1. INTRODUCTION
Withania coagulans, known as Rishyagandha, belong to the genus Withania of the Solanaceae family; due to Withania’s rich pharmacological characteristics, it is one of India’s most well-known genera of medicinal plants [1]. However, only two of the 23 species of Withania recognized, Withania somnifera, and W. coagulans, are of substantial economic importance [2]. W. coagulans is a grayish-white shrub common in East India, Nepal, and Afghanistan; in India, it occurs in Punjab, Rajasthan, Simla, Kumaun, and Garhwal [3]. W. coagulans have been suggested for treating several illnesses in traditional medical systems, including anti-tumor, anti-hyperglycemic, anti-diabetic, hepatoprotective, cardiovascular, immunosuppressive, hypolipidemic, free radical scavenging, and anti-depressant properties in the central nervous system [4–6]. In addition, W. coagulans has distinctive properties, making them a unique phytochemical.
While the global acceptance of plant-based herbal therapy is increasing, the adoption of false substitution and adulteration is rising. The use of non-authentic herbal materials not only reduces the efficacy of herbal medicine but also creates safety issues. Despite its significant therapeutic value and widespread use, investigations on the authenticity of W. coagulans market samples are rare. Since W. coagulans and W. somnifera market samples show resemblance; in these circumstances, non-morphological DNA barcoding is quite useful for species identification and authentication [7].
DNA barcoding represents one of the most molecular biological marker-based techniques for determining target plant species in a short time. The goal of DNA barcoding, which is a widely used technology, is to identify various plant species utilizing nucleotide sequences accurately. Paul Hebert and colleagues were the first to propose using short DNA sequences known as DNA barcodes to identify biological organisms [8]. DNA barcoding studies can be used to authenticate plant species. The Plant Working Group, Consortium for the Barcode of Life (CBOL), recommends using the chloroplast genes matK, rbcl, and the intergenic spacer trnH-psbA, in addition to the nuclear region ITS, for DNA barcoding in plants [8–10]. All plant tissues contain DNA, a macromolecule more stable than RNA. Therefore, DNA-based markers are preferable for accurately identifying medicinal plants [11].
The present study aimed to identify differences in the gene sequences of two samples of W. coagulans grown in Gujarat and Mumbai by amplifying the psbA-trnHf, rpoB, rbcl, matK, and ITS gene markers from each sample, taking into account that environmental factors affect the physical characteristics of most plants [12]. Studies goal is to find the ideal barcode to identify and authenticate the plant. The sequences acquired will also be used to build a phylogenetic tree and to see its interrelationships with other Solanaceae plant species. It also aims to discover the best genetic marker for W. coagulans barcoding studies.
2. MATERIALS AND METHODS
2.1. Plant Sample Collection and Authentication
Plant sample 1 (WCNB1) of W. coagulans was obtained from the ICAR-Directorate of Medicinal Aromatic Plants Research in Anand, Gujarat, India (22°35’57.2”N 72°55’58.9” E). The seeds were collected from ICAR-Gujarat and were grown at The Institute of Science, Mumbai; it was referred to as plant sample 2 (WCNB2). The voucher herbarium specimen prepared was deposited to the Department of Botany, Shivaji University, Kolhapur, for authentication.
2.2. DNA Isolation
DNA isolation was carried out by taking 1 g of cleaned plant fresh leaf sample crushed in mortar and pestle using liquid nitrogen. 2% cetyltrimethylammonium bromide (CTAB), 100mMTris- pH 8.0, 20mM EDTA, and 1.7 M NaCl with 0.3% (v/v) β-mercaptoethanol were used as a lysis buffer for isolation of total genomic DNA. The pellet obtained was suspended in 30 μl TE buffer. The quality of the isolated DNA was analyzed on 0.8% agarose gel electrophoresis, and the quantity was checked using a NanoDrop spectrophotometer. The extracted DNA was stored chilled at –20°C [13].
