In recent decades, the rapid development of manufacturing industries has accelerated the rate of water pollution. One of the severe pollution sources in Vietnam is wastewater from traditional craft villages, especially those engaged in food production. According to statistics from the Ministry of Agriculture and Rural Development (2019), there are approximately 4,575 craft villages nationwide, of which 13.59% are involved in food production. This process requires a large amount of water, most of which is then directly discharged into the environment. Without an appropriate treatment, wastewater from craft villages can even accumulate in drainage systems and undergo anaerobic decomposition, causing air pollution. Furthermore, it also penetrates the soil, polluting the land and degrading groundwater quality. The pollution source of vermicelli production wastewater is primarily derived from the discharge of starch and easily biodegradable organic matter during the soaking stage after rice immersion. Parameters such as chemical oxygen demand (COD), biochemical oxygen demand (BOD), total nitrogen content, total phosphorus content, etc., exceed the permissible standards. In addition, vermicelli production wastewater also causes a very unpleasant odor.
In recent years, various physical, chemical, and biological methods such as precipitation, oxidation, membrane filtration, electrochemical treatment, and particularly biodegradation have been widely applied in wastewater treatment [1,2]. Generally, the combination of all three methods yields the most cost-effective water quality. Biological wastewater treatment is an efficient and economical method that does not cause secondary pollution to the environment. Biological wastewater treatment methods based on the use of microorganisms to degrade organic and some inorganic pollutants in water generate non-toxic and eco-friendly products. According to Crini and Lichtfouse [2], many microorganisms are able to produce a wide range of extracellular enzymes with high-level biological degradation, absorption, or accumulation of non-degradable organic compounds such as NH4+, NH3, and phosphate.
In vermicelli production wastewater, starch in the form of rice flour is the primary raw material that contributes the most to pollution. On the other hand, the high levels of residual phosphorus in wastewater, especially orthophosphate, are the main factor causing eutrophication in the aquatic environment. Therefore, the biological phosphorus removal is a treatment technique that utilizes microorganisms to convert the soluble phosphate accumulated in wastewater into an insoluble form of intracellular polyphosphate (Poly-P). In this study, strain JN459 accumulated phosphorus aerobically and released phosphorus anaerobically, and Poly-P granules increased in the cells during the aerobic period and decreased during the anaerobic period [3].
Therefore, isolation and selection of bacterial strains that produce enzymes to degrade starch and absorb phosphate in the form of Poly-P granules represent a promising research direction and provide a foundation for diversifying the pool of bacteria capable of wastewater treatment in the future.
2. MATERIALS AND METHODS
2.1. Isolation of Bacterial Strains from Wastewater of the Vermicelli Production Process
2.1.1. Sample collection
Wastewater samples were collected at the discharge points of two vermicelli production factories in Long Tuyen Ward (10°01’15.4”N 105°42’56.5”E) and Long Hoa Ward (10°02’12.4”N 105°44’31.5”E), Binh Thuy District, Can Tho City, Vietnam. Samples collected from Long Hoa Ward were designated with the letter “Y,” while those from Long Tuyen Ward were labeled with the letter “P.” Upon collection, the wastewater samples were stored at 4°C and immediately subjected to isolation procedures.
Samples were collected from three locations within the wastewater area, with 100 mL taken from each site and then homogenized in a sample container. The samples were preserved at 4°C in a specialized storage box and transported to the laboratory. The sample collection for microbial isolation was kept within 24 h.
2.1.2. Bacterial isolation
Ten mL of the wastewater sample was transferred into 90 mL of distilled water in a sterilized triangular flask. Subsequently, the solution was diluted 1,000 times with sterilized distilled water. 50 μL of the diluted solution was evenly spread onto the surface of the nutrient agar (NA) plates. After 24 h, colonies with different morphological characteristics were transferred to NA medium [4]. The subculturing process was repeated until pure bacterial colonies were obtained. The samples were then stored in test tubes at 4°C for further experiments.
2.2. Selection of Bacterial Strains with the Ability to Degrade Starch
2.2.1. Investigation of the starch degradability using the agar well diffusion method
The bacterial strains were cultured in a nutrient broth (NB) medium with shaking at 150 rpm at room temperature until a bacterial density of 106 CFU/mL measured at 600 nm of wavelength (0.5 McFarland standard) was obtained. After incubation for 2 days, the bacterial suspensions were centrifuged at 13,000 rpm for 15 min at 4°C to remove cells. Five wells (diameter of 6 mm) were created on a starch agar (SA) medium using the punching method [5]. Subsequently, 100 μL of the supernatant from each bacterial strain was pipetted into three wells on the plate, while the remaining two wells were added with 100 μL of distilled water and 100 μL of NB enrich media as negative controls, respectively. To visualize the degradation zones, the agar plates were stained with iodine. The starch degrading ability was indicated by the difference between the diameter of the halo zone (mm) and the well recorded after 48 h.
2.2.2. Determination of reducing sugar content using the acid dinitrosalicylic (DNS) method
Standard glucose concentrations used in this experiment ranged from 0 mg/mL to 1.2 mg/mL. All tubes were prepared with the appropriate concentration, with 2 mL of DNS reagent added to 1 mL of standard glucose solution and thoroughly vortexed. The mixture was then heated in a water bath for 5 min and allowed to cool. Absorbance was measured at 540 nm wavelength.
