1. INTRODUCTION
Rice (Oryza sativa L.) is one of the most important cereal crops in the world and directly feeds considerably more people than any other crop [1]. More than three billion population worldwide consume rice grain daily, which provides 20% of the world’s nutritional energy [2]. The most harmful disease called blast, caused by an anamorphic fungus Magnaporthe oryzae is mainly common in rice fields, where it significantly lowers grain quality and productivity [3]. Increasing rice’s resistance to M. oryzae is found to be a practical and cost-efficient process for preventing blast disease in rice plants [4].
Plants have developed complex defense mechanisms to fend off disease invasion during evolution. Plant cells contain a group of receptor molecules known as protein recognition receptors (PRRs) [5] that immediately perceive pathogen-associated molecular patterns (PAMPs) and switch on the defense mechanism called PAMP-triggered immunity (PTI) to combat the contagious pathogens [6,7]. PTI serves as the first line of inducible protection against a variety of pathogens and is thought to be a preserved and ancient form of plant immune system [8].
The great pathogenic diversity in M. oryzae frequently causes the quick deterioration of resistant cultivars, whereas traditional rice breeding programs require about 10 years to group numerous blast resistance genes into a single rice variety through repeated crossing and backcrossing [9]. The establishment of a wide range of resistance through PTI enhancement has thus been recommended to be a successful strategy for developing rice blast-resistant types [10]. With the development of RNA interference-based breeding, Rice transcription factor expression is downregulated and has been shown to be an alternate method for improving rice blast resistance [11]. However, RNAi gene expression differs among transgenic plants; therefore, it takes a lot of transgenic plants to find candidates where the transgene is strongly carried out to many generations. In addition, RNAi-derived plants were typically considered transgenic plants and were subjected to stringent processes for regulation.
Recent studies have demonstrated the efficacy of sequence-specific nucleases (SSNs), including clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated Cas9 (CRISPR/Cas9), transcription activator-like effector nucleases (TALENs), and zinc finger nucleases (ZFNs), for editing the genome of plants [12]. With the use of these genome editing technologies, desirable genes can be switched out selectively [12]. In particular, CRISPR/Cas9 has been demonstrated to be the most sophisticated and successful SSN for genome editing in rice yet [13]. With the aid of CRISPR technology, several genes in rice namely OsERF922 [14], OsHAK1 [15], Badh2 [16], TMS5 [17], and OsRR22 [18] genes have been successfully knocked out and obtained the desired phenotype. This technique presents a novel approach to rice breeding. The study’s initial finding reveals how CRISPR/Cas9 technology was used to specifically modify the OsHDT701 gene to increase blast resistance.
2. MATERIALS AND METHODS
2.1. Plant Germination
In our laboratory, we cultivated the superior Indica rice cultivar RP Bio-226. All transgenic plants as well as wild-type RP Bio-226 plants were grown in the fields at Hyderabad under typical growing circumstances in a greenhouse between 28°C and 35°C. Rice callus was grown in a regular nutritional medium for 25 days following germination on a 96-well plate to test for blast resistance at the callus stage [18]. They were put in a growth chamber with 70% relative humidity for 10 h at 20°C during the night time and 14 h at approximately 30°C during the day time under natural sunlight.
2.2. Construction of CRISPR Vector
The sgRNA expression vector (pRGEB31gRNA) and the plant expression gene cassette with Cas9 (pRGEB31CRISPR/Cas9) were purchased from Addgene company [19]. A suitable target sequence was chosen 20 bases upstream region of the protospacer adjacent motif (PAM), in accordance with the design guidelines of the CRISPR/CASs technique [20]. A BLAST scan of the sequences, including PAM, against the rice genome was conducted to establish the targeted sequences’ targeting specificity in the rice genome. The target region containing gRNA-forward/gRNA-reverse primers was synthesized by the Bioserve Company (https://bioserve.in/) and both primers were hybridized by annealing [Table 1].
Table 1: Primer sequence of sgRNA (forward and reverse) with restriction sites.
| PRIMER | SEQUENCE |
|---|---|
| sgRNA–F | 5’ GGTCTC GAGGGAACTTGTCAGCTGAG GAGACC 3’ |
| sgRNA–R | 3’ CCAGAG CTCCCTTGAACAGTCGACTC CTCTGG 5’ |
*The green color nucleotide sequences on both ends of the target gene are the restriction sites of the Bsa1 enzyme.
