1. INTRODUCTION
Methyl orange (MO) is a sulfonated azo dye extensively utilized in textile dyeing facilities and as a pH indicator in biochemical applications [1]. Despite its utility, MO and its metabolites pose significant environmental and health risks due to their resistance to biodegradation and potential mutagenic, carcinogenic, and teratogenic effects [2]. The presence of MO in industrial effluents necessitates effective decolorization and detoxification before discharge into the environment to mitigate these hazards [3]. Physicochemical methods, such as filtration, adsorption, precipitation, chemical oxidation, coagulation, electrolysis, and photodegradation, have been employed to remove MO from wastewater [4–7]. However, these methods often fall short due to high costs, energy consumption, reagent requirements, insufficient decolorization, and the potential generation of harmful byproducts [8–10]. Advanced oxidation processes, although effective, are similarly cost-prohibitive and energy-intensive [11]. In contrast, biological methods using microorganisms offer a promising alternative due to their cost-effectiveness, non-toxic byproducts, and potential for complete mineralization of dyes [12–15].
Microbial dye removal generally involves either the degradation of dyes by microbial cells or the adsorption of dyes onto microbial biomass [16]. Unlike adsorption, which retains the dye’s original structure, microbial degradation breaks down the dye into smaller, less harmful fragments, potentially transforming them into carbon dioxide, water, and inorganic salts [17]. Various microorganisms, including fungi, bacteria, yeast, and algae, have been shown to decolorize and mineralize azo dyes, with bacterial decolorization often being more rapid and hence more favored [15,18]. Locally isolated Rhodococcus strain UCC 0004 demonstrated complete removal of 0.5 g/l MO after 2 days and exhibited significantly higher removal efficiency and substrate affinity when immobilized, indicating its potential as a biological tool for efficient wastewater treatment [19]. However, there is a notable research gap regarding the efficacy of gram-positive bacteria in degrading dyes like MO. This study addresses this gap by isolating and characterizing bacterial strains capable of efficiently degrading MO. The novelty of this research lies in identifying and optimizing Bacillus cereus J4, a gram-positive bacterium, for the effective bioremediation of MO. The main focus of this study included isolating bacterial strains from industrial effluents, optimizing the decolorization conditions using Plackett–Burman (PB) design and response surface methodology (RSM), and evaluating the efficacy of Bacillus cereus J4 in MO degradation. As demonstrated in previous research on Rhodococcus strain UCC 0010 for Congo red decolorization, RSM effectively identified optimal conditions, resulting in nearly 100% decolorization, an 83% reduction in incubation time, and an 18% increase in decolorizing activity compared to conventional optimization methods. These findings underscore the advantages of RSM in enhancing bioremediation processes [20]. The present study will contribute to sustainable development goals (SDGs) by promoting clean water (SDG 6) through the development of efficient bioremediation techniques, fostering responsible consumption and production (SDG 12) by reducing industrial pollutants, and supporting life below water (SDG 14) by mitigating the adverse impacts of dye pollutants on aquatic ecosystems [21–23]. By advancing microbial bioremediation strategies, this research aims to provide sustainable solutions for the treatment of industrial effluents, thereby enhancing environmental health and safety.
2. MATERIALS AND METHODS
2.1. Chemicals and Dye
The chemicals used are of prescribed analytical grade. MO was procured from Sigma Aldrich in Bangalore, India. Nutrient media [nutrient agar (NA) and nutrient broth] and nitrogen sources and carbon sources were obtained from HI Media (Mumbai, India).
2.2. Isolation of Bacteria from Collected Samples
The bacterial colonies were isolated from the industrial dye effluents, wet sludge, dry sludge, and soil samples by following the serial dilution method. For the isolation of bacterial colonies, 1 ml (v/v) liquid or 1 g (w/v) of solid was put into the sterile water and serial dilution was carried out up to the 10−1 to 10−10. Then, 100 µl of the diluted sample was transferred onto NA medium, followed by incubation at 37°C for 24 hours (Navyug-107 Incubator Bacteriological). A loopful of selected colonies was streaked on fresh NA plates to obtain pure colonies. The process is repeated till pure cultures are obtained [24].
2.3. Culture Conditions
The pure bacterial cultures were preserved on slants of NA Medium and stored in the refrigerator (4°C) and glycerol stock solution at −20°C for long-term preservation according to Pearce et al. [25].