For bacterial samples: 1 mL of supernatant from the selected bacterial strains (at a concentration of 107 CFU/mL) was mixed with 9 mL of liquid SA medium. The mixture was shaken at 150 rpm at room temperature. The same procedure was performed for the negative control. After 2, 4, 6, and 8 days of incubation, samples were collected to determine the concentration of reducing sugar produced at each time point. Samples were centrifuged at 5,000 rpm for 15 min, and 1 mL of the supernatant was transferred to a new tube with the addition of 2 mL of DNS reagent. The mixture was boiled for 5 min and allowed to cool for 5 min at room temperature. Absorbance was measured at 540 nm wavelength using a spectrometer (GENESYS 50, Thermo Fisher Scientific), and the concentration of reducing sugar produced in the sample was determined using the standard curve equation.
Standard curve: y = 0.5304× −0.0097 with (R2 = 0.976).
Where y: The OD540 nm value; ×: The concentration of reducing sugar produced (mg/mL).
2.2.3. Selection of bacterial strains with the ability to accumulate Poly-P
After isolation, bacterial strains were cultured on phosphate-removing agar medium [6], comprising 20 g/L glucose, 37.5 mg/L KH2PO4 (equivalent to 39.14 mg/L P2O5), 0.02 g/L peptone, 0.01 g/L MgSO4.7H2O, 0.02 g/L NH4Cl, 0.005 g/L CaCl2, and 0.5 mL to initially assess their ability to accumulate Poly-P. Subsequently, 1 mL of bacterial suspension after culture (at a density of 106 CFU/mL) was transferred into 5 mL of liquid Poly-P-accumulating medium and cultured with shaking at 150 rpm for 6 days at room temperature. The Poly-P content accumulated was determined according to the methods previously described by Wang et al. [6] and Eixler et al. [7].
The standard curve for P2O5 was y = 0.0177× −0.0051 with an R2 value of 0.999.
Where y is the OD880 nm value; × is the P2O5 concentration (mg/L). A standard P2O5 solution was used from 0.5 mL to 2.5 mL.
Poly-P extraction: After 6 days of cultivation, centrifugation was performed at 12,000 rpm for 10 min to collect the biomass. The biomass was then dissolved in 4 mL of 0.2 M NaOH and incubated for 20 h to extract Poly-P. The mixture was filtered through filter paper to remove cell debris, resulting in a filtrate containing Poly-P. The filtrate was divided into 2 parts:
Part 1: Measurement was conducted to determine the amount of free phosphate (PP1) in the solution using the molybdate blue method (colorimetric detection at 880 nm wavelength).
Part 2: Hydrolysis was carried out using 1 M HCl at 100°C for 10 min. The hydrolyzed solution was then cooled, and the phosphate content (PP2) was measured using the same method as in Part 1.
The concentration of intracellular Poly-P (mg/L) was determined by subtracting PP1 from PP2.
2.2.4. Identification of bacterial strains with potential for treating wastewater from the vermicelli production process
The selected bacterial strain with potential for wastewater treatment was identified using the polymerase chain reaction (PCR) amplification method combined with sequencing of the 16S rRNA gene.
The PCR reaction included DNA template, Taq polymerase enzyme, primer sequences 1492R (5’–TACGGTTACCTTGTTACGACT–3’) and 27F (5’AGAGTTTGATCCTGGCTC–3’), dNTPs, and MgCl2. The thermal cycling conditions in a PCR reaction involved the denaturation of the target DNA strand and primers at 95°C for 5 min, followed by heating to approximately 98°C for 1 min, annealing at 55°C for about 30 s, and finally, extension at approximately 72°C for 5 min.
Subsequently, the sequencing of the 16S rRNA gene segment was performed. After obtaining the sequencing results, an analysis was conducted using the basic local alignment search tool (BLAST) on the National Center for Biotechnology Information (NCBI) system to compare them with similar published sequences. Sequences were collected from the BLAST results of two bacterial strains in the study, compared with the 16S rRNA gene database of bacteria and archaea from the NCBI database. Multiple-sequence alignment was performed using the Clustal W algorithm in BioEdit version 7.2.1. The MEGA X software [8] was used to construct the phylogenetic tree based on the maximum likelihood method, applying the general time reversible (GTR) model (GTR + G + I) for strain Y9 and the Kimura 2-parameter model [9] for strain Y11. The reliability of the phylogenetic tree was tested by the bootstrap method with 1,000 replicates. Wautersiella enshiensis and Metabacillus herbersteinensis were used as outgroup species.
2.2.5. Data analysis
The experiments were conducted in triplicate. The results are presented as mean values ± standard deviation. The experimental data were analyzed using Minitab 16 software (Minitab Inc., State College, PA, USA) to assess the differences between treatments with a significance level of 5%.
