Bacillus sp. KDO13; in contrast, we report a purified 28 kDa fibrinolytic enzyme from a non-food source, i.e., Coringa soils for the very first time. Furthermore, in our study, clot dissolution of 72.96 ± 0.16% was observed after 48 h of incubation, which was consistent with other reports showing that B. amyloliquefaciens strain KJ10 isolated from soya bean paste had a clot dissolution of 28 ± 1.8% [37]. B. subtilis LD-8547 isolated from Douchi only showed a clot resolution of 38.4% [38]. The use of commercially available streptokinase showed clot lysing activity between 62.2% and 70.8% [24]. B cereus SRM-001, isolated from the blood-containing soil of a chicken coop, showed that almost 70–80% of the blood clot was dissolved by the fibrinolytic enzyme [39]. The strong thrombolytic activity of the protease demonstrated by the in vitro blood clot lysis experiment supports its potential application as a bioactive molecule for thrombolytic treatment, but the stability of the enzyme, its amino acid composition, and some in vivo studies need to be further investigated.
5. CONCLUSION
The fibrinolytic drugs available on the market generally have low selectivity and undesirable side effects such as internal hemorrhage and neurotoxicity. Microbial-based fibrinolytic enzymes have proven to be safe in this aspect. Therefore, the search for microbial fibrinolytic enzymes has been intense over the past decade. Therefore, current research is focused on the development of new fibrinolytic drugs with improved therapeutic properties and selectivity. In this study, we report the purification of a fibrinolytic enzyme from B. subtilis AIBL_AMSB2_M7E32 isolated from Coringa mangrove soils. The enzyme was purified in a four-step purification process that yielded a purification of 43.33-fold with a specific activity of 6368.20 (U/mg). The molecular weight of the isolated fibrinolytic enzyme was calculated to be 28 kDa, and it appeared homogeneous on SDS-PAGE. The zymographic analysis also showed the fibrinolytic activity of the purified enzyme. When its activity to lyse blood clots in blood samples was evaluated, the purified enzyme showed a lysis percentage of 72.96 ± 0.16a% after 48 h of incubation. Further studies need to be conducted to confirm the performance of the enzyme in terms of stability and in vivo activity.
6. ACKNOWLEDGMENTS
The author is thankful to CHRIST (Deemed-to-be University) for providing lab facilities. The author SB is extremely grateful to the Department of Science and Technology, KSTePS, Government of Karnataka, for providing fellowship.
7. AUTHOR CONTRIBUTIONS
SB participated in the concept and research design, data collection, data analysis, interpretation of results, and writing the manuscript. AKM contributed to the idea and design of the experimental work and participated in the critical revision of the article. Both authors approved the manuscript.
8. FINANCIAL SUPPORT AND SPONSORSHIP
There is no funding to report.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Surma S, Banach M. Fibrinogen and atherosclerotic cardiovascular diseases—Review of the literature and clinical studies. Int J Mol Sci 2022;23(1):193.
2. Malik A, Rehman FU, Shah KU, Naz SS, Qaisar S. Hemostatic strategies for uncontrolled bleeding: A comprehensive update. J Biomed Mater Res B Appl Biomater 2021;109(10):1465–77.
3. Advitiya, Khasa YP. The evolution of recombinant thrombolytics: Current status and future directions. Bioengineered 2017;8(4):331–58.
4. Fleming M, Melzig MF. Serine-proteases as plasminogen activators in terms of fibrinolysis. J Pharm Pharmacol 2012;64(8):1025–39.
5. Smitha KV, Pradeep BV. Optimization of physical and cultural conditions of fibrinolytic enzymes from Bacillus altitudinis S-CSR 0020. J Pure Appl Microbiol 2018;12(1):343–54.
6. Zhou Y, Chen H, Yu B, Chen G, Liang Z. Purification and characterization of a fibrinolytic enzyme from marine Bacillus velezensis Z01 and assessment of its therapeutic efficacy in vivo. Microorganisms 2022;10(5):843.