2.4. Inoculum Preparation
A colony of the isolated strain (Bacillus cereus J4) was added to the nutrient broth and incubated at 37°C under shaking conditions (120 rpm) until the optical density (OD)600 reached 0.6–0.8. Next, the inoculum was transferred aseptically into the dye solutions for further experimentation, i.e., screening of dye-removing bacterium.
Prepared the 50 ml nutrient broth in 250 ml borosilicate glass flask containing dye solution concentration 10 mg/ml for each pure isolate, then inoculated the media with the 24 hours inoculum and incubated at 37°C for 24 hours. For successive experiments, ambient culture conditions were used. Aliquots (2.5 ml) were taken at the regular time of intervals and centrifugated at 6,000 rpm for 20 minutes (ThermoFisher) to segregate the bacterial biomass, which interfered with measurement. The OD of the supernatant was noted. The maximum wavelength of the tested dye was calculated at a wavelength of 465 nm using a spectrophotometer. Standard curve of (MO) dye conc. versus OD was noted and used to compute the decrease in dye concentration. The following equation was used to get the removal percentage applying absorbance to examine decolorization activity [25]. The percent of decolorization was determined by the decrease in absorbance and visual appearance.
where A° is the initial absorbance and A is the final absorbance after treatment.
2.5. Identification and Characterization of Microorganisms
Several pure bacterial isolates were obtained out of which 11 bacterial colonies were selected and were designated as J1, J2. J3, J4 J5, J6, J7, J8, J9, J10, and J11. Out of 11 isolates, 1 isolate were selected based on maximum decolorization of dye in minimum times of hours and morphological, biochemical, and molecular characterizations (16SrRNA) were confirmed via MTCC, IMTECH Chandigarh, India. The 16S rRNA sequencing of Bacillus cereus J4 were submitted to NCBI GenBank and assigned the accession number OQ392442, identifying them as Bacillus cereus J4.
2.6. Factors Affecting Bacterial Decolorization
Optimization of an operating system is necessary to obtain the maximum rate of decolorization of azo dyes MO. Decolorization performance can be enhanced by using different media compositions and supplementing them with various carbon and nitrogen sources. Different physical parameters, i.e., initial dye concentration, inoculum%, incubation time, temperature, pH, and shaking rate [26] were optimized by using OFAT (one factor at a time) experiments as these factors (parameters) greatly influence the microbial decolorization efficiency. The composition of wastewater from textiles can vary daily and seasonally and typically includes organic compounds, nutrients, sulphur compounds, salts, and various toxic substances [25]. It is possible that the existence of these compounds could impede the process of dye decolorization. Therefore, the impact of each factor on the dye removal must be determined before the treatment of the industrial effluent by biological means.
2.7. Optimization of Physicochemical Parameters Using the PB Method and RSM
PB design: The PB design was used to identify significant factors (one variable at a time) responsible for the removal of MO. RSM was employed to ascertain the best response for the decolorization of MO [27].
The Expert software (Version 7.5.7.1, Stat-Ease) was utilized to enhance the decolorization of MO with Bacillus cereus J4. Initially, The PB design, also known as fractional factorial design, [28] was employed to represent the relative significance of various environmental aspects on the dye (MO) removal with bacterial strain. For every variable, eight chosen individual factors were evaluated at both the highest (+) and lowest (-) levels in 11 combinations arranged in accordance with the (PB) design (Table 1). All experiments were performed in triplicate and averages of the dye decolorization were taken as responses. Each variable’s primary effect was ascertained using the following formula [29].
Exi = ({Mi+ - Mi-}) / N
Exi stands for the variable major effect in this case.
Mi+ and Mi- represent the percentage of MOdecolorization in trials where independent variables (xi) were present in low and high concentrations, respectively, and N represents the number of trials divided by 2.
2.8. Screening Using the PB Design
A PB design along with three central points was used to assess the effects of the culture conditions [30]. The eight physical factors examined were initial dye concentration (10–100 mg/l), inoculum % (0.5 to 3%v/v), incubation time (8–72 hours.), pH (4–9), temperature (25°C–50°C), and agitation speed (0 to 200 rpm). The chemical parameters that were analyzed included extra carbon sources, such as glucose, lactose, sucrose, and fructose, as well as extra nitrogen sources, such as ammonium chloride, ammonium sulfate, peptone, potassium nitrate, and urea, from 0.0% to 3.0% (w/v) concentration. The best results were obtained from lactose and urea (data not included), so they were used throughout the entire study. Based on the findings of experiments where one variable was changed at a time (COVT), all independent factors were set at their fixed values [27]. Based on PB analysis, four key factors were selected for the central composite design (CCD).
