1. INTRODUCTION
Vietnamese ginseng, commonly referred to as Ngoc Linh ginseng and scientifically known as Panax vietnamensis Ha et Grushv., is a valuable ginseng species native to Vietnam. Vietnamese ginseng, a herbaceous perennial that is a member of the Araliaceae family, was first discovered at Ngoc Linh mountain in Gia Lai-Kontum, Vietnam, in 1973 [1,2]. Its native distribution is limited to the Ngoc Linh and Hoang Lien Son mountain ranges, and they are cultivated in two conservation areas in the provinces of Quang Nam and Kon Tum on the Ngoc Linh mountain’s slopes [3,4]. Vietnamese ginseng was utilized by ethnic minorities as traditional medicine for the enhancement of physical strength and treatment of a variety of illnesses before it was discovered [1,2,5].
Vietnamese ginseng has been shown to have obvious pharmacological effects in the treatment of anti-stress, anti-oxidation, anti-inflammatory, anti-aging, and anti-cancer [6,7]. Saponins are the main group of biologically active compounds in Vietnamese ginseng, with more than 52 different derivatives [7-9]. Due to their significant medicinal and commercial worth, as well as overexploitation, they are included in the Vietnam Red Book as a rare plant species that must be preserved [10].
Saponins are glycosides of triterpenoids or steroids, of which the triterpenoid saponins are dominant in Vietnamese ginseng [11,12]. Triterpenoid saponins offer numerous other advantages outside of defending plants from pathogenic bacteria and herbivores, such as enhancing health through immunomodulation, hypocholesterolemia, anti-coagulation, anti-cancer, hepatoprotection, anti-inflammatory properties, antioxidants, and many more [13,14].
Terpenoids are the largest group of plant secondary metabolites and are biosynthesized by two pathways in plants: The mevalonate (MVA) pathway in the cytosol and the non- MVA pathway (MEP) in the plastids [15]. The biosynthesis of terpenoids by the MEP begins with two acetyl-CoAs that are condensed to form acetoacetyl-CoA under the catalysis of acetoacetyl-CoA thiolase (AACT, EC 2.3.1.9). Acetoacetyl-CoA is then catalyzed to form 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) by HMGCoA synthase (HMGS, EC 2.3.3.10). HMG-CoA is catalyzed to form mevalonic acid by HMG-CoA reductase (HMGR, EC 1.1.1.34), followed by enzymatically catalyzed reactions to form other precursors, and finally triterpenoid saponins. Recent research on the MEP has primarily focused on discovering novel pathways [16], identifying and characterizing important enzymes, and improving their expression in plant cells and tissue culture [17-19]. Genes from the Panax genus that encode key enzymes in the MEP have also been studied [Table 1]. HMGR is the rate-limiting enzyme, which is one of the key elements involved in the catalytic reactions of terpenoid biosynthesis in the MEP in plants [20]. However, the literature review showed that the HMGR genes from P. vietnamensis have not yet been cloned and their structures have not been analyzed. The use of sodium chloride or sodium carbonate as abiotic elicitors in Vietnamese ginseng cultures in vitro has not yet been reported. Although some studies have indicated that the elicitors can improve the expression levels of genes involved in the production of triterpenoid saponin in plant cells and tissues [21-25].
Table 1: Identification of some key enzymes in the MVA pathway of Panax genus.
| S. No | Gene | Enzymes and EC number | Panax species (GenBank) |
|---|---|---|---|
| 1 | AACT | Acetyl-CoA C-acetyltransferase EC: EC 2.3.1.9 | P. vietnamensis Ha et Grushv. (MZ272018.1) and P. notoginseng (KJ804173.1). |
| 2 | HMGS | 3-hydroxy-3-methylglutaryl coenzyme-A synthase EC: 2.3.3.10 | P. notoginseng (KJ804167.1; KP702301.1), P. ginseng (GU565098.1) |
| 3 | HMGR | 3-hydroxy-3-methylglutaryl coenzyme-A reductase EC: 1.1.1.34 | P. notoginseng (KP702300.1, KJ804166.1, KJ578757.1, KP702301.1), P. ginseng (KM386694.1, KM386695.1, GU565097.1, KJ939263.1), P. quinquefolius (FJ755158.2) |
| 4 | FPS | Farnesyl diphosphate synthase EC: 2.5.1.10 | P. ginseng (DQ087959.1), P. vietnamensis (MZ272019.1), P. notoginseng (MK757455.1, KF597527.1, KJ804175.1, KC953034.1, KC524468.1), P. quinquefolius (GQ401664.1), P. japonicas (KP684141.1), P. sokpayensis (JZ822887.1) |
| 5 | SS | Squalene synthase EC: 2.5.1.21 | P. quinquefolius (KT869136.1, GU997681.1, AM182457.1, AM182456.1, KC524471.1, KC524469.1), P. notoginseng (KT123897.1, DQ186630.1, KC953032.1, KC422650.1, KT123898.1), P. sokpayensis (KT936528.1), P. ginseng (EU502717.1, GU183406.1, GQ468527.2, AB010148.1, KP689323.1, KP689322.1, KP689321.1, KP689320.1, KP689319.1, KP689318.1, KP689317.1, KP689316.1, KP689315.1, KP689314.1, KJ939264.1), P. japonicas (KP890782.1) |
| 6 | SE | Squalene epoxidase EC: 1.14.14.17 | P. notoginseng (DQ457054.1, KT123898.1, JX625132.1, DQ386734.1, KC953033.1, KC422651.1, KT123897.1), P. vietnamensis Ha et Grushv. (MW258700.1, MW258699.1, MW258698.1), P. sokpayensis (KT936529.1), P. vietnamensis var. fuscidiscus (KJ946469.1, KJ946468.1, KJ946467.1) P. ginseng (FJ393274.2, AB122078.1, AB003516.1), P. japonicas (MK603118.1). |
| 7 | PPDS | Protopanaxadiol synthase EC: 14.14.121 | P. sokpayensis (MF682462.1), P. notoginseng (KJ995703.1), P. ginseng (JX036031.1), P. vietnamensis Ha et Grushv. (ON693838) |
| 8 | PPTS | Protopanaxatriol synthase EC: 1.14.14.121 | P. sokpayensis (MF682463.1), P. ginseng (JX036031.1), P. quinquefolius (KC190491.1), P. notoginseng (KF935232.1), P. vietnamensis Ha et Grushv. (ON693839) |
P. vietnamensis: Panax vietnamensis, P. notoginseng: Panax notoginseng, P. sokpayensis: Panax sokpayensis, P. quinquefolius: Panax quinquefolius, P. ginseng: Panax ginseng
In the previous studies, we identified three isoforms of squalene epoxidase (SE) in the MEP of Vietnamese ginseng [19]. In the present work, we first reported on molecular cloning, characteristic analysis, 3D structural modeling, and annotation of the HMGR from Vietnamese ginseng. The study also wanted to improve the expression level of the key gene in the adventitious root culture of this medicinal species using salt elicitation, which had not been done in any previous studies.