2.3. PCR Amplification and Sequencing
Polymerase chain reaction (PCR) is used to amplify five genetic markers from W. coagulans: Two intergenic spacer regions (trnH-psbA, rpoB-trnCGAR), ribulose 1, 5 bisphosphate (rbcl), one nuclear region Inter Transcribed Spacer (ITS) in a 25 μl reaction mixture, and maturase kinase (matK) in a 50 μl reaction mixture. The reaction mixture of 25 μl contained 100 ng DNA templet, 0.5 U DreamTaq DNA Polymerase enzyme (Thermo Fischer), 10 mM dNTPs, 2.5 mM MgCl2, 10X DreamTaq Buffer, and 0.5 μ M forward and reverse primers (Eurofins, India). The list of forward and reverse primers, annealing temperature (Tm), and cycling conditions used in this study is mentioned [Table 1]. A thermal cycler (Applied BioSystems) was used for the PCR reaction. The amplified product was observed for quality using 1.5% agarose gel electrophoresis (1x TAE buffer and 0.5 μg/mL ethidium bromide). All samples were cleaned using the Qiagen Gel Extraction kit to remove extra primer dimers, templates, and dNTPs. The samples were then transported to Eurofins India Pvt. Ltd. for Sanger sequencing.
Table 1: List of primers and cycling conditions for DNA amplification.
| DNA marker | Primer sequence | Tm in degree | Cycling conditions |
|---|---|---|---|
| psbA-trnHf | |||
| Forward primer | 5’ GTTATGCATGAACGTAATGCTC 3’ [20] | 58° C | 95°-5 min, |
| Reverse primer | 5’ CGCGCATGGTGGATTCACAATCC 3’ [20] | ||
| rpoB-trnCGAR | |||
| Forward primer | 5’ CKACAAAAYCCYTCRAATTG 3’ [21] | 58° C | |
| Reverse primer | 5’ CACCCRGATTYGAACTGGGG 3’ [21] | ||
| rbcl | |||
| Forward primer | 5’ ATGTCACCACAAACAGAGACTAAAGC 3’ [8] | 58° C | |
| Reverse primer | 5’ GTAAAATCAAGTCCACCRCG 3’ [8] | ||
| ITS | |||
| Forward primer | 5’ TCCGTAGGTGAACCTGCGG 3’ [22] | 58° C | |
| Reverse primer | 5’ TCCTCCGCTTATTGATATGC 3’ [22] | ||
| matK | |||
| Forward primer | 5’ CGATCTATTCATTCAATATTTC 3’ [23] | 54° C | 95°-5 min, |
| Reverse primer | 5’ TCTAGCACACGAAAGTCGAAGT 3’ [23] |
2.4. DNA Sequence Data Analysis
The nucleotide sequences from W. coagulans were compared to sequences found in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) for similarity match. The sequences obtained were then submitted to NCBI GenBank. Their accession numbers are cited [Table 2]. Published sequences were retrieved for additional analysis. Then, multiple sequence alignment was performed with retrieved sequences, and Finch TV was used to edit them. MEGA 11 (Molecular Evolutionary Genetic Analysis) software was used to evaluate parameters including genetic distance, parsimony informative sites, conserved sites, and variables sites [14]. Maximum likelihood and Tamura-Nei Model were used to build a phylogenetic tree with 1000 bootstrap replications. As out-groups, taxa from the Ipomoea and Convolvulus genes were utilized.
Table 2: GenBank accession numbers of submitted sequences of Withania coagulans in NCBI [24].
| Species name | Accession number of psbA-trnHf | Accession number of rpoB-trnCGAR | Accession number of rbcl | Accession number of ITS | Accession number of matK |
|---|---|---|---|---|---|
| Withania coagulans sample 1 (WCNB1) | OQ406250 | OQ406252 | ON369434 | MW205772 | MW729349 |
| Withania coagulans sample 2 (WCNB2) | OQ406251 | OQ406253 | 0N369435 | MW205773 | MW729350 |
3. RESULTS
3.1. Results of PCR and DNA Barcode Sequence Analysis
In the present study, the ten sequences obtained from the two samples of W. coagulans grown in Mumbai and collected from ICAR-Anand met the requirements for successful sequencing. The five DNA barcodes psbA-trnHf, RpoB-trnCGAR, rbcl, ITS, and matK were efficiently amplified using the primers employed; as a result, agarose gel electrophoresis revealed a sharp DNA band, which, when purified, produced successful sequencing results. The identified ten nucleotide sequences for W. coagulans have all been submitted to the NCBI Gene Bank database. All DNA markers produced different base-pair lengths with indels/deletions were found after distance and phylogenetic analysis.