2.9. Statistical Analysis Using RSM
Design of experiments (DOE): A CCD was used in this study, which is a standard RSM design. RSM facilitates the measurement of the correlations between critical input factors and one or more measurable responses. The three main steps in this optimization process include conducting statistically planned trials, assessing the model’s suitability, and figuring out the coefficients in a response-predictive mathematical model [31].
Y = (X1, X2 . . . Xn)
The initial dye concentration (A), initial pH (E), temperature (D), and extra nitrogen source (urea) (H) are the four important factors employed to raise the percentage decolorization. Table 1 illustrates how the coded numbers +1, 0, and −1 correspond to the upper and lower levels of the variables. Based on practical results, a range of input parameters is selected. This suggests that the total number of tests needed for each of the four parameters is N = 2n + 2n + nC = 24 + 2 × 4+6 = 30
Usually, the CCD comprises nC central run and two (2n) axial runs with two (2n) factorial runs.
3. RESULTS AND DISCUSSION
3.1. Characterization of Isolated Strains
In the present study, a total number of 11 morphologically different isolates (J1 to J11) were screened for MO removal efficiency (Fig. 5). Among these isolates, only two (J4 and J10) were able to decolorize MO within 48 hours. To determine the best isolate for further studies, additional screening and biochemical tests were conducted. Both J4 and J10 showed significant decolorization within 48 hours. However, J4 demonstrated a higher removal efficiency, achieving 98% decolorization compared to J10. Furthermore, J4 exhibited a faster growth rate under laboratory conditions, which is crucial for large-scale applications as it allows for quicker processing times and potentially lower costs. Also, J4 was more robust and easier to maintain in culture, showing consistent performance across multiple tests and conditions. This reliability is essential for reproducibility and scalability in practical applications. Based on these criteria, J4 was selected as the best isolate for further studies due to its superior performance in MO removal, faster growth rate, and ease of maintenance. This comprehensive selection process ensured that J4 was not only effective in the initial decolorization screening but also reliable and efficient for potential practical applications in bioremediation
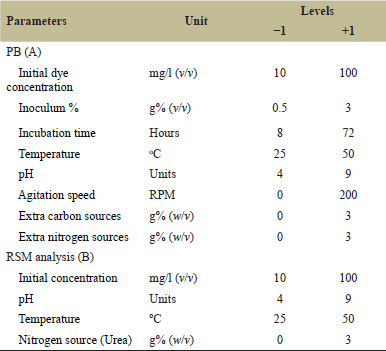 | Table 1. Real and coded values of parameters utilized for PB (A) and RSM analysis (B). [Click here to view] |
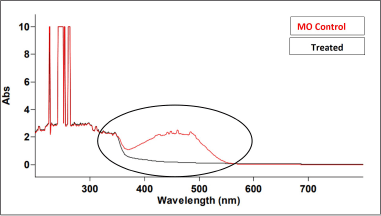 | Figure 1. UV spectra of MO before and after degradation by B. cereus J4. [Click here to view] |
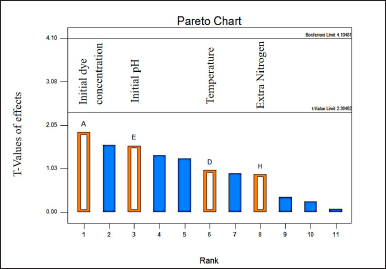 | Figure 2. Pareto Chart depicting the results of PB analysis, highlighting the significant contributions of initial dye concentration, initial pH, temperature, and nitrogen source to the decolorization efficiency of MO dye. [Click here to view] |