2. MATERIALS AND METHODS
2.1. Plant Material
Vietnamese ginseng that is 6 years old was provided by Mr Tran Ut (Director of the Center for Developing Ngoc Linh Ginseng and Medicinal Herbs, Quang Nam Province, Vietnam) and identified by the Institute of Ecology and Biological Resources of Vietnam. The plants were collected according to the relevant guidelines and regulations of Vietnam. The in vitro adventitious roots of Vietnamese ginseng were prepared by Prof. Duong Tan Nhut (Tay Nguyen Institute of Scientific Research, Vietnam Academy of Science and Technology) and subcultured on Schenk and Hildebrandt (SH) medium [26], supplemented with 30 g/L sucrose, 4 mg/L indolebutyric acids, and 0.5 mg/L kinetin. In vitro cultures were incubated at 25 ± 2°C with a shaking speed of 100 rpm in the dark [25].
2.2. Molecular Cloning
The specific primers for PCR amplification of the PvH_HMGR gene were designed by the Primer 3 web tool based on the predicted nucleotide sequences of this gene [19]. The primers were then specifically checked by the Primer-BLAST tool against the non-redundant (NR) database limited to Vietnam ginseng. The following primers (5’-3’) of forward ATGGAC GTTCGCCGGCGAC and reverse GTAATTAGGAAGAGAGCTTGGAGACATC were used for cloning. Total RNA was extracted from the root tissues by the Gene JET Plant RNA Purification Kit (Thermo Scientific, USA). The putative PvH_HMGR gene was amplified by the RT-PCR method. The PCR reactions were performed under the following conditions: 94°C for 5 min; followed by 30 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 2 min; and a final extension at 72°C for 10 min. The PCR products were subcloned into a pGEM T-easy vector (Promega, USA) and then sequenced by the Sanger method (FirstBase, Malaysia).
2.3. Bioinformatic Analysis
The full-length sequence of the putative PvH_HMGR gene was subjected to a BLAST search against the NR databases to find out the close homologs. In addition, the HMGR gene was predicted using databases from the P. vietnamensis (SRA accession number: PRJNA665343) [19] and P. vietnamensis var. fuscidiscus (SRA accession number: SRR1198998) [12]. Based on nrBLAST results, the best templates and predicted PvH HMGR sequences were multi-aligned by the CLC Sequence Viewer (Qiagen) to identify the presence of conserved motifs. The introns and exons were predicted by comparing the cloned cDNA and predicted DNA sequences of the PvH_HMGR gene. The transmembrane helices of enzymes were predicted using the DeepTMHMM server. The hydrophobicity of PvH_HMGR enzymes was analyzed by ProtScale using the amino acid scale of Kyte-Doolittle. The secondary structures of enzyme sequences were predicted using the LOMET and I-TASSER tools. The structural and functional domains in the deduced PvH_HMGR enzyme molecule were identified by the SMART and HMMSCAN tools. The enzyme structure model was analyzed and compared by aligning the predicted models from the various servers (PHYRE2, SWISS-MODEL, I-TASSER, and AlphaFold 2.0) and simulated with Pymol v2.5.2. The phylogenetic tree was generated by MEGA X following the neighbour-joining method (NJ) with 1000 bootstrap replicates and visualized by iTOL. The signal peptide of the enzyme was predicted by SignalP 6.0 based on Neural Networks and Hidden Markov models databases of eukaryotes with the best confidence value. The theoretical isoelectric point (pI) was estimated by the Compute pI/MW application from Expasy. Furthermore, the 500 bp of promoter regions upstream of the translation start site of the PvH_HMGR gene were extracted from the draft assembly genome’s predicted PvH_scaffold0913 (756124-759846 nt), which was predicted and annotated by various bioinformatics tools, including PlantPAN 3.0, TSSPlant, Plantprom, PlantCARE, and CpG islands.