Following that, the sgRNA-expressing cassette pRGEB31sgRNA was cloned with the target region having chimeric primers using a BsaI site [21]. The hybridized molecule is now called an insert. In comparison with the non-targeted region in the PAM or PAM-proximal area, the targeted specific sequence differs by one base. Through overlapping polymerase chain reaction (PCR), the sgRNA-expression cassette was created. The 5′ ends of the primers were inserted with the sgRNA’s target-specific sequence (target OsHDT701).
2.3. Development of Recombinant Vector
The total length of the CRISPR vector is 15 kb whereas the size of the designed sgRNA is 32 base pairs. Both the CRISPR vector and the sgRNA were subjected to restriction digestion with the BSA1 restriction enzyme. Once the plasmid and the sgRNA were properly digested with the enzyme, a ligation reaction was set up with the vector and insert DNA. The transformation was carried out and the ligated product was then transferred to the top 10 Escherichia coli competent cells through the heat shock method. The plasmid was isolated and run on an electrophoresis unit and a PCR was set as initial denaturation – 94°C for 5 min, cyclic denaturation – 94°C for 30 s, annealing –56°C for 30 s, elongation – 72°C for 30 s, and final elongation – 72°C for 10 min with the universal M13 forward and M13 reverse primers for the recombinant plasmid confirmation [Table 2].
Table 2: Primer used for PCR analysis of recombinant bacterial colonies.
| S. No. | Construct name | Selectable marker | Forward and reverse primer sequence | Annealing temperature | Amplicon size |
|---|---|---|---|---|---|
| 1. | Escherichia coli harboring | M13 | F-5’- CCC AGT | 56°C | 32bp |
| pRGEB31- | CAC GAC GTT | ||||
| HDT701-sgRNA | GTA AAA CG-3’ | ||||
| R- 5’- AGC GGA | |||||
| TAA CAA TTT CAC | |||||
| ACA GG-3’ |
PCR: Polymerase chain reaction.
2.4. Rice Genetic Transformation with Agrobacterium
Using the heat shock method, the Cas9/sgRNA-expressing binary vector PRGEB31 was introduced into the Agrobacterium tumefaciens strain LBA 4404. It was done to transform the embryogenic calli from the RP Bio-226 Indica rice variety using an Agrobacterium. In a nutshell, kanamycin-resistant calli were selected using the kanamycin-containing medium. After that, the selected calli were placed on regeneration media to help transgenic plants grow [15]. After 60–90 days of culture, the transgenic seedlings are transplanted onto a grassland area in the course of the rice harvesting period.
2.5. Identification of Mutated Transgenic Plants
To find the mutation at the desired location, genomic DNA was extracted from the transgenic plants’ leaves using the CTAB procedure. Utilizing PCR mastermix, the isolated DNA with a concentration of 25 ng was employed as the template to conduct PCR. With the help of a particular pair of designed primers that surround the intended target position, PCR was carried out with the initial denaturation –94°C for 5 min, cyclic denaturation – 94°C for 30 s, annealing – 62°C for 30 s, elongation – 72°C for 30 s, and final elongation – 72°C for 10 min for amplifying the genomic area that contains the CRISPR-Cas9 target site. The amplified product was directly sequenced using the Sanger sequencing method. The sequence obtained from both the transgenic as well as the wild-type plants was compared to find out the mutations. The term “homozygote mutation” was used to describe mutations with typical sequencing chromatograms.
With the aid of agarose gel electrophoresis and PCR analysis, plants were separated into T-DNA-free and Cas9-free plants. RPBio-226 DNA as a (+) control and sterile distilled H2O as a (−) control was employed, together with the pRGEB31CRISPR/Cas9 plasmids and the transgenic plants. T-DNA-free plants were those that tested negative for Cas9.
2.6. Green House Trial for Blast Resistance
A blast test was conducted to assess the blast disease in plants at the seedling stage. In the greenhouse, we assessed the blast resistance of 4-week-old wild type and mutated homozygous plants and these 4 weeks plants were subjected to freshwater treatment. Analyzing the plant height, leaf size, leaf shape, leaf length, panicle length, and the leaf response to the blast resistance was measured.
3. RESULTS AND DISCUSSION
3.1. CRISPR/Cas9 Designing
A 20 basepair nucleotide sequence in OsHDT701’s exon number one was selected as the mutation’s targeted region to create a sudden gene change that surely targets OsHDT701 gene sequence in RPBio-226 cv. of rice [Figure 1a]. The gene’s coding region had a predicted Cas9 cleavage site that was 28 bp downside of the start point (ATG) [Figure 2]. The twofold plasmid called Cas9-OsHDT701-gRNA [Figure 1b] was created with the help of the CRISPR-Cas9 gene cassette reported by Ma et al. [22]. Through Agrobacterium-mediated transformation, the vector was employed to modify the rice variety RPBio-226. The plant DNA was isolated and amplified by the DNA that contains the targeted region to assess the effectiveness of OsHDT701 in gene editing. Six kanamycin-resistant transgenic RPBio-226 plants yielded RPBio-226 mutants that could be identified using site-specific PCR analysis and the Sanger sequencing method. Sequencing revealed that there could be a deletion of a single nucleotide.