3.2. DNA Sequence Data Analysis
NCBI BLAST analysis of all five DNA barcodes obtained from W. coagulans (sample 1 and sample 2) with the retrieved sequences of some Solanaceae family taxa was aligned, including other Withania species. Ipomoea carnea, Ipomoea batatas, and Convolvulus arvensis were used as out group taxa. W. coagulans sample 1 and sample 2 comparative studies is noted [Table 3], and W. coagulans sample 1 and sample 2 comparative studies [Table 4] with other sequences retrieved from NCBI databases is also analyzed.
Table 3: Sequence length and (G + C) % content of barcode regions in Withania coagulans species [14].
| Species name | psbA-trnHf | rpoB-trnCGAR | rbcl | ITS | matK | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | %GC | Length (bp) | %GC | Length (bp) | %GC | Length (bp) | %GC | Length (bp) | % GC | |
| Withania coagulans WCNB1 | 525 | 27.5 | 549 | 41.8 | 684 | 44 | 695 | 66.9 | 805 | 33.3 |
| Withania coagulans WCNB2 | 549 | 27.5 | 385 | 41.5 | 546 | 44 | 696 | 62.6 | 783 | 32.8 |
Table 4: Characteristic features of five DNA markers among Withania coagulans sample 1 and sample 2 [14].
| Withania coagulans sample 1 and sample 2 | Pairwise distance average mean | Number of conserved sites | Number of variables | Number of parsimony informative sites | Number of singletons |
|---|---|---|---|---|---|
| psbA-trnHf | 0.000 | 509/525 | None | None | None |
| rpoB-trnCGAR | 0.132 | 327/376 | 44/376 | None | None |
| Rbcl | 0.000 | 507/507 | None | None | None |
| ITS | 0.041 | 650/744 | 32/744 | None | None |
| matK | 0.029 | 739/767 | 22/767 | None | None |
The psbA-trnHf gene sequences from W. coagulans samples 1 and 2 (WCNB1 and WCNB2), respectively, showed the least amount of length variation (24 bp) and the lowest amount of (G+C) content (27%), in comparison to matK, ITS, rpoB, and psbA [Table 5], which did not exhibit any pairwise distance average mean. Sequences found 509 conserved sites out of 525 with no variables, parsimony informative sites, and singleton base. The aligned sequences of different species showed base replacement according to statistical analysis of nucleotide pair frequencies, and the transitional rate 14 was lower than the transversional rate 18 in psbA. It was discovered that psbA has the best ability to distinguish between Solanaceae species.
Table 5: Characteristic features of five DNA markers among Solanaceae species and out-group taxa [14].
| Solanaceae species and out-group taxa | Number of taxa selected | Pairwise distance average mean | Number of conserved sites | Number of variables | Number of parsimony informative sites | Number of singletons |
|---|---|---|---|---|---|---|
| psbA-trnHf | 14 | 0.135 | 368/525 | 149/525 | 59/525 | 89/525 |
| rpoB-trnCGAR | 10 | 0.033 | 305/376 | 71/376 | 60/376 | 11/376 |
| Rbcl | 15 | 0.028 | 469/507 | 38/507 | 33/507 | 05/507 |
| ITS | 14 | 0.122 | 488/744 | 224/744 | 111/744 | 109/744 |
| matK | 16 | 0.071 | 625/767 | 142/767 | 110/767 | 32/767 |
RpoB-trnCGAR gene sequences from W. coagulans WCNB1 and WCNB2 (549 bp and 385 bp) showed significant length variation of 164 base pairs and 41.8% and 41.5% (G+C) content, respectively, with 0.132 pairwise distance average mean. Sequences found 327 conserved sites out of 376 base pairs with 44 variables. After alignment, WCNB1 and WCNB2 showed changes in the sequences from positions 1 to 7, and different gaps were found between positions 13 and 39. Purines replaced by purines predominated over purines replaced by pyrimidines when selected sequences were aligned for analysis.
Rbcl was found to be a highly conserved genetic marker in the Solanaceae family. The species Iponeace carnea showed gaps from positions 1-303; therefore, except for the out-group species Ipomea carnea, all 15 taxa were chosen for sequence alignment of the rbcl gene exhibited 100% similarity in their sequences. Nonetheless, the sequence from position 303 to 507 for Ipomoea carnea showed perfect similarity with other chosen taxa, demonstrating that no insertions or deletions were found in this genetic marker.