2.4. Elicitation and Semi-quantitative RT-PCR
The adventitious root of Vietnamese ginseng was treated with NaCl (0.05, 0.1, 0.15, and 0.2%) and Na2CO3 (0.25, 0.5, 0.75, and 1%) [24] at the beginning or after 20 days of culture. After 56 days of culture [25], including the time for elicitation, the root biomass was collected, and it was then immediately frozen with liquid nitrogen in preparation for total RNA extraction using the GeneJET Plant RNA Purification Kit (ThermoFisher, USA). The first-strand cDNA was synthesized by the RevertAid First-Strand cDNA Synthesis Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. The oligonucleotide primers (5’-3’) GGTCTCCAAAGGCGTCCA (forward) and CCAAGGCTGCCACATTA-GTC (reverse) were used for gene expression analysis. Two microliters of the first-strand cDNA were then used for PCR amplification with the following conditions: 94°C for 5 min; followed by 35 cycles of 94°C for 30 s, 58°C for 45 s, and 72°C for 2 min; and a final extension at 72°C for 10 min. The GAPDH gene was utilized as the internal control gene for measuring and reducing the errors between the samples in RT-PCR expression analyses [12].
3. RESULTS
3.1. Molecular Cloning of PvH_HMGR
The putative PvH_HMGR gene’s cDNA sequence (1.773 bp long), which was obtained by RT-PCR amplification, was compared to related genes from other Panax species. The results showed that this sequence is 100% similar to the coding DNA sequence of the predicted gene from the assembled genome of Vietnamese ginseng. In addition, it was inferred from the comparison that the full-length PvH_HMGR gene may also include four exons and three introns. The deduced amino acid sequence of the PvH_HMGR gene consists of a polypeptide chain of 590 amino acids with an estimated molecular weight (MW) of about 63.11 kDa.
3.2. Characterization of PvH_HMGR Enzyme
The structural and functional domain analysis of the protein encoded by the PvH_HMGR gene indicated that it is the enzyme HMG-CoA reductase with a conserved domain (cd00643) from amino acids (aa) 203–580, and the active site was presented at 269 and 575 aa. Furthermore, the PvH_HMGR also contains two HMG-CoA binding motifs (EMPVGYVQIP and TTEGCLVA) as well as two NADP(H)-binding motifs (GTVGGG and DAMGMNM). The result of transmembrane helix analysis indicated PvH_HMGR has two transmembrane domains (43–57 and 78–96 aa) and no signal peptide [Figures 1 and 2]. The subcellular localization analysis of the PvH_HMGR revealed that its membrane domain contributes to the endoplasmic reticulum (ER) morphogenesis in Vietnamese ginseng cells. Also shown by the analysis was the presence of typical hydrophobic regions in the PvH_HMGR-deduced amino acid sequences. With a high localization probability of 0.6923, these regions, which contain the RRR ER-retention motif, play a crucial role in anchoring enzymes