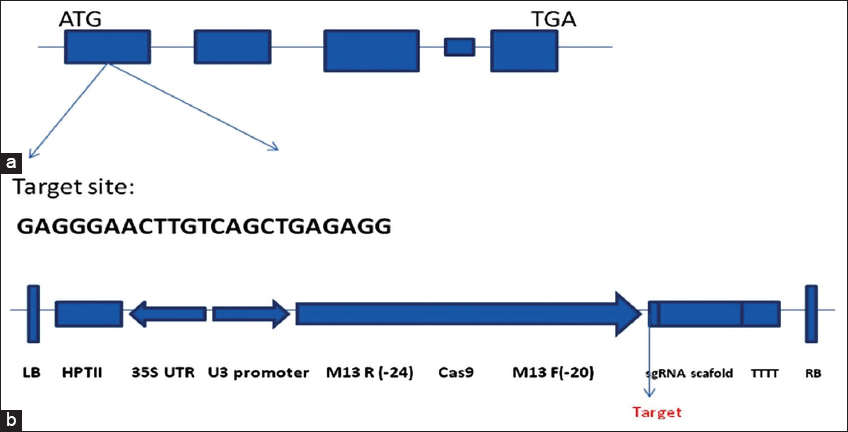 | Figure 1: OsHDT701 gene alteration was caused through CRISPR-Cas9 in the RP Bio-226 rice variety. (a) Structural representation of OsHDT701 gene targeting sequence. (b) Structural representation of transfer-DNA region of the CRISPR-Cas9 vector pRGEB31 targeting construct along with the sgRNA targeting OsHDT701 gene. [Click here to view] |
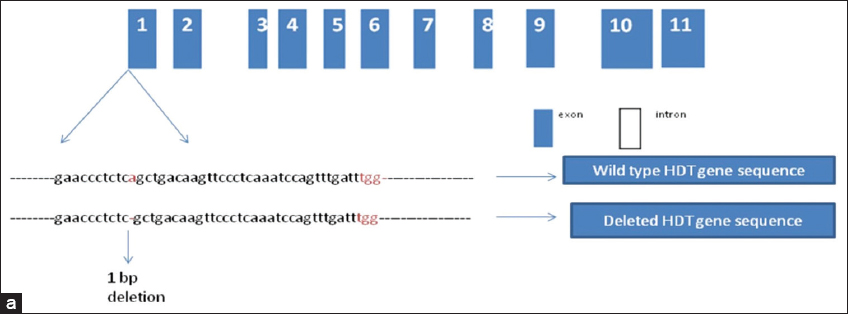 | Figure 2: The target gene’s specifics. (a) Structural representation of the OsHDT gene and the targeted position on the 1st exon to develop sgRNA. [Click here to view] |
The homozygous mutant lines had shown one base pair deletion at the 174th position in the OsHDT701 gene. Allele mutation caused by the OsHDT701 gene was observed in the mutant lines, but the transferred DNA was not present. As a result of this finding, it can be demonstrated that CRISPR/Cas9 has the ability to edit the rice blast gene to produce a mutant rice plan.
3.2. Confirmation of Recombinant Clone
The recombinant plasmid was isolated and when run on the 1.0% agarose gel electrophoresis yielded more than a 15 kb fragment (vector 15kb + insert 32 bp) [Figure 3]. This implies that both the vector as well as the insert have properly ligated along with the restriction sites in the recombinant clone [Figure 4]. A PCR reaction was set up with M13 forward and M13 reverse primers to double-check and confirm the recombinant clone on 1% agarose gel [Figure 5].