The following barcode study used the ITS gene sequences from samples 1 and 2 (695 and 696 bp, respectively), with a one base pair variance and 66.9% and 62.6% (G+C) content in each, which is the highest GC content among all the DNA barcodes selected for studies. After ITS was aligned, it was found that WCNB1 and WCNB2 had a variety of gaps from positions 50 to 62, 106 to 112, 23 to 34, 47 to 82, 503 to 504, 532 to 534, 600 to 603, 637 to 638, 649 to 653, and 676 to 686. In addition, four single nucleotide gaps were found at locations 144, 164, 173, and 205. In the statistical analysis of nucleotide pair frequencies, transitional pairings 37 are more numerous than transversional pairs 21.
MatK gene sequence lengths for samples 1 (805 bp) and 2 (783 bp), with 22bp variation and 33.3% (G+C) content and 32.8% (G+C) content, for sample 1 and 2 respectively. In the Solanaceae family, matK also demonstrated a high level of conservation with 27 single gaps in the sequences of all chosen taxa. Furthermore, in matK, the transitional results revealed 22 base pair variations using multiple sequence alignment in both samples of W. coagulans.
3.3. Phylogenetic Analysis
A phylogenetic tree was constructed with Maximum Likelihood analysis using Mega 11 software. It showed a higher resolution for all five DNA barcodes. DNA marker sequences for W. coagulans (sample 1 and 2) were collected, and a BLAST search was conducted. The selected sequences showed ≥90% similarities from the Solanaceae family and out-group taxa were then aligned in MEGA 11 using multiple sequence comparison by Log-Expectation. Using the Maximum Likelihood approach with 1000 bootstrap replications and the Tamura-Nei model, the nucleotide sequences of W. coagulans were utilized for evolutionary studies.
In contrast to other species of the Solanaceae family, the phylogenetic analysis of psbA [Figure 1] shows that it successfully recognizes W. coagulans. The W. coagulans sample 1(WCNB1) and sample 2 (WCNB2) had the same branch length of 0.00004 and originated from the same branch point belonging to a monophyletic group. All Withania species exhibit the most significant degree of similarity. Ipomoea carnea, Ipomoea batatas, and Convolvulus arvensis are out-group taxa belonging to a separate clade showing another monophyletic cluster with a branch length of 0.01950, which aids in establishing a distinction between the species of the Solanaceae family and the out-group taxa chosen. The tree with the highest log probability (–1439.68) is presented.
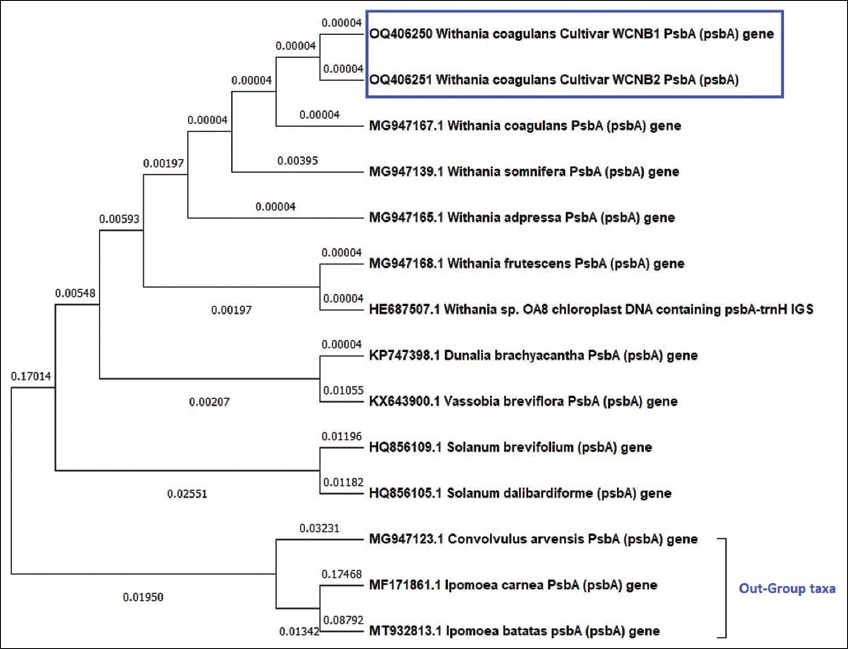 | Figure 1: Maximum likelihood phylogenetic analysis using psbA genetic marker among Withania coagulans samples (WCNB1 and WCNB2), Solanaceae species, and out-group taxa. [Click here to view] |
Phylogenetic analysis of rpoB [Figure 2] W. coagulans samples 1 and 2 was shown to be in two distinct clusters with branch points of 0.006727 and 0.072480, respectively, indicating a significant difference. The tree with the most excellent log probability (–889.38) is displayed. W. somnifera species are most similar to W. coagulans sample 1. Whereas W. coagulans sample 2 revealed a high similarity with the NCBI-retrieved W. coagulans rpoB gene sequence. The three out-group taxa were present in two separate monophyletic clades in the middle of the tree. To distinguish between various Withania species, it was found that rpoB as a DNA marker was insufficient.