3. RESULTS AND DISCUSSION
3.1. Isolation of Bacterial Strains from Wastewater from the Vermicelli Production Process
A total of 32 bacterial strains were isolated from 2 wastewater samples obtained from 2 vermicelli production factories in Can Tho City, Vietnam. The isolated bacterial strains exhibited yellow, creamy white, or opaque white colors; convex or raised elevation; colony morphologies of entire or lobate margins; with bacterial colony sizes ranging from 0.5 to 3.5 mm. All bacterial strains tested positive for the catalase test [Table 1 and Figure 1].
Table 1: Morphological characteristics of bacterial colonies, cells, and biochemical characteristics of isolated bacterial strains.
| No. | Bacterial strain | Morphological characteristics | Biochemical testing | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Colors | Morphology | Buoyancy | Margin | Colony size (mm) | Gram | Oxidase/catalase | |||
| Colony | Cell | ||||||||
| 1 | Y1 | Creamy white | Round | Coccus | Convex | Entire | 1.5–2.0 | + | −/+ |
| 2 | Y2 | Opaque white | Round | Rod-shaped | Convex | Entire | 2.0–2.5 | − | +/+ |
| 3 | Y3 | Opaque white | Round | Coccus | Raised | Lobate | 0.5–1.1 | − | +/+ |
| 4 | Y4 | Opaque white | Round | Rod-shaped | Convex | Lobate | 0.5–1.0 | − | −/+ |
| 5 | Y5 | Opaque white | Round | Coccus | Convex | Entire | 0.5–1.0 | − | +/+ |
| 6 | Y6 | Opaque white | Round | Coccus | Convex | Entire | 0.5–1.0 | − | +/+ |
| 7 | Y7 | Opaque white | Round | Coccus | Convex | Entire | 0.7–1.2 | − | +/+ |
| 8 | Y8 | Opaque white | Round | Coccus | Raised | Lobate | 1.0–1.5 | + | −/+ |
| 9 | Y9 | Yellow | Round | Rod-shaped | Raised | Entire | 0.5–1.0 | − | +/+ |
| 10 | Y10 | Opaque white | Round | Rod-shaped | Convex | Entire | 1.5–2.0 | + | +/+ |
| 11 | Y11 | Opaque white | Round | Rod-shaped | Raised | Erose | 3.0–3.5 | + | +/+ |
| 12 | Y12 | Opaque white | Round | Coccus | Convex | Lobate | 0.5–1.0 | − | −/+ |
| 13 | Y13 | Opaque white | Round | Coccus | Convex | Entire | 1.0–1.5 | − | +/+ |
| 14 | Y14 | Opaque white | Round | Rod-shaped | Raised | Erose | 0.5–1.0 | + | +/+ |
| 15 | Y15 | Opaque white | Round | Coccus | Raised | Lobate | 0.5–1.0 | + | +/+ |
| 16 | Y16 | Opaque white | Round | Coccus | Raised | Lobate | 1.5–2.0 | − | +/+ |
| 17 | Y17 | Opaque white | Round | Coccus | Convex | Entire | 1.5–2.0 | − | +/+ |
| 18 | P1 | Yellow | Round | Rod-shaped | Convex | Entire | 1.0–1.5 | + | +/+ |
| 19 | P2 | Yellow | Round | Coccus | Convex | Entire | 1.2–1.5 | − | +/+ |
| 20 | P3 | Opaque white | Round | Coccus | Convex | Entire | 0.5–1.0 | + | −/+ |
| 21 | P4 | Creamy white | Round | Rod-shaped | Convex | Entire | 1.0–1.5 | + | +/+ |
| 22 | P5 | Creamy white | Round | Rod-shaped | Convex | Entire | 0.5–1.0 | − | +/+ |
| 23 | P6 | Opaque white | Round | Coccus | Convex | Entire | 0.5–1.0 | − | +/+ |
| 24 | P7 | Yellow | Round | Coccus | Convex | Entire | 0.7–1.2 | − | +/+ |
| 25 | P8 | Opaque white | Round | Coccus | Convex | Entire | 1.0–1.5 | − | +/+ |
| 26 | P9 | Creamy white | Round | Coccus | Convex | Entire | 1.0–1.5 | - | +/+ |
| 27 | P10 | Yellow | Round | Coccus | Convex | Entire | 0.5–1.0 | + | +/+ |
| 28 | P11 | Yellow | Round | Coccus | Convex | Entire | 0.5–1.0 | − | +/+ |
| 29 | P12 | Yellow | Round | Coccus | Raised | Entire | 0.5–1.1 | − | +/+ |
| 30 | P13 | Creamy white | Round | Rod-shaped | Convex | Entire | 0.5–1.0 | − | +/+ |
| 31 | P14 | Opaque white | Round | Coccus | Convex | Entire | 0.5–1.0 | − | +/+ |
| 32 | P15 | Creamy white | Round | Rod-shaped | Convex | Entire | 1.0–1.5 | + | −/+ |
(+): Positive; (−): Negative; letter “Y” indicates samples collected from Long Hoa Ward, letter “P” indicates samples collected from Long Tuyen Ward.