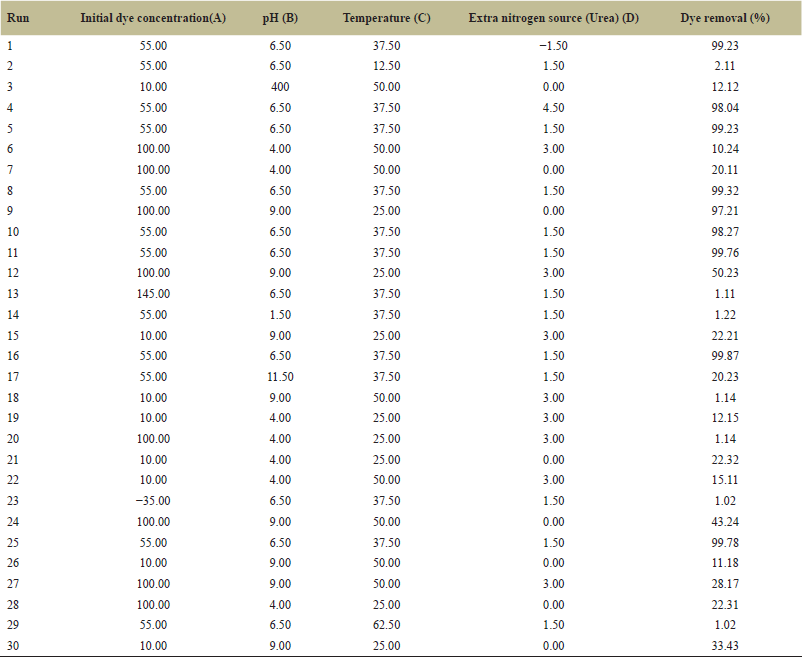 | Table 2. Experimental runs for decolorization of dye MO using CCD. [Click here to view] |
3.2. Morphological and Biochemical Identification
The isolated bacterial strain J4 exhibited distinct morphological and biochemical characteristics, forming dull creamy colonies on NA plates. Microscopic examination revealed that J4 is a small, rod-shaped, non-motile, gram-positive bacterium. Biochemical tests showed that J4 tested positive for the methyl red test and oxidase activity, while it tested negative for the Voges-Proskauer test, citrate utilization, urea hydrolysis, indole production, and catalase activity. To ensure the accurate identification, 16S rRNA gene sequencing and a phylogenetic tree revealed 100% similarity with Bacillus cereus, thus confirming that strain J4 is Bacillus cereus, with an accession number of OQ392442 Figure 6 [32]. Bacillus cereus J4 demonstrated a remarkable ability to fully decolorize MO dye within 48 hours of incubation. UV-visible spectroscopy was employed to monitor the decolorization process, wherein the initial UV-visible spectrum of MO displayed a characteristic absorption peak at around 464 nm, corresponding to its azo bond chromophore. During the incubation with Bacillus cereus J4, a significant decrease in the intensity of this peak was observed, indicating the breakdown of the azo bond. Additionally, the appearance of new peaks in the UV-visible spectrum suggested the formation of intermediate degradation products. The disappearance of the original peak and the emergence of new peaks confirmed that the dye was chemically degraded rather than simply being adsorbed onto the bacterial cells (Fig. 1) [26,33]. After a 12-hour incubation period with the dye, methanol extracts of the cell pellets showed no absorbed dye, reinforcing that dye decolorization was due to the breakdown of dye molecules rather than surface adsorption. This comprehensive characterization, using both traditional biochemical tests and modern molecular techniques, underscores the potential of Bacillus cereus J4 for bioremediation applications involving azo dye degradation [34].
3.3. Statistical Optimization of Physicochemical Parameters Affects the Dye-Removal Process
Factors affecting the removal of MO were screened using the (PB) method. The major impacts in PB design are often significantly confused by two-factor interactions. The Pareto chart (Fig. 2) screening results of PB analysis with physical and chemical factors indicated that the initial concentration (A), initial pH (E), temperature (D), and nitrogen source (H) contributed the most to the maximum decolorization of dye MO. A significant p-value (<0.05) was also observed in removing the dye. The overall model for the PB design was significant, A number higher than four indicates that the signal is satisfactory for the system.