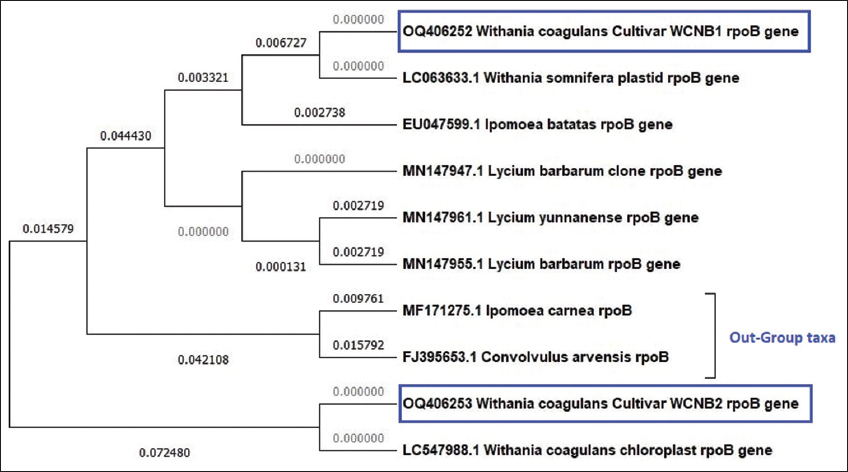 | Figure 2: Maximum likelihood phylogenetic analysis using rpoB genetic marker among Withania coagulans samples (WCNB1 and WCNB2), Solanaceae species, and out-group taxa. [Click here to view] |
Phylogenetic evaluation of rbcl [Figure 3] nucleotide sequences showed a well-resolved tree with multiple clades, whereas W. coagulans both samples belonged to a monophyletic group. In contrast, other Withania species belonged to a separate cluster, including adpressa, frutescence, and somnifera from the same branch point, showing higher similarities in the genus. Furthermore, the out-group displayed a different cluster. Finally, the tree with the most significant log probability is displayed (–967.83).
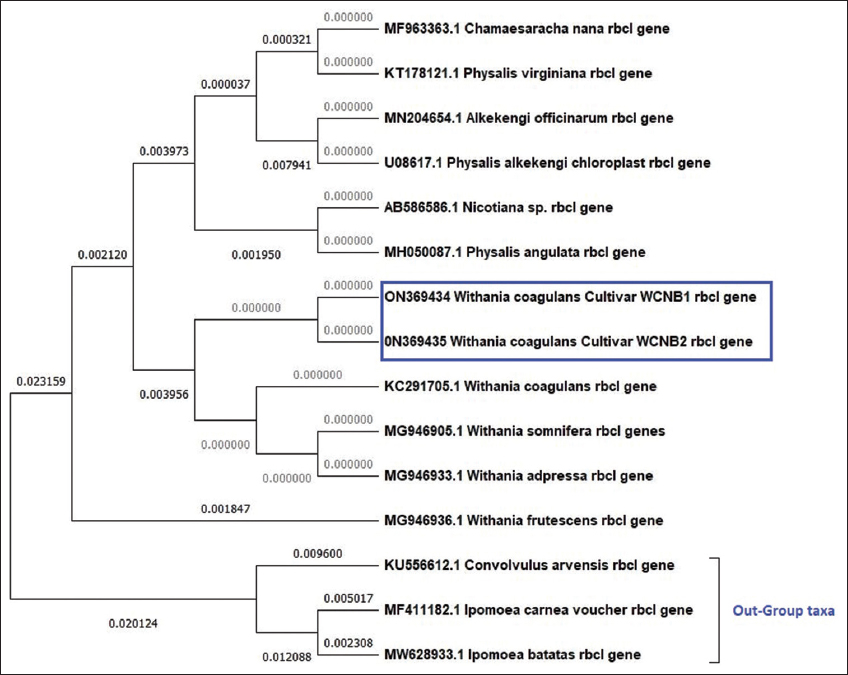 | Figure 3: Maximum likelihood phylogenetic analysis using rbcl genetic marker among Withania coagulans samples (WCNB1 and WCNB2), Solanaceae species, and out-group taxa. [Click here to view] |
ITS phylogenetic analysis [Figure 4] revealed, W. coagulans WCNB1 and WCNB2 belonged to a non-monophyletic sister clade below clade 1, representing W. somnifera sequences, showing exact ancestral origin. The sequence divergences of the ITS marker differentiate due to the tree making different clusters in the Solanaceae family and the out-group taxa Ipomoea carnea and Convolvulus arvensis displaying a separate group. The tree was shown with the highest log likelihood (–2621.07).