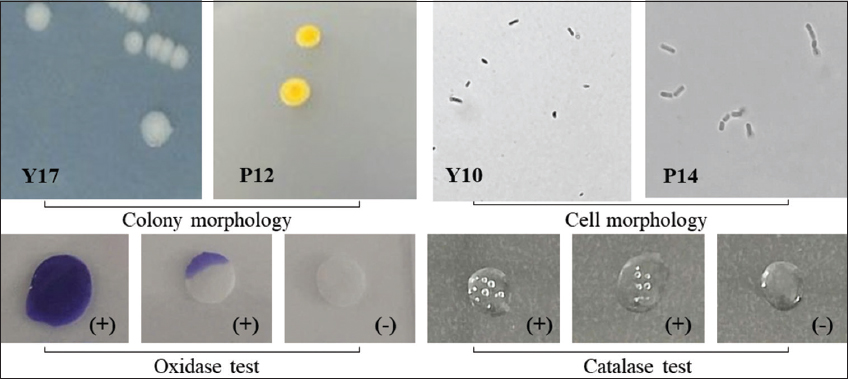 | Figure 1: Morphological and biochemical characteristics of the isolated bacterial strains. [Click here to view] |
3.2. Selection of Bacterial Strains with Starch Degradability
3.2.1. Starch-degrading ability using the agar well diffusion method
The results of the survey on starch-degrading ability indicated that 22 out of 32 bacterial strains were identified to be capable of starch degradation using the agar well diffusion method. The specific diameter of the starch-degrading ability is presented in detail in Table 2 and Figure 2.
Table 2: Diameter of starch-degrading zones (mm) on SA medium of isolated bacterial strains.
| Bacterial strains | Diameter of starch-degrading zones (mm) | Bacterial strains | Diameter of starch- degrading zones (mm) |
|---|---|---|---|
| P1 | 21.33bc±0.58 | P12 | 24.33b±0.58 |
| P2 | 27.33a±0.58 | P13 | 13.67efg±0.58 |
| P3 | 16.33d±0.58 | P14 | 21.67c±0.58 |
| P4 | 12.67g±0.58 | P15 | 11.67gh±0.58 |
| P5 | 12.67g±0.58 | Y1 | 15.00def±1.00 |
| P6 | 9.67hi±0.58 | Y4 | 13.33fg±0.58 |
| P7 | 13.67efg±0.58 | Y7 | 7.67i±1.53 |
| P8 | 16.33d±0.58 | Y8 | 10.33h±0.58 |
| P9 | 16.33d±0.58 | Y9 | 22.33bc±0.58 |
| P10 | 15.67de±0.58 | Y11 | 9.67hi±0.58 |
| P11 | 9.67hi±0.58 | Y12 | 13.33fg±0.58 |
Values are the averages of 3 replications; values within the same column followed by the same letter do not significantly differ at the 5% level by Tukey’s test.
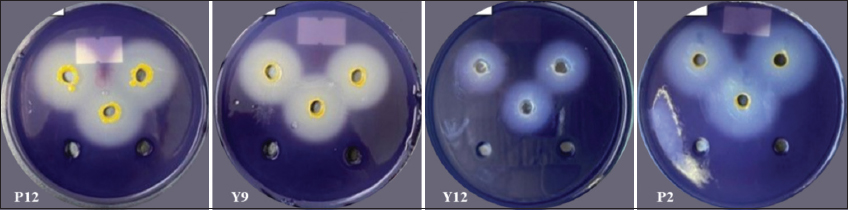 | Figure 2: Starch-degrading ability on the agar plate. [Click here to view] |
The results showed that the majority of isolated bacterial strains could degrade starch after 2 days, as indicated by the halo diameters ranging from 7.67 to 27.33 mm [Table 2 and Figure 2]. The starch degradation ability observed in this study was similar to the findings of Mai et al. [10] who reported the starch degradation capacity of bacterial strains isolated from the gut of Holotrichia parallela, Lumbricus terrestris, and organic waste with halo diameters ranging from 5.7 mm to 23.4 mm when stained with Lugol’s solution. The survey results on the starch degradation ability of bacterial strains isolated from vermicelli wastewater in this study were higher than those isolated from decomposed straw and solid waste in the previous survey by Dang and Tran [11], where the results ranged from 9.66 to 13.81 mm. Vermicelli wastewater is a starch-rich environment that provides favorable conditions for starch-utilizing bacteria to thrive and compete effectively. In contrast, decomposed straw and solid waste contain lower starch levels but are rich in lignocellulose, promoting bacterial adaptation to the degradation of more complex compounds. Consequently, vermicelli wastewater typically has a higher starch-degrading bacteria density than decomposed straw and solid waste.
The bacterial strains P2, P12, and Y9 all exhibited strong starch degradation abilities [Figures 2 and 3] compared to the remaining 19 bacterial strains. Various environmental conditions, such as pH, temperature, and bacterial density, influenced the extracellular enzyme production of bacteria, especially pH. The pH of the bacterial culture medium changed during the bacterial growth process, affecting both enzyme production and enzyme activity. The pH of the environment may vary depending on the carbon source utilization, growth rate, and growth phase of each bacterial strain [12]. Specifically, pH of the environment increases as bacteria utilize nutrients from the environment, hydrolyzing nutrients such as peptone, yeast extract, amino acids, releasing amino groups into the environment, leading to alkaline pH; whereas the pH of the environment tends to decrease due to the production of organic acids during bacterial growth and development, leading to an acidic pH [13]. According to Ramin and Allison [14], increased enzyme production by bacteria will reduce the efficiency of carbon source utilization from nutrients in the environment, leading to a decrease in growth rate and enzyme synthesis. On the other hand, each enzyme has an optimal pH range. When the pH value exceeds this range, it will slow down the activity of the enzyme. A high pH value can denature enzymes, significantly affecting their ability to degrade substrates. At each stage of the bacterial growth cycle, enzymes are synthesized with different concentrations and activities. If enzymes are obtained too early, the concentration and activity of the enzymes will be low. Conversely, if the enzyme collection time is prolonged, the excessive release of enzymes will be affected by external environmental factors, thus reducing the enzyme activity or disrupting the natural structure of enzymes.