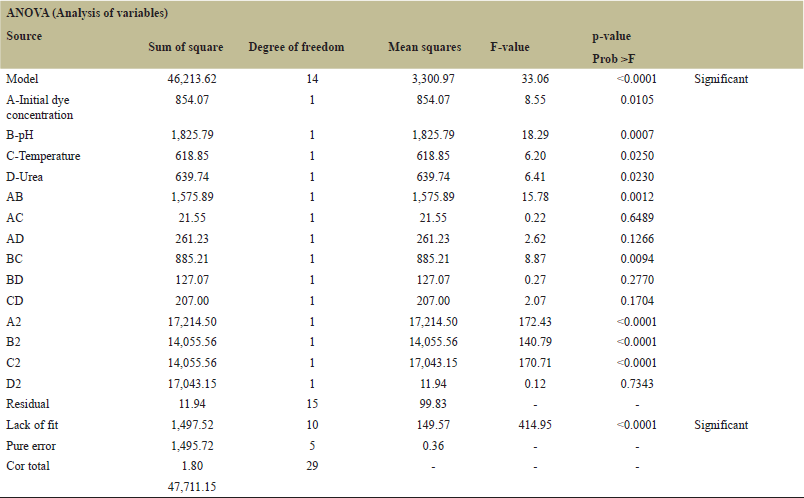 | Table 3. ANOVA table. [Click here to view] |
3.4. Statistical Optimization Using RSM
The impact of the four significant variables on the removal of MO was predicted using a CCD. Using the MO decolorization as a response, RSM was carried out using DOE software. However, the initial dye concentration initial pH, temperature, and additional nitrogen source values had significant positive p values in the PB design, but were disregarded during the CCD design process due to the increased cost and power consumption of controlling these factors on an industrial scale for wastewater treatment. Table 2 shows the list of experiments generated by the software, along with the corresponding figures for dye removal percentage for each experiment involving the decolorization of MO. To fit the responses, a regression evaluation has been completed. The data were fitted with a quadratic model (p < 0.001), as recommended by the software. Additionally, a square root conversion was suggested, resulting in R2 = 0.9686, R2 Adj = 0.9393, R2 Pre = 0.8194, and an adequate precision of 16.96. To evaluate the model’s suitability, coefficients (R2) and adjusted R2 (R2 Adj) are usually computed. Larger R2 values indicate the proportion of response variations in the model [35]. Therefore, our R2 (0.9894) and R2 Adj (0.9393) values in this instance are satisfactory.
Based on ANOVA (Table 3), the combined impact of pH and initial dye concentration was determined to be the most significant (p < 0.0012) in the decolorization of MO. The combined relationship between temperature and pH was also important (p < 0.0094). The significance value of the nitrogen source and the initial dye concentration together is comparatively low when compared to other conjugates, but the overall model is significant (p < 0.0001). Every single factor has been determined to be significant (p < 0.0001), with the initial pH being the most influential (p < 0.0007) in the decolorization process (f value = 18.29). The three-dimensional responses and the contour diagrams of the previously discussed conjugates are displayed in Figures 3 and 4 illustrate the predicted versus actual values plot.
The pH and initial dye concentration effects are depicted, along with their correlation, in Figure 3A. At the lowest values (10 mg/l and 4) for initial dye concentration and pH, the dye removal percentage was nearly 39%. However, there was no noticeable difference in the dye removal percentage of 40.3% when the initial dye concentration was raised while maintaining a constant pH. At a certain pH, the initial dye concentration rises to a specific point. However, because pH is essential to bacterial physiology, MO elimination reached approximately 85.5% when pH increased towards an alkaline value. This could be due to the fact that MO decolorization increased when the pH was neutral, which is the ideal environment for bacterial survival and function.
Under constant initial dye concentration, urea was observed to partially improve the removal of MO in Figure 3B. The removal of MO increased by almost 10% from the previous value (71%) when the nitrogen concentration increased from 0.5% to 3%. This suggests that an additional nitrogen source may enhance the removal process by enhancing the activities of the bacteria, promoting optimal physiology and survival at a neutral pH, and ultimately speeding up the decolorization of MO.
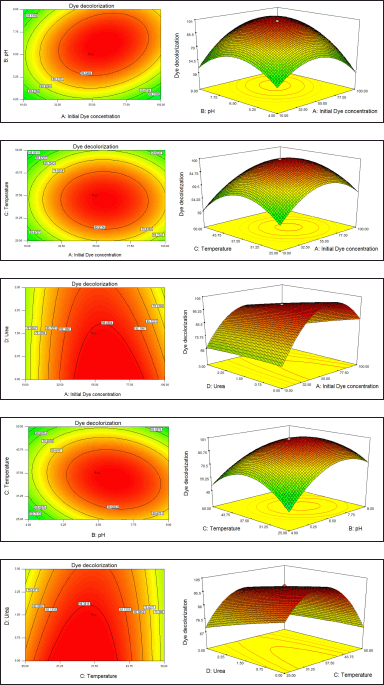 | Figure 3. (a) Contour plots correlated to response surface (3D) plots for the interaction of pH versus initial dye for the de-colorization of MO using B. cereus J4. (b) Contour plots correlated to response surface (3D) plots for the interaction of temperature versus initial dye for the de-colorization of MO using B. cereus J4. (c) Contour plots correlated to response surface (3D) plots for the interaction of urea versus Initial dye for the de-colorization of MO using B. cereus J4. (d) Contour plots correlated to response surface (3D) plots for the interaction of temperature versus pH for the de-colorization of MO using B. cereus J4. (e) Contour plots correlated to response surface (3D) plots for the interaction of urea versus temperature for decolorizing MO using B. cereus J4. [Click here to view] |
Temperature was found to be a significant factor in relation to the initial dye concentration in Figure 3C. Initially, there was very low (less than 39%) removal of MO at a temperature of 25°C–30°C with an initial dye concentration of 10 mg/l. When the initial dye concentration was 55.0 mg/l and the temperature was 37.5°C, the rate of dye removal increased to 89.5%. Since bacteria could not grow at either the lowest or highest temperatures, this could be the result of their inability to develop at lower temperatures.