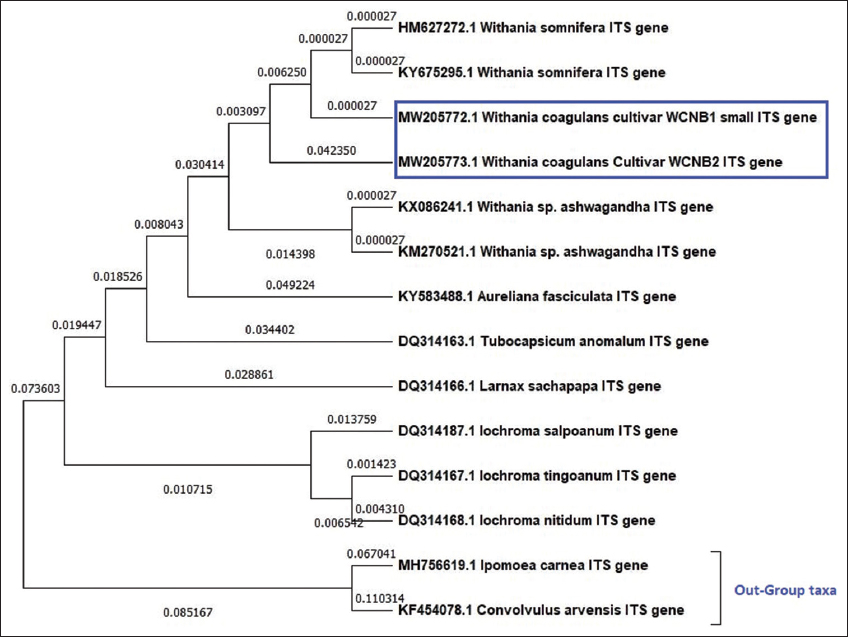 | Figure 4: Maximum likelihood phylogenetic analysis using ITS genetic marker among Withania coagulans samples (WCNB1 & WCNB2), Solanaceae species, and out-group taxa. [Click here to view] |
The phylogenetic analysis of the matK sequences [Figure 5] showed a tree with just two clusters, the first of which encompassed all the Solanaceae species, including coagulans and frutescens, the out-group of which formed a monophyletic cluster. The fact that WCNB1 and WCNB2 are clearly distinctly located in the first cluster causes them to exhibit divergences. The tree with the highest log likelihood (–1740.62) is shown.