7. Majumdar S, Sarmah B, Gogoi D, Banerjee S, Ghosh SS, Banerjee S, et al. Characterization, mechanism of anticoagulant action, and assessment of therapeutic potential of a fibrinolytic serine protease (Brevithrombolase) purified from Brevibacillus brevis strain FF02B. Biochimie 2014;103:50–60.
8. Raj A, Khess N, Pujari N, Bhattacharya S, Das A, Rajan SS. Enhancement of protease production by Pseudomonas aeruginosa isolated from dairy effluent sludge and determination of its fibrinolytic potential. Asian Pac J Trop Biomed 2012;245–51.
9. Chen H, Takahashi S, Imamura M, Okutani E, Zhang Z-g, Chayama K, et al. Earthworm fibrinolytic enzyme: anti-tumor activity on human hepatoma cells in vitro and in vivo. Chin Med J 2007;120(10):898–904.
10. Sun MZ, Liu S, Greenaway FT. Characterization of a fibrinolytic enzyme (ussurenase) from Agkistrodon blomhoffii ussuriensis snake venom: Insights into the effects of Ca2+ on function and structure. Biochim Biophys Acta Proteins Proteomics 2006;1764(8):1340–8.
11. Huang MX, Ye Y, Chen YX, Han YL. Partial purification and characterization of fibrinolytic enzymes from yellow mealworm. Int J Pept Res Ther 2012;18(2):153–61.
12. Salzet M. Anticoagulants and inhibitors of platelet aggregation derived from leeches. FEBS Lett 2001;492(3):187–92.
13. Bedade D, Pawar S. Chapter 26 – Downstream processing of biotechnology products. In: Bhatt AK, Bhatia RK, Bhalla TC, editors. Basic biotechniques for bioprocess and bioentrepreneurship [Internet]. Academic Press; 2023: p. 377–90.
14. Wang SH, Zhang C, Yang YL, Diao M, Bai MF. Screening of a high fibrinolytic enzyme producing strain and characterization of the fibrinolytic enzyme produced from Bacillus subtilis LD-8547. World J Microbiol Biotechnol 2008;24(4):475–82.
15. Chandramohan M, Yee CY, Kei Beatrice PH, Ponnaiah P, Narendrakumar G, Samrot AV. Production, characterization and optimization of fibrinolytic protease from Bacillus pseudomycoides strain MA02 isolated from poultry slaughter house soils. Biocatal Agric Biotechnol 2019;22:101371.
16. Sharma C, Osmolovskaya, Singh R. Microbial fibrinolytic enzymes as anti-thrombotics: Production, characterisation and prodigious biopharmaceutical applications. Pharmaceutics 2021;13(11):1880.
17. Milne JJ. Scale-up of protein purification: Downstream processing issues. Methods Mol Biol 2017;1485:71–84.
18. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54.
19. Cupp-Enyard C. Sigma’s non-specific protease activity assay - Casein as a substrate. J Vis Exp 2008 Sep 17;(19):899.
20. Vijayaraghavan P, Arun A, Vincent SGP, Arasu MV, Al-Dhabi NA. Cow dung is a novel feedstock for fibrinolytic enzyme production from newly isolated Bacillus sp. IND7 and its application in in vitro clot lysis. Front Microbiol 2016;7:361.
21. Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Nat Protoc 2006;1(4):1852–8.
22. Velusamy P, Pachaiappan R, Christopher M, Vaseeharan B, Anbu P, So J-S. Isolation and identification of a novel fibrinolytic Bacillus tequilensis CWD-67 from dumping soils enriched with poultry wastes. J Gen Appl Microbiol 2015;61(6):241–7.
23. D’Souza DH, Bhattacharya S, Das A. Fibrinolytic protease from Bacillus cereus S46: Purification, characterization, and evaluation of its in vitro thrombolytic potential. J Basic Microbiol 2020;60(8):661–8.