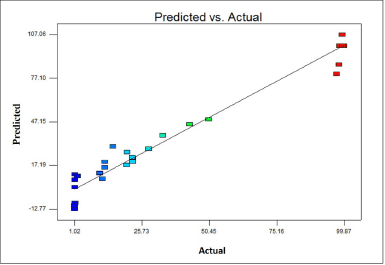 | Figure 4. Illustrates the comparison between predicted and actual values for the variables under study. [Click here to view] |
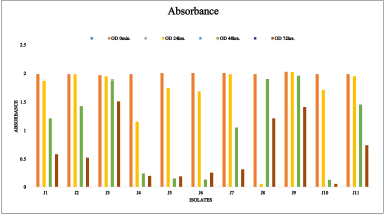 | Figure 5. Screening for MO removal efficiency of different isolates (J1 to J11). [Click here to view] |
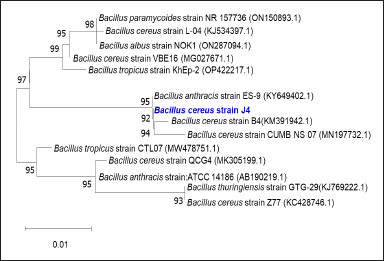 | Figure 6. Phylogenetic tree of isolated bacterium (J4) capable of decolorizing MO dye. [Click here to view] |
Figure 3D shows the relation between pH and temperature. Meanwhile, the effect of the interaction of pH and temperature had no significant difference on the dye removal efficiency of Bacillus cereus J4 on MO dye. However, the effect of temperature in the interaction with pH at 37°C enhanced the dye removal of the MO dye.
Figure 3E shows the relation between urea and temperature. A significant effect of extra nitrogen source (urea) at 37°C Temperature on dye decolorization of MO. This shows that extra nitrogen sources (urea) are acting as removal enhancers by bacterial activities.
The whole study revealed that initial dye concentration, pH, temperature, and extra nitrogen source (urea) play a key role in the MO removal process. The initial aim of this study was to remove as much MO dye as possible from water using Bacillus cereus J4 in an affordable system. Considering these factors, the following criteria have been chosen to optimize dye removal conditions: initial dye concentration within a specific range.
Initial pH, temperature, and extra nitrogen: all within range; ANOVA table showed that the optimal model was generated at 50 mg/l initial dye concentration, pH 7, 37°C, and 2% extra nitrogen (urea). This model demonstrates an 88.93% removal of MO from an aqueous solution with a desirability of 1. The practical efficacy of the model was confirmed by the 93.05% dye removal obtained in the validation experiment.
In this study, eleven bacterial strains were isolated from the Textile Industrial area in Panipat, Haryana, India. Bacillus cereus J4 can easily remove MO from aqueous solution up to 89.8%. We observed that several physical and chemical parameters significantly influence the rate of MO decolorization. Initial dye concentration, temperature, extra nitrogen sources (urea), and pH are the key variables for MO decolorization, according to statistical analysis for physicochemical factors. Therefore, Bacillus cereus J4 may be a useful agent for the future safe release of MO from industrial effluents into the environment. Similarly, a study aimed to isolate and characterize bacterial strains capable of decolorizing and degrading azo dyes used in textile production from activated sludge systems treating textile wastewater. Two bacterial strains, Acinetobacter (ST16.16/164) and Klebsiella (ST16.16/034), demonstrated strong decolorization and degradation abilities across a range of temperatures and pH levels, showing potential for use in treating industrial wastewaters containing azo dyes [24].
4. CONCLUSION
This study successfully isolated and characterized B. cereus J4, which exhibited exceptional efficiency in decolorizing MO dye, achieving up to 98% removal within 48 hours under optimized conditions. The morphological, biochemical, and molecular identification confirmed its classification as Bacillus cereus, providing a solid foundation for its application in industrial wastewater treatment. Statistical optimization using PB and RSM highlighted key factors influencing dye removal, including initial dye concentration, pH, temperature, and nitrogen source (urea). The findings underscore the importance of these parameters in enhancing Bacillus cereus J4’s decolorization capabilities. Future studies should focus on scaling up this process for practical applications, exploring its efficacy in real-world textile effluent scenarios, and further optimizing operational conditions to maximize efficiency and sustainability in dye removal processes.