to the ER. The PvH_HMGR enzyme was also shown to contain a large number of hydrophilic domains, including ones from aa 34 to 56, 68-104, 180-187, 218-223, 233-255, 283-296, 326-332, 344-351, 417-424, 433-460, 463-473, 486-492, 522-532, and 546-569. The secondary structure of PvH_HMGR contains 42.88% of alpha-helices with 38.14% of random coils, 14.41% of extended strands, and 4.58% of beta-turns. In addition, the average theoretical isoelectric point (pI) of the enzymes was also estimated at 6.66. The deduced amino acid of PvH_HMGR was searched in the STRING database 2021 to investigate the protein-protein interaction network. The analysis showed that PvH_HMGR is similar to 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform 1, with various percentages (72.4–88%) identified in some plants.
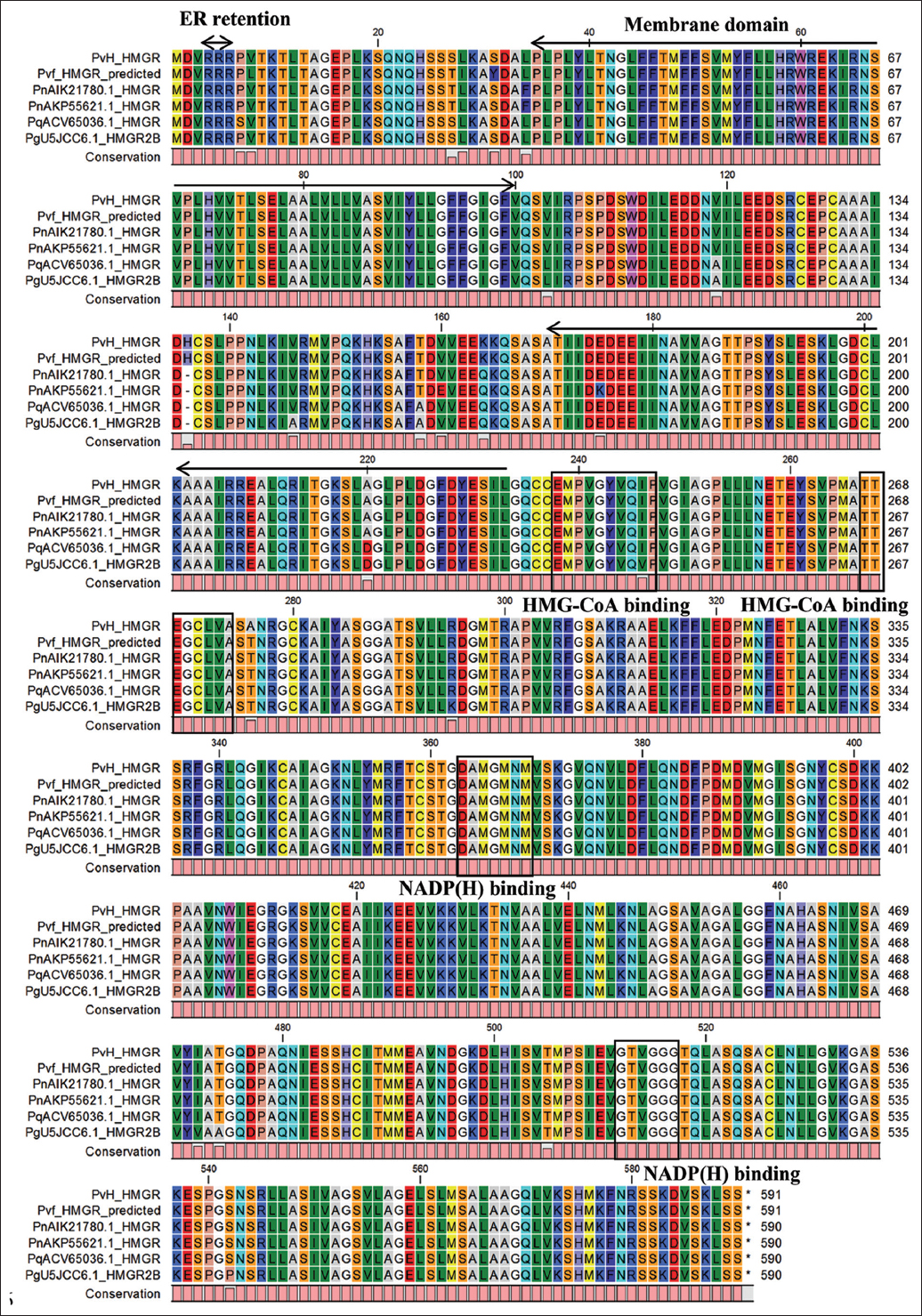 | Figure 1: Multiple alignments of PvH_HMGR deduced amino acid sequences with Panax genus homologous HMGRs. P. vietnamensis var fucisous (PvfHMGR), P. quinquefolius (ACV65036), P. notoginseng (AIK21780.1, AKP55621.1), and P. ginseng (U5JCC6.1). Two putative HMGR-CoA-binding sites and two NADP(h)-binding sites were indicated with a thin square box, and the motif responsible for ER retention was highlighted with a bold square box. [Click here to view] |
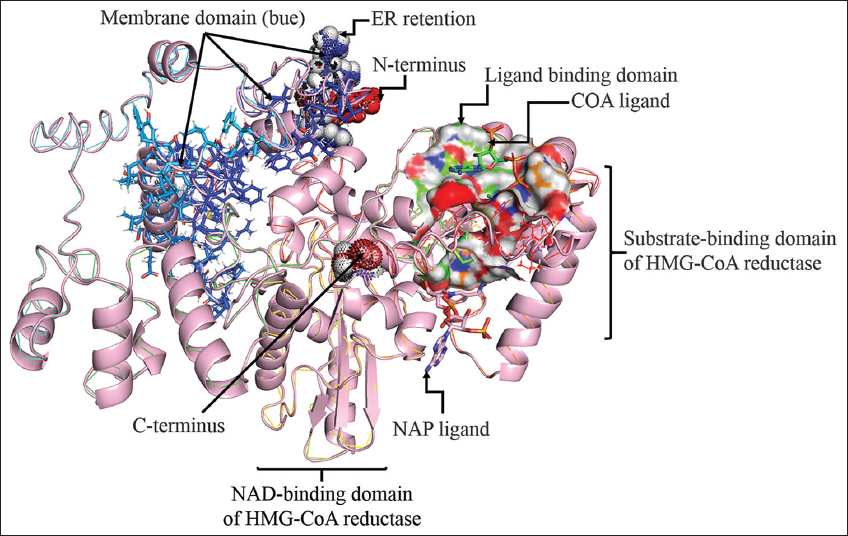 | Figure 2: 3D structure prediction of PvH_HMGR with various annotations such as membrane domain (blue), ER retention, C-terminus, N-terminus, substrate-binding domain, beta-sheet, alpha-helix, and ligand binding domains (COA and NAD ligand). [Click here to view] |
3.3. Homology-modeling of the PvH_HMGR Structure
PvH_HMGR’s 3D protein structure was predicted by several servers and software, including PHYRE2, I-TASSER, and AlphaFold-2. Specifically, PHYER2 modeled 394 residues of PvH_HMGR (67%), with 100.0% confidence, using the single highest scoring template of human HMG-CoA reductase (PDB No.: 1HWJ). Besides, I-TASSER predicted the protein structure (70% coverage) using the best template of human HMG-CoA reductase (PDB No.: 1DQ8) with the highest Z-score significance (4.61). AlphaFold-2 assigns a confidence score to each residue based on a pLDDT value ranging from 0 to 100. The results of Alphafold-2 structure prediction revealed that 70% of residues have very high confidence (pLDDT value >90) and approximately 3.9% have low confidence (residue positions 50–70), which are found at the N and T-termini. In addition, the predicted binding site of PvH_HMGR to some ligands such as HR2, COA, ADP, and NAP is depicted in Figure 2. Furthermore, the active site of the enzyme was predicted at residues 269 and 575. The predicted homology model with high similarity to human HMG-CoA reductase would provide important information for further studies on the biological function and kinetics of this enzyme.