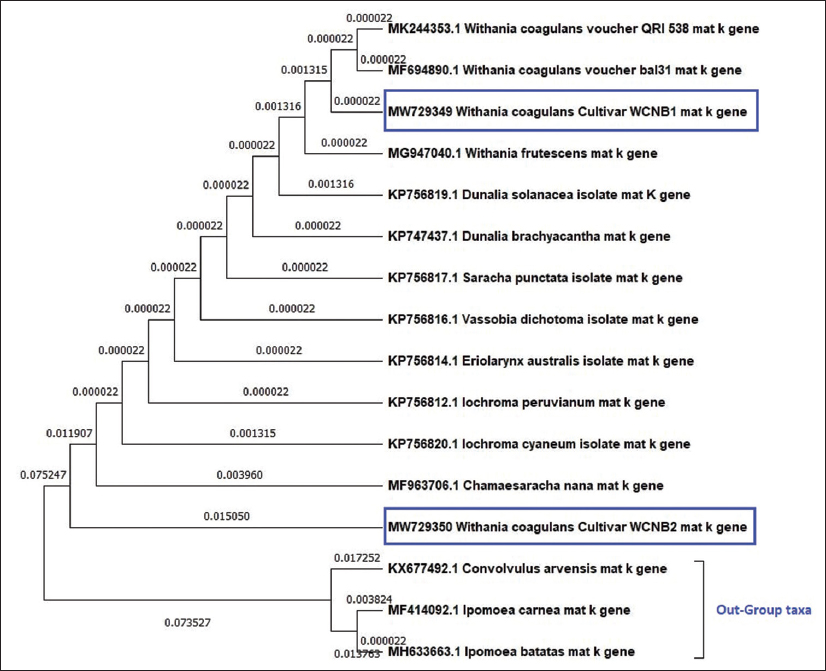 | Figure 5: Maximum likelihood phylogenetic analysis using matK genetic marker among Withania coagulans samples (WCNB1 and WCNB2), Solanaceae species, and out-group taxa. [Click here to view] |
4. DISCUSSION
Understanding the evolutionary history of many significant plants depends on accurately identifying and assessing biologically significant plant species and their families [12]. DNA barcoding soon became a reliable method for identifying species, spotting adulteration, quality control, and ecological assessments. However, it cannot replace the conventional taxonomic identification technique, but it will be easier to understand phylogeny and its origins with the help of this information. The purity of the DNA is crucial for PCR-based amplification. Even closely related species may need distinct DNA isolation procedures because metabolites in medicinal plants can occasionally affect DNA integrity during isolation [15]. The main principle of plant species identification is the sequencing variation from the reference sequence and phylogenetic reconstruction [16]. To barcode the W. coagulans plant in the present study, we used the chloroplast trnH-psbA, rpoB, rbcl, ITS, and matK region. The chloroplast genomes of angiosperms contain characteristics conserved across and among many plant lineages and have almost identical gene content and organization [17–19]. The amplification and sequencing rates have been high enough to be considered for five selected genetic loci as DNA barcodes.
In the present study, multiple sequence alignment and phylogenetic analysis showed that compared to the other three possible barcodes, rbcl, and psbA sequences have a low rate of nucleotide evolution due to the presence of highly conserved regions in them. In both the WCNB1 and WCNB2 samples, rbcl demonstrated complete conservation. Furthermore, compared to the Solanaceae family species, it showed the most significant similarity. However, the out-group taxa had some differences and belonged to a different group on the phylogenetic tree. With an average mean distance of 0.000 between WCNB1 and WCNB2, psbA demonstrated significant levels of conservation and the highest similarity among the W. coagulans species, matK sequence analysis revealed 22 variable sites for W. coagulans, and rpoB showed the highest number of 44 variable sites.
The best genetic markers for identifying the W. coagulans plant were psbA and rbcl. It revealed no genetic differences even when samples were grown in ICAR-Gujarat, and The Institute of Science, Mumbai. However, rpoB, ITS, and matK differed in both samples due to 22-44 bp variations. The results of this work indicate that the above-mentioned indigenous plant species can be amplified, identified, and differentiated successfully using universal primers (rbcl+psbA) for DNA barcoding. “matK can also be used due to fewer variations in the evolution of genes. Over 90% of flowering plants can be distinguished using the (rbcl+matK) pair combination,” according to CBOL 5. However, more conserved genomic areas could find and used to identify adulteration of therapeutic material. Once the barcodes have evaluated, the barcode markers psbA, rbcl, and matK can be combined to create a barcode appropriate for identifying W. coagulans. W. coagulans samples can be separated using the present study’s (rbcl+psbA) combination.
As per the Log-likelihood values by MEGA 11 in the phylogenetic analysis, it was observed that the rpoB marker showed the highest probability value (–889.38) than rbcl (-967.83). The other three markers showed the lowest values. Hence, it was proved to have the rpoB gene with fewer possible mutations in the evolutionary studies.
5. CONCLUSION
The current work examined two samples of W. coagulans produced at ICAR-Anand and the Institute of Science, Mumbai for potential evolutionary relationships using modern DNA barcode technologies and five genetic markers. This research is the first to evaluate molecular markers based on identifying and classifying W. coagulans from two sites in India cultivated from the same seed samples. The current work uses DNA barcoding techniques and offers essential results for establishing the phylogeny and relationship between several Solanaceae family species. Genetic markers rbcl and psbA show the highest conservations, whereas matK, ITS, and rpoB can be discriminating markers in the Withania genus and the Solanaceae family. More DNA barcode investigations are required to describe W. coagulans from all possible growth-friendly environments. This research will help gather basic information and provide details on the species molecular taxonomy.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This research does not involve any animal or human subjects for ethical approvals.