5. ACKNOWLEDGMENT
The authors would like to thank to Department of Microbiology, School of Bioengineering and Biosciences, Lovely Professional University Phagwara, Punjab, India. Additionally, we are grateful to Haryana Test House and Consultancy Services, Sec. 29 Part II Huda Panipat, for supplying the research facilities.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Ibrahim A, El-Fakharany EM, Abu-Serie MM, ElKady MF, Eltarahony M. Methyl orange biodegradation by immobilized consortium microspheres: experimental design approach, toxicity study and bioaugmentation potential. Biology 2022;11(1):76.
2. Haque MM, Hossen MN, Rahman A, Roy J, Talukder MR, Ahmed M, et al. Decolorization, degradation and detoxification of mutagenic dye methyl orange by novel biofilm producing plant growth-promoting rhizobacteria. Chemosphere 2024;346:140568; doi: 10.1016/j.chemosphere.2023.140568
3. Kishor R, Purchase D, Saratale GD, Saratale RG, Ferreira LFR, Bilal M, et al. Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. J Environ Chem Eng 2021;9(2):105012.
4. Irki S, Ghernaout D, Naceur MW, Alghamdi A, Aichouni M. Decolorizing methyl orange by Fe-electrocoagulation process—a mechanistic insight. Int J Environ Chem 2018;2:18–28.
5. Akter S, Islam MS. Effect of additional Fe2+ salt on electrocoagulation process for the degradation of methyl orange dye: an optimization and kinetic study. Heliyon 2022;8(8):e10176 [Online]. Available via https://www.cell.com/heliyon/fulltext/S2405-8440(22)01464-5 (Accessed 16 July 2024)
6. Akter S. Optimization and kinetic study of methyl orange dye degradation using catalytic electrocoagulation. 2021 [Online]. Available via http://lib.buet.ac.bd:8080/xmlui/handle/123456789/5966 (Accessed 16 July 2024)
7. Ahmad A, Mohd-Setapar SH, Chuong CS, Khatoon A, Wani WA, Kumar R, et al. Recent advances in new generation dye removal technologies: novel search for approaches to reprocess wastewater. RSC Adv 2015;5(39):30801–18.
8. Katheresan V, Kansedo J, Lau SY. Efficiency of various recent wastewater dye removal methods: a review. J Environ Chem Eng 2018;6(4):4676–97.
9. Roy M, Saha R. Dyes and their removal technologies from wastewater: a critical review. Intell Environ Data Monit Pollut Manag 2021:127–60.
10. Cao Y, Chen X, Feng S, Wan Y, Luo J. Nanofiltration for decolorization: membrane fabrication, applications and challenges. Ind Eng Chem Res 2020;59(45):19858–75; doi: 10.1021/acs.iecr.0c04277
11. Gilmour CR. Water treatment using advanced oxidation processes: application perspectives. The University of Western, Ontario, Canada, 2012 (Accessed 16 July 2024).
12. Tang KHD, Darwish NM, Alkahtani AM, AbdelGawwad MR, Karácsony P. Biological removal of dyes from wastewater: a review of its efficiency and advances. Trop Aquat Soil Pollut 2022;2(1):59–75.
13. Alsukaibi AK Various approaches for the detoxification of toxic dyes in wastewater. Processes 2022;10(10):1968.
14. Pinheiro LRS, Gradíssimo DG, Xavier LP, Santos AV. Degradation of azo dyes: bacterial potential for bioremediation. Sustainability 2022;14(3):1510.
15. Yadav A, Kumar P, Rawat D, Garg S, Mukherjee P, Farooqi F, et al. Microbial fuel cells for mineralization and decolorization of azo dyes: recent advances in design and materials. Sci Total Environ 2022;826:154038.
16. Kapoor RT, Danish M, Singh RS, Rafatullah M, Abdul Khalil HPS. Exploiting microbial biomass in treating azo dyes contaminated wastewater: mechanism of degradation and factors affecting microbial efficiency. J Water Process Eng 2021;43:102255.
17. Wang T, Tang X, Zhang S, Zheng J, Zheng H, Fang L. Roles of functional microbial flocculant in dyeing wastewater treatment: bridging and adsorption. J Hazard Mater 2020;384:121506.