3.4. The Core Promoter of PvH_HMGR Prediction
Several putative cis-elements were identified in the partial promoter of PvH_HMGR [Figure 3]. One MYC element was identified in the promoter. In addition, motifs involved in hormonal regulation, including ABRE, ABRE3a, and the TCA element, were identified in the promoter regions of the gene. These cis-elements are potentially responsive to abscisic acid and salicylic acid. The PlantCARE and PLACE tools revealed the presence of the CATTTG motif, which is the binding site for the MYC transcription element and has a role in jasmonic and abscisic acid signaling. The promoter contains an anaerobic induction element (ARE), a wound-responsive element (motif), and a stress-responsive element (TC-rich repeats), suggesting that the PvH_HMGR gene may be involved in various stress responses in Vietnamese ginseng. Besides, CpG sequence clusters have not been detected in the predicted promoter region, suggesting a potential lack of inhibition of gene expression due to cytosine methylation.
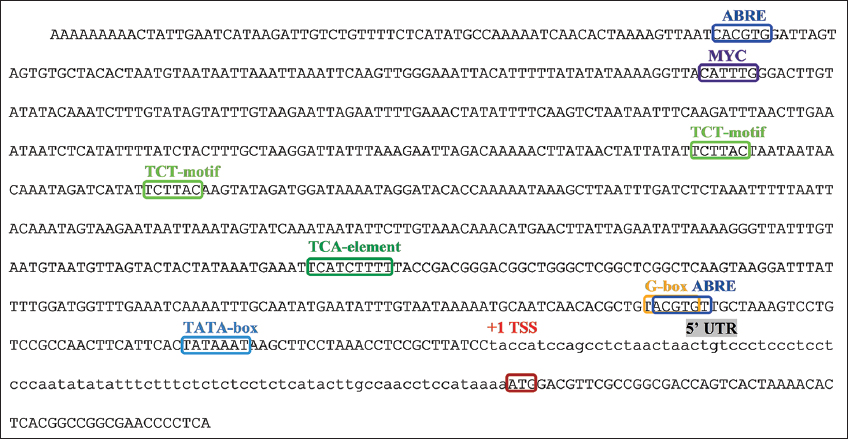 | Figure 3: Prediction of an important cis-regulatory element found in the promoter of PvH_HMGR gene. Red frame: translation initiation site, low letter DNA sequence: 5’ UTR, TSS: transcriptional start site, green frame: TATA-box, dark blue frame: ABRE cis-element involved in the response to abscisic acid, yellow frame: G -box cis-element involved in the response to light, dark green frame: TCA cis-element-probably involved in the response to salicylic acid, light green frame: TCT-motif involved in the response to light, purple frame: MYC motif. [Click here to view] |
3.5. Phylogenetic Analysis
The evolutionary relationships of the PvH_HMGR in Vietnamese ginseng with corresponding genes from some Panax species and various organisms (bacteria, archaea, fungi, plants, and animals) were aligned in MUSCLE and neighbor-joining trees generated by the MEGA X program with 1000 bootstraps [Figure 4]. The phylogenetic analysis results show that a large group of plant HMGRs originated from a more recent common ancestor with animal HMGRs than the other organisms. It may be suggested that the HMGR genes of the plant group diverged at a later period than the corresponding genes of groups such as archaea, bacteria, fungi, and animals. The HMGRs of the Panax genus formed a common cluster within the plant’s group and represented the most recently diverged lineage within this group. Specifically, the PvH_HMGR has a high similarity with corresponding HMGRs to P. vietnamensis var. fuscidiscus (99.7%), P. notoginseng (98.98%), P. quinquefolius (98.64%), and P. ginseng (97.80%).
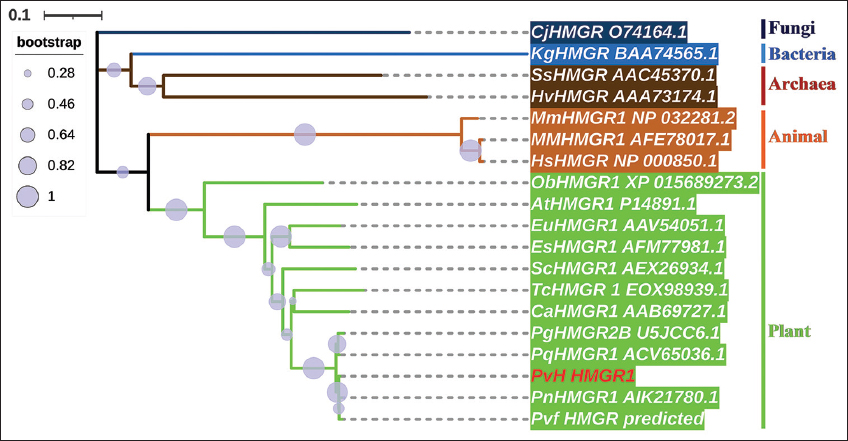 | Figure 4: A phylogenetic tree of PvH_HMGR and various organisms. Plants (greenish-yellow: PvH: P. vietnamensis, Pvf: P. vietnamensis var fuscidiscus, Pg: P. ginseng, Pn: P. notoginseng, Pq: P. quinquefolius, Ca: Camptotheca acuminata, Sc: Solanum chacoense, Tc: Theobroma cacao, Es: Eleutherococcus senticosus, Eu: Eucommia ulmoides, At: Arabidopsis thaliana, Ob: Oryza brachyantha), animals (yellowish-orange: Hs: Homo sapiens, MM: Macaca mulatta, Mm: Mus musculus), fungi (light navy: Cj: Cyberlindnera jadinii, Pm: Paraphaeosphaeria minitans), achaea (antique bronze: SS: Saccharolobus solfataricus, Hv: Haloferax volcanii), and bacterium (blue color: Kg: Kitasatospora griseola). [Click here to view] |
3.6. Effect of Elicitors on the Expression of PvH_HMGR
The present study indicated that the addition of various concentrations of NaCl and Na2CO3 to create abiotic stress significantly improved the expression of the PvH_HMGR gene in adventitious roots of Vietnamese ginseng compared with the untreated control sample. In general, Na2CO3 had a stronger effect on the expression of PvH_HMGR than NaCl.