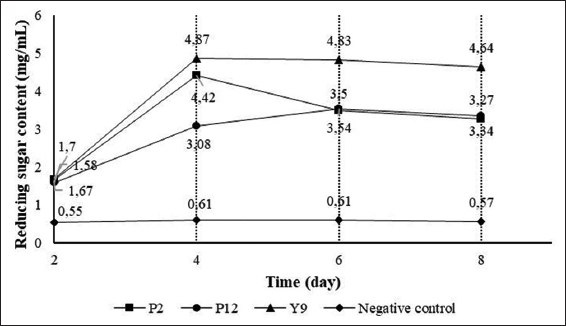 | Figure 3: Levels of reducing sugar (mg/mL) produced by isolated bacterial strains. [Click here to view] |
3.2.2. Determination of reducing sugar content by using the DNS method
The three bacterial strains P2, P12, and Y9 showed a high capability of starch degradation after 2 days of agitated cultivation, so these three bacterial strains were investigated for their reducing sugar production. Overall, the results revealed variation in the reducing sugar content among the samples at the 4 surveyed time points. The reducing sugar content produced by the three bacterial strains tended to increase from day 2 to day 8. Specifically, during the period from day 2 to day 8, the highest and lowest reducing sugar content were produced by the Y9 strain (1.70–4.64 mg/mL) and P12 strain (1.58–3.34 mg/mL) [Figure 3].
However, in the experiment investigating the starch-degrading ability on the plates, the Y9 bacterial strain did not exhibit the highest diameter of starch degradation. This could be explained by the fact that the NB medium did not contain starch substrates, thereby not stimulating bacteria to produce a significant amount of amylase enzymes for starch degradation. Conversely, in the liquid SA medium, which contains abundant nutrients and provides substrates, bacteria are more stimulated to produce enzymes. Hence, the amount of reducing sugar produced by the Y9 bacterial strain reached the highest level and showed statistically significant differences compared to the other bacterial strains.
In addition to the influence of environmental conditions on the ability to hydrolyze starch into sugar, bacteria may have reused the produced reducing sugar as a carbon source, resulting in fluctuations in reducing sugar production. The amount of reducing sugar produced in this study was higher than the findings of Tran et al. [15] regarding the KTXA1 bacterial strain, which had a reducing sugar content of 2.99 mg/mL after 2 days of observation.
3.3. The Results of Poly-P Accumulation Capability
The results showed that 11 out of 32 bacterial strains could accumulate Poly-P, with phosphate content in the form of P2O5 ranging from 0.06 to 1.79 mg/L. Among them, the Y11 and Y10 strains exhibited the highest and the lowest accumulated Poly-P content, at 1.79 mg/L and 0.06 mg/L, respectively, which was statistically significant compared to the other bacterial strains [Table 3].
Table 3: Polyphosphate content (mg/L) accumulated in bacterial cells.
| No. | Bacterial strains | Phosphate content in bacterial cell fluid (mg/L) | Accumulated poly-P content in bacterial cells (mg/L) |
|---|---|---|---|
| 1 | P1 | 8.97 | 0.32c±0.07 |
| 2 | P2 | 7.75 | 1.05b±0.03 |
| 3 | P11 | 8.84 | 0.36c±0.09 |
| 4 | P12 | 9.03 | 0.23c±0.06 |
| 5 | Y4 | 9.10 | 0.28c±0.00 |
| 6 | Y9 | 8.74 | 0.23c±0.06 |
| 7 | Y10 | 9.27 | 0.06c±0.00 |
| 8 | Y11 | 7.61 | 1.79a±0.31 |
| 9 | Y12 | 9.74 | 0.34c±0.00 |
| 10 | Y13 | 8.54 | 1.24b±0.06 |
| 11 | Y17 | 8.78 | 0.23c±0.06 |
| Initial phosphorus content (mg/L) | 10.40 | ||
Values in the same column followed by the same letter are not significantly different at the 5% level using Tukey’s test.