10. DATA AVAILABILITY
This research publication includes all data gathered and analyzed, and the sequences are available online in the NCBI database.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Rohit J, Sumita K, Kothari SL. Phytochemistry, pharmacology, and biotechnology of Withania somnifera and Withania coagulans:A review. J Med Plants Res 2012;6:5388-99. [CrossRef]
2. Hemalatha S, Kumar R, Kumar M. PHCOG REV.:Plant review Withania coagulans Dunal:A review. Pharmacognosy Rev 2008;2:351-8.
3. Rageeb MD, Bhat NG, Usman MD. Evaluation of antidiabetic activity of Withania coagulans L. Dunal. Int J Pharm Sci Res 2023;14:981-7.
4. Maurya R. Chemistry and pharmacology of Withania coagulans:An ayurvedic remedy. J Pharm Pharmacol 2010;62:153-60. [CrossRef]
5. Gupta V, Keshari BB. Withania coagulans Dunal. (paneer doda):A review. Int J Ayurvedic Herb Med 2013;3:1330-6.
6. Ojha S, Alkaabi J, Amir N, Sheikh A, Agil A, Fahim MA, et al. Withania coagulans fruit extract reduces oxidative stress and inflammation in kidneys of streptozotocin-induced diabetic rats. Oxid Med Cell Longev 2014;2014:201436. [CrossRef]
7. Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci 2003;270:313-21. [CrossRef]
8. Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, et al. ADNA barcode for land plants. Proc Natl Acad Sci U S A 2009;106:12794-7. [CrossRef]
9. Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A 2005;102:8369-74. [CrossRef]
10. Kress WJ, Erickson DL. DNA barcodes:Methods and protocols. Methods Mol Biol 2012;858:3-8. [CrossRef]
11. Wattoo JI, Saleem MZ, Shahzad MS, Arif A, Hameed A, Saleem MA. DNA barcoding:amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species. Adv Life Sci 2016;4:3-7.
12. Shinwari ZK, Jan SA, Khalil AT, Khan A, Ali M, Qaiser M, et al. Identification and phylogenetic analysis of selected medicinal plant species from Pakistan:DNA barcoding approach. Pak J Bot 2018;50:553-60.
13. Anerao J, Jha V, Shaikh N, Shivalkar A, Nityanand A, Sawant D, et al. DNA barcoding of important fruit tree species of agronomic interest in the genus Garcinia L. from the Western Ghats. Genet Resour Crop Evol 2021;68:3161-77. [CrossRef]
14. Tamura K, Stecher G, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [CrossRef]
15. Khanuja SP, Shasany AK, Darokar MP, Kumar S. Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Biol Report 1999;17:74. [CrossRef]
16. Altschul S, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST:A new generation of protein database search programs. Nucleic Acids Res 1997;25:3389-402. [CrossRef]
17. Abdullah, Mehmood F, Shahzadi I, Waseem S, Mirza B, Ahmed I, et al. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae):Comparative analyses and identification of mutational hotspots. Genomics 2020;112:581-91. [CrossRef]
18. Amiryousefi A, Hyvönen J, Poczai P. The chloroplast genome sequence of bittersweet (Solanum dulcamara):Plastid genome structure evolution in Solanaceae. PLoS One 2018;13:e0196069. [CrossRef]
19. Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn:Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 2006;7:439. [CrossRef]
20. Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot 2003;28:723-37.
21. Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, et al. The tortoise and the hare II:Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 2005;92:142-66. [CrossRef]
22. Bruns TD, Lee SB, Taylor JW, Bruns TD, Lee SB, Taylor JW. Amplification and Direct sequencing of fungal ribosomal RNA genes for phylogenetics acid phosphatase NFSTC lecture view project low template DNA collection view project;1990:315-22. [CrossRef]
23. Cuenoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 2002;89:132-44. [CrossRef]
24. National Center for Biotechnology Information (NCBI). Bethesda (MD):National Library of Medicine (US), National Center for Biotechnology Information;1988. Available from:https://www.ncbi.nlm.nih.gov/ [Last accessed on 2023 Feb 08].