24. Prasad S, Kashyap RS, Deopujari JY, Purohit HJ, Taori GM, Daginawala HF. Development of an in vitro model to study clot lysis activity of thrombolytic drugs. Thromb J 2006;4(1):14.
25. Diwan D, Usmani Z, Sharma M, Nelson JW, Thakur VK, Christie G, et al. Thrombolytic enzymes of microbial origin: A review. Int J Mol Sci 2021;22(19):10468.
26. Scott JE, Williams KP. Validating identity, mass purity and enzymatic purity of enzyme preparations. In: Markossian S, Grossman A, Brimacombe K, Arkin M, Auld D, Austin C, et al., editors. Assay Guidance Manual. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004.
27. Xin X, Ambati RR, Cai Z, Lei B. Purification and characterization of fibrinolytic enzymes from a bacterium isolated from soil. 3 Biotech 2018;8(2):90.
28. Zhou Y, Chen H, Yu B, Chen G, Liang Z. Purification and characterization of a fibrinolytic enzyme from marine Bacillus velezensis Z01 and assessment of its therapeutic efficacy in vivo. Microorganisms 2022;10(5):843.
29. Agrebi R, Haddar A, Amidet N, Jellouli K, Manni L, Nasri M. BSF1 fibrinolytic enzyme from a marine bacterium Bacillus subtilis A26: Purification, biochemical and molecular characterization. Process Biochem 2009;44(11):1252–9.
30. Wang CT, Ji BP, Li B, Nout R, Li PL, Ji H, et al. Purification and characterization of a fibrinolytic enzyme of Bacillus subtilis DC33, isolated from Chinese traditional Douchi. J Ind Microbiol Biotechnol 2006;33(9):750–8.
31. Chen PT, Chiang C-J, Chao Y-P. Medium Optimization for the production of recombinant nattokinase by Bacillus subtilis using response surface methodology. Biotechnol Prog 2007;23(6):1327–32.
32. Peng Y, Huang Q, Zhang R-h, Zhang Y-z. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douchi, a traditional Chinese soybean food. Comp Biochem Physiol B Biochem Mol Biol 2003;134(1):45–52.
33. Mican J, Toul M, Bednar D, Damborsky J. Structural biology and protein engineering of thrombolytics. Comput Struct Biotechnol J 2019;17:917–38.
34. Yao Z, Kim JA, Kim JH. Properties of a fibrinolytic enzyme secreted by Bacillus subtilis JS2 isolated from saeu (small shrimp) jeotgal. Food Sci Biotechnol 2018;27(3):765–72.
35. Jeong YK, Park JU, Baek H, Park SH, Kong IS, Kim DW, et al. Purification and biochemical characterization of a fibrinolytic enzyme from Bacillus subtilis BK-17. World J Microbiol Biotechnol 2001;17(1):89–92.
36. Lee S-K, Bae D-H, Kwon T-J, Lee S-B, Lee H-H, Park J-H, et al. Purification and characterization of a fibrinolytic enzyme from Bacillus sp. KDO-13 isolated from soybean paste. J Microbiol Biotechnol 2001;11:845–52.
37. Rajaselvam J, Benit N, Alotaibi SS, Rathi MA, Srigopalram S, Biji GD, et al. In vitro fibrinolytic activity of an enzyme purified from Bacillus amyloliquefaciens strain KJ10 isolated from soybean paste. Saudi J Biol Sci 2021;28(8):4117–23.
38. Yuan J, Yang J, Zhuang Z, Yang Y, Lin L, Wang S. Thrombolytic effects of Douchi fibrinolytic enzyme from Bacillus subtilis LD-8547 in vitro and in vivo. BMC Biotechnol 2012 Jul 2;12:36.
39. Narasimhan MK, Chandrasekaran M, Rajesh M. Fibrinolytic enzyme production by newly isolated Bacillus cereus SRM-001 with enhanced in-vitro blood clot lysis potential. J Gen Appl Microbiol 2015;61(5):157–64.