The results indicated that the remaining phosphate content in the bacterial cell culture medium decreased compared to the initial phosphate concentration (10.40 mg/L). This could be explained by the fact that not only did the bacteria accumulate the Poly-P granules inside the cells, but they also utilized phosphate for their metabolic needs, such as cell membrane formation or energy absorption. In addition, Dorofeev et al. [16] reported that besides the ability to accumulate Poly-P as intracellular storage, Poly-P also plays an essential role in other processes such as regulating the activity of certain enzymes and controlling gene expression. Many studies demonstrated that the bacteria that are capable of accumulating phosphate in the form of Poly-P possess a set of characteristic genes for the accumulation and uptake of phosphorus, such as ppk1, ppk2, ppx2, ppgk, pap, and ppnk. For instance, Microlunatus phosphovorus JN459 has been shown to have the ability to accumulate a large amount of Poly-P, which is crucial for removing dissolved phosphorus in wastewater. JN459 strain can accumulate Poly-P under aerobic conditions (orthophosphates in the environment are transported into bacterial cells through the PstSCAB pathway) and absorb the accumulated Poly-P granules under anaerobic conditions, thanks to two characteristic genes, ppgk and ppnk, which encode for PPGK and PPNK proteins, respectively, which utilize the accumulated Poly-P granules for adenosine triphosphate and energy provision for bacterial cell growth [3].
3.4. Identification of Potential Bacterial Strains for Wastewater Treatment from Vermicelli Production Processes
After sequencing the 16S rRNA gene segment, the gene sequences of bacterial strains Y9 and Y11 were compared to gene sequences in the NCBI gene bank using the BLAST tool. The results revealed that bacterial strains Y9 and Y11 were identified as Chryseobacterium daecheongense and Bacillus velezensis, respectively, with a similarity of 100%.
The genetic relationship of bacterial strain Y9 with Chryseobacterium species is illustrated in the phylogenetic tree [Figure 4]. Based on clustering and genetic similarity, the results indicated that strain Y9 has a close genetic relationship with C. daecheongense. The reliability of this clustering is supported by a bootstrap value of 100%. Meanwhile, the genetic distances of strain Y9 to Chryseobacterium wanjuense, Chryseobacterium formosense, and Chryseobacterium chungangensis are 0.044, 0.041, and 0.042, respectively.
 | Figure 4: Maximum likelihood phylogenetic tree of the bacterial strain Y9 based on 16S rRNA gene. [Click here to view] |
According to Kim et al. [17], C. daecheongense CPW406T isolated from freshwater sediment exhibited rod-shaped morphology, was Gram-negative, formed orange-yellow pigmentation on NA medium, and showed positive results for catalase and oxidase tests. In addition, it demonstrated the ability to degrade starch, all of which aligns with the descriptions in the study. The bacterial strain C. daecheongense is relatively new, with limited in-depth research available. Recent studies have indicated that some strains of the Chryseobacterium genus have pathogenic potential for humans and animals. However, the pathogenicity of most of these bacterial strains remains limitedly studied, and the toxicity has not been accurately determined. To ascertain toxicity, studies are needed on hemolytic activity, identification of virulent enzymes produced by bacteria, and testing bacterial resistance/susceptibility to antibiotics [18]. Some recent studies have applied Chryseobacterium species in wastewater treatment. For instance, Layly et al. [19] isolated Chryseobacterium gleum strains from a palm oil production plant that are capable of lipid degradation, while Chryseobacterium hufense was able to absorb nitrogen and was applied in treating nitrogen-polluted wastewater [20].
The genetic relationship between bacterial strain Bacillus Y11 and other Bacillus species is illustrated through clustering on the phylogenetic tree [Figure 5] Bacillus spp. Y11 is grouped in the same clade as Bacillus amyloliquefaciens, showing complete genetic similarity. In addition, the genetic distance of strain Y11 compared to other Bacillus species, such as B. velezensis, Bacillus subtilis, Bacillus nakamurai, Bacillus stercoris, Bacillus rugosus, and Bacillus cabrialesii is only 0.001, reflecting a close genetic relationship with these species. The clustering is also confirmed to be reliable with a bootstrap value of 99%. The close genetic relationship among these species has also been reviewed in genetic diversity analyses based on the 16S rRNA gene [21,22].