The semi-quantitative RT-PCR result showed that increasing the NaCl concentration up to 0.1 % increased the expression of PvH_HMGR about 2.71 times more than the untreated sample [Figure 5]. At this concentration, there is the little observable inhibitory effect on the growth of Vietnamese ginseng adventitious roots. This result is quite similar to the study [21] on P. ginseng, which added 0.1% NaCl to enhance the saponin content. In addition, Gupta’s study [24] also showed that 0.1% NaCl increased the biosynthesis of steviol glycoside in Stevia rebaudiana. For the treatment with Na2CO3, the results showed the highest expression of PvH_HMGR at a concentration of 0.075%, reaching 3.28 times higher than that of the untreated control. Besides, Na2CO3 was found to inhibit adventitious root growth at a concentration higher than 0.05%. In general, for other tested concentrations of elicitors, the PvH_HMGR expression was reduced compared to the above ideal concentration.
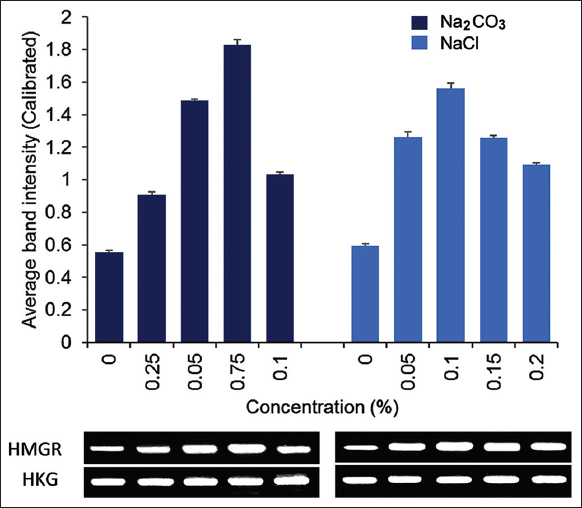 | Figure 5: Semi-quantitative RT-PCR expression analysis of HMGR in adventitious root Vietnamese ginseng under untreated and treated NaCl and Na2CO3 at various concentrations. The GAPDH gene was used as the reference gene. The data on the chart were expressed as the mean of three replicates with standard error bars. [Click here to view] |
4. DISCUSSION
HMGR is a key regulatory enzyme with an important rate-limiting function in the ginsenoside and cholesterol biosynthesis pathways in plants, yeasts, and animals [27-29]. At present, HMGR genes have been cloned, characterized, and expression analyzed in the Panax genus, including P. ginseng [30], P. quinquefolius [31], P. notoginseng [32], and other plants such as Centella asiatica L. [33], Sanghuangporus baumii [34], and Withania somnifera [35]. Vietnamese ginseng is a precious ginseng that is proven to have a high content of ginsenoside, have anti-inflammatory effects, improve memory, be anti-stressing, and support anti-cancer [1,12]. However, at the present, there have not been any reports on HMGR genes in Vietnamese ginseng. This research showed the isolation of the cDNA sequences’ structural and functional features and the identification of their promoter sequences in Vietnamese ginseng. The results indicated that PvH_HMGR is a functional ortholog of P. vietnamensis var. fuscidiscus and P. ginseng and that PvH_HMGR plays a similar regulatory role in the production of triterpene ginsenosides in ginseng plants. Besides, this study also shows that the PvH_HMGR feature of Vietnamese ginseng is similar to that of the active sites responsible for binding HMG-CoA and NADPH2 among Panax species, including P. ginseng, P. notoginseng, and P. quinquefolius. The predicted homology modeling of PvH_HMGR, showing high similarity to human HMG-CoA reductase, provided important information on enzyme characterization for further studies of the enzyme’s biological function and kinetics. The phylogenetic tree analysis showed that PvH_HMGR has more similarities with the corresponding genes from P. vietnamensis var. fuscidiscus and P. notoginseng than other species.
Plant cell biotechnology is considered a preferred solution for the production of large quantities of valuable compounds in a short period because bioactive metabolites accumulate slowly in medicinal plants under natural conditions [36,37]. A better understanding of the key genes involved in secondary metabolism pathways, as well as the effects of elicitors on their activity, will allow for more effective biosynthesis of bioactive compounds using plant tissues and cell cultures [38]. The elicitors (chitosan, yeast extract, sodium chloride, sodium carbonate, methyl jasmonate, abscisic acid, or salicylic acid) have been studied to improve the expression of key genes related to plant secondary metabolism to enhance yields of secondary metabolite biosynthesis in plant cell cultures for a shorter time [21,24,39,40]. Continuing to investigate suitable and low-cost elicitors for improving high-value ginsenosides in Vietnamese ginseng in a short period of time is a necessary research direction. Up to now, there are no reports regarding the effect of salt (NaCl and Na2CO3) on the expression of key regulatory genes for saponin metabolism or ginsenoside production from root cultures of Vietnamese ginseng.