 | Figure 3: Confirmation of plasmid DNA and insert. Lane-1: 1 kb DNA ladder; Lane-λ: Hind III λ DNA marker; Lane-1: Plasmid DNA; Lane-2: Insert DNA. [Click here to view] |
 | Figure 4: Restriction digestion of plasmid and recombinant plasmid DNA on 3.5% gel. Lane-1: 1 kb DNA ladder; Lane-λ: Hind III λ DNA marker; Lane-1,2: Plasmid DNA; Lane-3,4: recombinant plasmid DNA. [Click here to view] |
 | Figure 5: Polymerase chain reaction (PCR) confirmation of recombinant plasmid. Lane-1: 50bp DNA ladder; Lane-1,2,3,4: PCR confirmation of insert DNA (32 base pairs). [Click here to view] |
3.3. Selection of Stable Homozygous and CAS Free Edited Lines
We created the sequence targeting primers Cas9p-forward and Cas9p-reverse [Table 3] and conducted PCR for mutant plants to evaluate the feasibility of generating rice lines retaining the necessary alterations in OsHDT701 gene without the T-DNA of the vector pRGEB31 [Figure 2a]. Cas9 PCR results that were negative indicated that there were no transgenes present. A majority of mutant plants obtained are T-DNA-free plants. These findings demonstrated that there is a possibility of obtaining a T-DNA-free homozygous mutant population. To identify the traits associated with blast resistance, we identified homozygous T-DNA-free mutant lines and named them RPBio-226 cas-1, RPBio-226 cas-2, RPBio-226 cas-3, and RPBio-226 cas-4. This finding suggests that the plants without transfer-DNA and with the necessary target sequence changes will be obtained by CRISPR/Cas9-induced gene mutations in the OsHDT701 gene, thus enhancing blast resistance in rice plants.
Table 3: Primer sequence of Cas 9 (forward and reverse).
| PRIMER | SEQUENCE |
|---|---|
| Cas9–F | 5’ AAGCTTTAGCTCCGCCTCCCACCT 3’ |
| Cas9–R | 3’ ACTAGTCTTGGCGGGGTGCTTGGC 5’ |
Developing desirable mutant phenotypes with traditional breeding methods is a difficult exercise that requires producing and screening hundreds of gene-mutated offspring [23]. Gene edition is a regulated target-specific procedure that enables DNA edition with molecular tools, such as ZFNs, TALENs, and CRISPR/Cas. In genome editing technology the binding of proteins and enzymes is different for ZFNs, Talens, and CRISPR. DNA binding protein and the enzyme Fok1 bind ZFNs and TALENS whereas the Cas9 enzyme binds CRISPR forming a CRISPR/Cas9 system. This CRISPR/Cas9 system is a RNA-dependent nuclease system. The whole method of CRISPR depends on the RNA sequence and this helps to guide the breakdown of nuclease enzyme. CRISPR nucleases and Cas9 nucleases are the two components of this system. A CRISPR nuclease sequence consists of repeats that are <50 base pairs in length and are separated by distinct spacers of comparable length. However, Cas9 nuclease is the CRISPR-associated protein that promotes the events of degrading the foreign DNA [24]. The foreign DNA is identified during the pathogen attack and the sequence is incorporated into the CRISPR sequence. A type of bacterial genome impression is developed as a result of this. The CRISPR sequence recognizes the foreign DNA and degrades with the assistance of the Cas9 enzyme [25] when attacked for the 2nd time. This entire DNA degradation process was assisted by the Cas9 enzyme. Given its capacity to cause double-strand breaks in the genome, the CRISPR-Cas9 system is a fantastic tool for use as genomic scissors during gene editing. The preparation of the CRISPR/Cas9 system in the laboratory requires.
a. 20bp guide RNA sequence that is complementary to the target gene sequence
b. A cas9 enzyme that imparts a double-strand break after PAM region (PAM sequence) [26].
The double-stranded CRISPR breaks can be repaired either using a low-fidelity non-homologs end joining (NHEJ) method or a more specialized homology-directed repair system [27]. Because NHEJ is error-prone, it can alter the target sequence by inserting or deleting (InDels), which causes a loss of function and results in genetically modified plants. Transformation, screening, and molecular characterization of the desirable trait will be done once the PAM sequence is identified and the guide RNA is synthesized [28].
A CRISPR/Cas9 mechanism developed with a Cas9 endonuclease and an RNA guide complex has shown significantly improved gene editing efficiency [20]. Although TALENs and CRISPR-Cas9 are both useful technologies for gene alteration, they each have unique benefits and drawbacks. CRISPR-Cas9 expressing gene constructs are substantially simpler in construction which takes just 2–3 days for completion when compared to TALENs which usually take 7–10 days for construction [29]. In addition, compared to TALENs, CRISPR/Cas9 causes a substantially greater incidence of mutations, the frequency of the mutation that was targeted using CRISPR-Cas9 ranges from a percentage of 21–66 for 11 rice genes [30]. Similarly, Ma et al. described that editing 46 target regions in the rice genome using CRISPR-Cas9 had an average mutational rate of 85%. Corn, wheat, rice, and soybeans [31] are just a few of the key crops that have benefited from the widespread usage of CRISPR/Cas9 technology thus far.