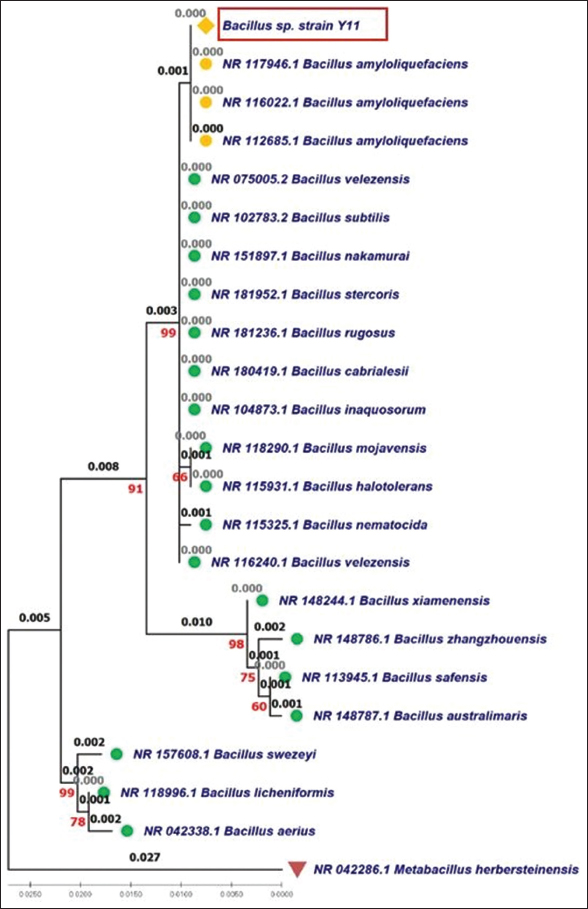 | Figure 5: Maximum likelihood phylogenetic tree of the bacterial strain Y11 based on 16S rRNA gene. [Click here to view] |
Bacteria belonging to Bacillus genus are rod-shaped and mostly non-pathogenic to humans (except for Bacillus anthracis and Bacillus cereus). Bacillus spp. proliferate rapidly under aerobic conditions and can survive by forming endospores even under anaerobic conditions. Moreover, they can survive for extended periods, even in nutrient-depleted environments or harsh conditions. Some Bacillus strains have been studied and used in various stages of wastewater treatment processes due to their ability to produce extracellular enzymes that degrade organic compounds such as starch, protein, cellulose, etc., and their capability to produce bioflocculants and accumulate Poly-P [23]. Layly et al. [19] isolated B. velezensis strains from a palm oil production factory capable of lipid degradation. B. cereus strains have been studied for optimal conditions to enhance lipase enzyme production for the removal of oil pollution in oil-producing plants, which could serve as an economical alternative for wastewater treatment [24]. The study by Agunbiade et al. [25] also demonstrated that B. velezensis strains can produce bioflocculants at over 90% efficiency. Three strains of Bacillus megaterium HT1, Bacillus licheniformis HT1, and B. subtilis HT1 have been proven to absorb and remove nitrogen sources in livestock wastewater by up to 85% [26]. The isolation and selection results in this study have contributed to diversifying bacterial sources capable of starch degradation while simultaneously accumulating phosphate as Poly-P from phosphate-contaminated wastewater sources. The present study has identified highly promising bacterial strains for treating wastewater by starch-based input sources.
The study isolated 22 bacterial strains capable of starch degradation from wastewater collected from the vermicelli production factory in Can Tho City. Among them, C. daecheongense Y9 and B. velezensis Y11 were selected for their potential application in treating starch-rich wastewater. Wastewater from vermicelli production contains a high concentration of starch, leading to elevated COD and BOD levels, as starch is an easily degradable organic compound that generates a large amount of organic matter in water. In addition, the high suspended solids content results from the incomplete dissolution of starch particles, which can cause sludge sedimentation in treatment systems. Moreover, this wastewater is rich in nutrients such as nitrogen (N) and phosphorus (P), posing a risk of eutrophication if not properly treated. Under anaerobic conditions, bacteria can ferment starch, producing H2S and NH3, which cause unpleasant odors. C. daecheongense Y9 exhibited a high sugar removal capacity, with reducing sugar degradation ranging from 1.70 to 4.46 mg/mL over 8 days. A high concentration of reducing sugars in microbial cultures may indicate polysaccharide degradation ability, sugar metabolism efficiency, or the accumulation of intermediate metabolic products. Meanwhile, B. velezensis Y11 demonstrated strong phosphate absorption ability, reaching 1.79 mg/L within 6 days, suggesting its potential for removing excess phosphate from wastewater. Poly-P accumulating organisms help reduce phosphate levels, preventing eutrophication and protecting aquatic ecosystems. This study suggests C. daecheongense Y9 and B. velezensis Y11 as potential treatment options for starch-rich wastewater, which may help control organic and nutrient pollution in aquatic environments.
4. CONCLUSION
Twenty-two out of thirty-two strains were screened based on their capacity for starch degradation, and eleven out of thirty-two bacterial strains could accumulate Poly-P isolated from the wastewater of the vermicelli production process in Can Tho City. Among these strains, Y9 demonstrated the strongest starch-degrading ability on the SA medium and produced the highest reducing sugar content, ranging from 1.70 mg/mL to 4.64 mg/mL after 8 days. In addition, strain Y11 showed the highest accumulation of Poly-P granules within cells on the phosphate-removing agar medium, reaching a maximum mass of 1.79 mg/L P2O5. These two potential wastewater treatment strains, Y9 and Y11, the results from 16S rRNA gene sequencing revealed that they were identified as C. daecheongense and B. velezensis. These strains have great potential in the treatment of wastewater containing starch and residual phosphorus.
4. Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
5. Funding
There is no funding to report.
6. Conflicts of interest
The authors report no financial or any other conflicts of interest in this work.
7. Ethical approvals
This study does not involve experiments on animals or human subjects.
8. Data availability
All the data is available with the authors and shall be provided upon request.
9. Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
10. Use of artificial intelligence (AI)-assisted technology
The authors declare that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Rathoure AK, Dhatwalia VK. Toxicity and Waste Management using Bioremediation. Hershey:IGI Global;2016.[CrossRef]
2. Crini G, Lichtfouse E. Wastewater treatment:An overview. In:Crini Green Adsorbents for Pollutant Removal-fundamentals and Design. Environmental Chemistry for a Sustainable World. Vol. 1., Ch. 1. Berlin:Springer;2018. 1-21.[CrossRef]
3. Zhong C, Jiafang F, Jiang T, Zhang C, Guangxiang C. Polyphosphate metabolic gene expression analyses reveal mechanisms of phosphorus accumulation and release in Microlunatus phosphovorus strain JN 459. FEMS Microbiol Lett 2018;365:1-8.[CrossRef]
4. Shivaji S, Chaturvedi P, Suresh K, Reddy GS, Dutt CB, Wainwright M,et al. Bacillus aerius sp. ., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int J Syst Evol Microbiol 2006;56:1465-73.[CrossRef]
5. Geetha K, Venkatesham E, Hindumathi A, Bhadraiah B. Isolation, screening and characterization of plant growth promoting bacteria and their efect on Vigna radita (L.) R. Wilczek. Int J Curr Microbiol Appl Sci 2014;3:799-899.
6. Wang DB, Li XM, Yang Q, Zeng GM, Liao DX, Zhang J. Biological phosphorus removal in sequencing batch reactor with single-stage oxic process. Bioresour Technol 2008;99:5466-73.[CrossRef]
7. Eixler S, Selig U, Karsten U. Extraction and detection methods for poly-P strorage in autotrophic planktonic organisms. Hydrobiologia 2005;533:135-43.[CrossRef]
8. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9.[CrossRef]
9. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20.[CrossRef]
10. Mai T, Che MN, Nguyen HH. Isolation, identification of starch degrading bacteria from organic wastes, guts of Holotrichia parallela and Lumbricus terestris. CTU J Sci 2019;55:57-64.
11. Dang QH, Tran TT. Isolation, screening and identification of effective microbial strains to make bio-organic fertilizer from organic solid waste. J Sci Technol Univ Danang 2022;20:56-61.
12. Clemente RS, Guijo MI, Nogales J, Blasco R. Carbon source influence on extracellular pH changes along bacterial cell-growth. Genes 2020;11:1292.[CrossRef]
13. Nilsesh S, Anil G. Isolation, characterization and identification of extracellular enzyme producer Bacillus licheniformis from municipal wastewater and evaluation of their biodegradability. Biotechnol Res Innov 2018;2:37-44.[CrossRef]
14. Ramin KI, Allison SD. Bacterial tradeoffs in growth rate and extracellular enzymes. Front Microbiol 2019;10:2956.[CrossRef]
15. Tran TG, Trinh HN, Nguyen TH, Vo DL, Do TK. Isolation and selection of starch degrading bacteria. CTU J Sci 2022;58:225-31.
16. Dorofeev AG, Nikolaev YA, Mardanov AV, Pimenov NV. Role of phosphate-accumulating bacteria in biological phosphorus removal from wastewater. Appl Biochem Microbiol 2020;56:1-14.[CrossRef]
17. Kim KK, Bae HS, Schumann P, Lee ST. Chryseobacterium daecheongense sp. ., isolated from freshwater lake sediment. Int J Syst Evol Microbiol 2005;55:133-8.[CrossRef]
18. Mwanza EP, Hugo A, Charimba G, Hugo CJ. Pathogenic potential and control of Chryseobacterium species from clinical, fish, food and environmental sources. Microorganisms 2022;10:895.[CrossRef]
19. Layly IR, Meryin A, Heianti I, Astuti RI. Identification of lipase producing bacteria from palm oil sewage sludge processing plant at Malimping, Banten, Indonesia. Biodiversitas 2021;22:4512-24.[CrossRef]
20. Kundu P, Pramanik A, Dasgupta A, Mukherjee S, Mukherjee J. Simultaneous heterotrophic nitrification and aerobic denitrification by Chryseobacterium sp. R31 isolated from abattoir wastewater. BioMed Res Int 2014;2014:436056.[CrossRef]
21. Adelskov J, Patel BK. A molecular phylogenetic framework for Bacillus subtilis using genome sequences and its application to Bacillus subtilis subspecies stecoris strain D7XPN1, an isolate from a commercial food-waste degrading bioreactor. 3 Biotech 2016;6:96.[CrossRef]
22. Fan B, Wang C, Song X, Ding X, Wu L, Wu H, et al. Bacillus velezensis FZB42 in 2018:The Gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol 2018;9:2491.[CrossRef]
23. Seo KW, Gu MB, Tsang YF, Choi YS, Chung J. Application of endospore-forming Bacillus species to food waste-recycling wastewater treatment:A focus on the fate of macromolecular nutrients. J Environ Chem Eng 2022;10:107584.[CrossRef]
24. Hassan SW, Latif HH, Ali SM. Production of cold-active lipase by free and immobilized marine Bacillus cereus HSS:Application in wastewater treatment. Front Microbiol 2018;9:2377.[CrossRef]
25. Agunbiade M, Oladipo P, Ademakinwa AN, Awolusi O, Adesiyan IM, Oyekola O, et al. Bioflocculant produced by Bacillus velezensis and its potential application in brewery wastewater treatment. Sci Rep 2022;12:10945.[CrossRef]
26. Dong NH, Viet NT, Hang DT, Hung PD, Duan TH. Ammonia oxidation capacity of Bacillus bacteria in swine wastewater after biogas treatment. Hue Univ J Sci Nat Sci 2022;131:77-87.[CrossRef]