In comparison to other concentrations, 0.1% NaCl and 0.075% Na2CO3 increased the expression of the HMGR gene in Vietnamese ginseng roots. The previous studies indicated that this concentration of NaCl also increased the saponin content in P. ginseng and the steviol glycoside content in S. rebaudiana [21,24]. Nevertheless, the report of Gupta showed that sodium carbonate at a lesser concentration (0.025%) enhanced the terpenoid glycoside content in S. rebaudiana species [24]. This may imply that sodium carbonate may affect gene activity in terpenoid metabolism differently depending on the species.
The findings suggested that sodium chloride (0.1%) and sodium carbonate (0.075%) might be used as suitable abiotic elicitors to improve the production of ginsenoside in the adventitious root culture of Vietnam ginseng.
5. CONCLUSION
PvH_HMGR, the key enzyme in the ginsenoside production pathway of Vietnamese ginseng, was successfully cloned and identified. The characteristics and 3D structure of PvH_HMGR were predicted. Their core promoter was also predicted concerning various stress responses in Vietnamese ginseng. Furthermore, abiotic stress (NaCl and Na2CO3) significantly improved the expression of PvH_HMGR for ginsenoside synthesis in Vietnamese ginseng adventitious root culture.
6. AUTHOR CONTRIBUTIONS
Loc NH, Tien NQD, and Nhut DT designed and performed the overall study. Tien NQD, Man LQ, Kha H, and Linh TL conducted molecular cloning and bioinformatics analysis. Tien NQD, Kha H, Anh TL conducted elicitor treatment and analyzed gene expression. Nhut DT provides research samples. Tien NQD and Loc NH wrote the final manuscript.
7. FUNDING
This work was financially supported by the National Foundation for Science and Technology Development (NAFOSTED) under Grant number 106.02-2018.49.
8. CONFLICTS OF INTEREST
All authors declared no conflicts of interest.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation
REFERENCES
1. Nguyen MD, Nham TN, Kasai R, Ito A, Yamasaki K, Tanaka O. Saponins from Vietnamese ginseng, Panax vietnamensis Ha et Grushv. collected in central Vietnam. I. Chem Pharm Bull (Tokyo) 1993;41:2010-4. [CrossRef]
2. Yamasaki K. Bioactive saponins in Vietnamese ginseng, Panax vietnamensis. Pharm Biol 2000;38(suppl 1):16-24. [CrossRef]
3. Le QU, Lay HL, Wu MC, Nguyen TH, Nguyen DL. Phytoconstituents and biological activities of Panax vietnamensis (Vietnamese Ginseng):A precious ginseng and call for further research-a systematic review. Nat Prod Commun 2018;13:1381-4. [CrossRef]
4. Vasyutkina EA, Adrianova IY, Reunova GD, Nguyen TP, Zhuravlev YN. A comparative analysis of genetic variability and differentiation in Panax vietnamensis Ha et Grushv. and P. ginseng CA Meyer using ISSR markers. Russ J Genet 2018;54:262-5. [CrossRef]
5. Van Le TH, Lee SY, Kim TR, Kim JY, Kwon SW, Nguyen NK, et al. Processed Vietnamese ginseng:Preliminary results in chemistry and biological activity. J Ginseng Res 2014;38:154-9. [CrossRef]
6. Dela Peña IJ, Kim HJ, Botanas CJ, de la Pena JB, Van Le TH, Nguyen MD, et al. The psychopharmacological activities of Vietnamese ginseng in mice:Characterization of its psychomotor, sedative-hypnotic, antistress, anxiolytic, and cognitive effects. J Ginseng Res 2017;41:201-8. [CrossRef]
7. Duc NM, Kasai R, Ohtani K, Ito A, Nham NT, Yamasaki K, et al. Saponins from Vietnamese ginseng, Panax vietnamensis Ha et Grushv. collected in central Vietnam. III. Chem Pharm Bull (Tokyo) 1994;42:634-40. [CrossRef]
8. Duc NM, Kasai R, Yamasaki K, Nham NT, Tanaka O. New dammarane saponins from Vietnamese ginseng. Stud Plant Sci 1999;6:77-82. [CrossRef]
9. Nguyen TH, Phuong TT. Vietnamese Ginseng (Panax vietnamensis Ha and Grushv.):Phylogenetic, phytochemical, and pharmacological profiles. Pharmacogn Rev 2019;13:59-62. [CrossRef]
10. Ministry of Science and Technology. Vietnam Red List Data Book, Part II. Plant. Hanoi:Natural Science and Technology Publishing House;2007.