18. Gopi T. Decolorization of azo dyes with Klebsiella pneumoniae under high saline conditions. Doctoral thesis, Nanyang Technological University, Singapore, 2021 [Online]. Available via https://dr.ntu.edu.sg/handle/10356/153445 (Accessed 16 July 2024)
19. Maniyam MN, Yaacob NS, Azman HH, Ab Ghaffar NA, Abdullah H. Immobilized cells of Rhodococcus strain UCC 0004 as source of green biocatalyst for decolourization and biodegradation of methyl orange. Biocatal Agric Biotechnol 2018;16:569–78; doi: 10.1016/j.bcab.2018.10.008
20. Sundarajoo A, Maniyam MN, Azman HH, Abdullah H, Yaacob NS. Rhodococcus strain UCC 0010 as green biocatalyst for enhanced biodecolourization of Congo red through response surface methodology. Int J Environ Sci Technol 2022;19(4):3305–22; doi: 10.1007/s13762-021-03400-4
21. Sutherland DL, McCauley J, Labeeuw L, Ray P, Kuzhiumparambil U, Hall C, et al. How microalgal biotechnology can assist with the UN Sustainable Development Goals for natural resource management. Curr Res Environ Sustain 2021;3:100050.
22. Donato K, Medori MC, Stuppia L, Beccari T, Dundar M, Marks RS, et al. Unleashing the potential of biotechnology for sustainable development. Eur Rev Med Pharmacol Sci 2023;27(6 Suppl):100–13.
23. Prasad MNV. Bioremediation, bioeconomy, circular economy, and circular bioeconomy—Strategies for sustainability. In: Bioremediation and bioeconomy, Elsevier, Amsterdam, The Netherlands, pp 3–32, 2024 [Online]. Available via https://www.sciencedirect.com/science/article/pii/B978044316120900025X (Accessed 17 July 2024)
24. Meerbergen K, Willems KA, Dewil R, Van Impe J, Appels L, Lievens B. Isolation and screening of bacterial isolates from wastewater treatment plants to decolorize azo dyes. J Biosci Bioeng 2018;125(4):448–56.
25. Pearce CI, Lloyd JR, Guthrie JT. The removal of colour from textile wastewater using whole bacterial cells: a review. Dyes Pigm 2003;58(3):179–96.
26. Kishor R, Purchase D, Saratale GD, Ferreira LFR, Hussain CM, Mulla SI, et al. Degradation mechanism and toxicity reduction of methyl orange dye by a newly isolated bacterium Pseudomonas aeruginosa MZ520730. J Water Process Eng 2021;43:102300.
27. Saha N, Samanta AK, Chaudhuri S, Dutta D. Characterization and antioxidant potential of a carotenoid from a newly isolated yeast. Food Sci Biotechnol 2015;24(1):117–24; doi: 10.1007/s10068-015-0017-z
28. Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika 1946;33(4):305–25.
29. Ghanem KM, Al-Fassi FA, Biag AK. Optimization of methyl orange decolorization by mono and mixed bacterial culture techniques using statistical designs. Afr J Microbiol Res 2012;6(12):2918–28.
30. Haaland PD. Experimental design in biotechnology. CRC Press, Boca Raton, FL, 2020 (Accessed 17 July 2024).
31. Dutta S, Bhattacharyya A, Ganguly A, Gupta S, Basu S. Application of response surface methodology for preparation of low-cost adsorbent from citrus fruit peel and for removal of methylene blue. Desalination 2011;275(1–3):26–36.
32. Premkrishnan BNV, Heinle CE, Uchida A, Purbojati RW, Kushwaha KK, Putra A, et al. The genomic characterisation and comparison of Bacillus cereus strains isolated from indoor air. Gut Pathog 2021;13(1):6; doi: 10.1186/s13099-021-00399-4
33. Shah MP, Patel KA, Nair SS, Darji AM. Microbial decolorization of methyl orange dye by Pseudomonas spp. ETL-M. Int J Environ Bioremediation Biodegrad 2013;1(2):54–9.
34. Ogugbue CJ, Sawidis T. Bioremediation and detoxification of synthetic wastewater containing triarylmethane dyes by Aeromonas hydrophila isolated from industrial effluent. Biotechnol Res Int 2011;2011:1–11; doi: 10.4061/2011/967925
35. Chatterjee S, Chatterjee T, Lim SR, Woo SH. Effect of the addition mode of carbon nanotubes for the production of chitosan hydrogel core–shell beads on adsorption of Congo red from aqueous solution. Bioresour Technol 2011;102(6):4402–9.