11. Van Le TH, Lee GJ, Vu HK, Kwon SW, Nguyen NK, Park JH, et al. Ginseng saponins in different parts of Panax vietnamensis. Chem Pharm Bull (Tokyo) 2015;63:950-4. [CrossRef]
12. Zhang GH, Ma CH, Zhang JJ, Chen JW, Tang QY, He MH, et al. Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics 2015;16:159. [CrossRef]
13. Da Silva Magedans YV, Phillips MA, Fett-Neto AG. Production of plant bioactive triterpenoid saponins:From metabolites to genes and back. Phytochem Rev 2021;20:461-82. [CrossRef]
14. Sawai S, Saito K. Triterpenoid biosynthesis and engineering in plants. Front Plant Science 2011;2:25. [CrossRef]
15. Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, et al. Inhibitors of the non-mevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 1999;285:1573-6. [CrossRef]
16. Muhlemann JK, Klempien A, Dudareva N. Floral volatiles:From biosynthesis to function. Plant Cell Environment 2014;37:1936-49. [CrossRef]
17. Xu J, Chu Y, Liao B, Xiao S, Yin Q, Bai R, et al. Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience 2017;6:1-15. [CrossRef]
18. Lan TT, Huy ND, Luong NN, Nghi NV, Tan TH, Quan LV, et al. Identification and characterization of genes in the curcuminoid pathway of Curcuma zedoaria Roscoe. Curr Pharm Biotechnol 2018;19:839-46. [CrossRef]
19. Tien NQ, Ma X, Man LQ, Chi DT, Huy NX, Nhut DT, et al. De novo whole-genome assembly and discovery of genes involved in triterpenoid saponin biosynthesis of Vietnamese ginseng (Panax vietnamensis Ha et Grushv.). Physiol Mol Biol Plants 2021;27:2215-29. [CrossRef]
20. Ma R, Fu B, Yang P, Teng X, Zhao D, Jiang R, et al. Sucrose induced HMGR to promote ginsenoside biosynthesis in the growth of wild cultivated ginseng (Panax ginseng). J Soil Sci Plant Nutr 2022;22:2255-65. [CrossRef]
21. Jeong GT, Park DH. Enhanced secondary metabolite biosynthesis by elicitation in transformed plant root system:Effect of abiotic elicitors. Appl Biochem Biotechnol 2006;129-32:436-46. [CrossRef]
22. Huang C, Zhong JJ. Elicitation of ginsenoside biosynthesis in cell cultures of Panax ginseng by vanadate. Process Biochem 2013;48:1227-34. [CrossRef]
23. Gengmao Z, Yu H, Xing S, Shihui L, Quanmei S, Changhai W. Salinity stress increases secondary metabolites and enzyme activity in safflower. Ind Crops Prod 2015;64:175-81. [CrossRef]
24. Gupta P, Sharma S, Saxena S. Effect of salts (NaCl and Na2CO3) on callus and suspension culture of Stevia rebaudiana for steviol glycoside production. Appl Biochem Biotechnol 2014;172:2894-906. [CrossRef]
25. Linh NT, Cuong LK, Tam HT, Tung HT, Luan VQ, Hien VT, et al. Improvement of bioactive saponin accumulation in adventitious root cultures of Panax vietnamensis via culture periods and elicitation. Plant Cell Tissue Organ Cult 2019;137:101-13. [CrossRef]
26. Schenk RU, Hildebrandt AC. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures.Can J Bot 1972;50:199-204. [CrossRef]
27. Liao Z, Tan Q, Chai Y, Zuo K, Chen M, Gong Y, et al. Cloning and characterisation of the gene encoding HMG-CoA reductase from Taxus media and its functional identification in yeast. Funct Plant Biol 2004;31:73-81. [CrossRef]
28. Burg JS, Espenshade PJ. Regulation of HMG-CoA reductase in mammals and yeast. Prog Lipid Res 2011;50:403-10. [CrossRef]
29. Kim YJ, Zhang D, Yang DC. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv 2015;33:717-35. [CrossRef]
30. Kim YJ, Lee OR, Oh JY, Jang MG, Yang DC. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol 2014;165:373-87. [CrossRef]
31. Wu Q, Sun C, Chen S. Identification and expression analysis of a 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from American ginseng. Plant Omics 2012;5:414-20.
32. Liu WJ, Lv HZ, He L, Song JY, Sun C, Luo HM, et al. Cloning and bioinformatic analysis of HMGS and HMGR genes from Panax notoginseng. Chin Herb Med 2016;8:344-51. [CrossRef]
33. Kalita R, Patar L, Shasany AK, Modi MK, Sen P. Molecular cloning, characterization and expression analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Centella asiatica L. Mol Biol Rep 2015;42:1431-9. [CrossRef]
34. Liu Z, Wang S, Xu X, Wang S, Sun T, Zou L. Molecular cloning and characterization of a gene encoding HMG-CoA reductase involved in triterpenoids biosynthetic pathway from Sanghuangporus baumii. Biotechnol Biotechnol Equip 2021;35:796-804. [CrossRef]
35. Akhtar N, Gupta P, Sangwan NS, Sangwan RS, Trivedi PK. Cloning and functional characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Withania somnifera:An important medicinal plant. Protoplasma 2013;250:613-22. [CrossRef]
36. DiCosmo F, Misawa M. Plant cell and tissue culture:Alternatives for metabolite production. Biotechnol Adv 1995;13:425-53. [CrossRef]
37. Wawrosch C, Zotchev SB. Production of bioactive plant secondary metabolites through in vitro technologies-status and outlook. Appl Microbiol Biotechnol 2021;105:6649-68. [CrossRef]
38. Chezem WR, Clay NK. Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 2016;131:26-43. [CrossRef]
39. Loc NH, Diem DT, Binh DH, Huong DT, Kim TG, Yang MS. Isolation and characterization of antioxidation enzymes from cells of zedoary (Curcuma zedoaria Roscoe) cultured in a 5-1 bioreactor. Mol Biotechnol 2008;38:81-7. [CrossRef]
40. Singh A, Dwivedi P. Methyl-jasmonate and salicylic acid as potent elicitors for secondary metabolite production in medicinal plants:A review. J Pharmacogn Phytochem 2018;7:750-7.