3.4. RPBio-226 rice Mutants Showed Increased Resistance to M. oryzae
Homozygous mutant lines were exposed to the fungal microorganism M. oryzae strain B157 during the sapling stage to identify the resistance toward the blast trait of the mutant plants in rice. Due to its highest pathogenic nature the isolate B157 being extremely powerful resisted the infection whereas the wild plants being extremely vulnerable, the leaves were almost dried with the fungal disease [Figure 6] [32]. However, in comparison to wild-type plants, all mutant rice lines had much fewer places where the pathogen had caused lesion formation. By quantifying the lesion regions, the differences were further assessed and it was shown the mutated plants had an improved resistance to the blast disease. At the tillering stage, pathogen-induced lesion lengths were likewise shorter in mutant rice lines than in wild-type plants, and all gene-mutated plants had significant differences from the non-mutated wild plants, according to the lesion length formed. This finding suggests a deletion mutation in the OsHDT701 gene increases M. oryzae resistance in mutated rice plants and that implies the OsHDT701 gene adversely controls rice’s blast resistance.
 | Figure 6: Identification of blast resistance in wild-type and cas-free edited homozygous mutant rice lines (RPBio-226 cas-1, RPBio-226 cas-2, RPBio-226 cas-3, and RPBio-226 cas-4). [Click here to view] |
3.5. The Major Agronomic Features were Unaffected in Induced RPBio-226 Mutant Rice
We examined visually the plant height, leaf size, shape, length, panicle length, and leaf response of all homozygous mutant lines [Figure 6] to ascertain if mutations in the OsHDT701 gene had an impact on agronomic variables. In terms of the agronomic traits examined, we hardly found any prominent difference when both the homozygous mutated plants and wild plants were compared under normal growing conditions.
4. CONCLUSION
New breeding strategies enable scientists to insert the target genes more accurately and quickly than traditional conventional breeding techniques. In this regard, CRISPR/Cas9-based gene editing technology acts as a game-changing technique. The usage of genome editing tools in the field of crop improvement to improve crop yield, nutrition value, disease resistance, etc. will be a focus in future research. It has been used actively over the past 5 years in numerous plant systems for functional research, biotic and abiotic stress management, and to enhance other crucial agronomic traits [33].
The OsHDT701 gene editing in rice was produced with excellent efficiency by the CRISPR/Cas9 method. Studies presented here demonstrate how the CRISPR-Cas9 system and the susceptible S gene can be used to develop fungus-pathogen resistance. The experiment also concludes that the mutant strains lack the T-DNA but carry the required change in the OsHDT701 gene. The resistance to M. oryzae was dramatically increased in T-DNA-free homozygous mutant lines. This study is a good illustration of how CRISPR-Cas9 genome editing plays an important role to increase rice blast resistance. Hence, CRISPR-Cas9 being a brand-new novel technology is an effective method for genome modification. Nonetheless, CRISPR/Cas9-based genome editing will become a popular and necessary technique for obtaining the most suitable edited lines in the field of agriculture that will aid in achieving the “no hunger” phase by feeding the world’s growing population.
5. ACKNOWLEDGMENT
The authors would like to thank Addgene Company for providing the gene construct and also express sincere gratitude to PRR Biotech Innovations Pvt. Ltd. Mehdipatnam, Hyderabad, Telangana, India, for providing the chemicals and laboratory facilities required to carry out the research work successfully.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kim YA, Moon H, Park CJ. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. Oryzae. Rice (N Y) 2019;12:67. [https://doi.org/10.1186/s12284-019-0325-7]
2. Mishra R, Joshi RK, Zhao K. Genome editing in rice:Recent advances, challenges, and future implications. Front Plant Sci 2018;9:1361. [https://doi.org/10.3389/fpls.2018.01361]
3. Srivastava D, Shamim M, Kumar M, Mishra A, Pandey P, Kumar D, et al. Current status of conventional and molecular interventions for blast resistance in rice. Rice Sci 2017;24:299-321. [https://doi.org/10.1016/j.rsci.2017.08.001]
4. Boyd LA, Ridout C, O'Sullivan DM, Leach JE, Leung H. Plant-pathogen interactions: Disease resistance in modern agriculture. Trends Genet 2013;29:233-40. [https://doi.org/10.1016/j.tig.2012.10.011]
5. Kachroo A, Vincelli P, Kachroo P. Signaling mechanisms underlying resistance responses: What have we learned, and how is it being applied? Phytopathology 2017;107:1452-61. [https://doi.org/10.1094/PHYTO-04-17-0130-RVW]
6. Tyagi S, Mulla SI, Lee KJ, Chae JC, Shukla P. VOCs-mediated hormonal signaling and crosstalk with plant growth promoting microbes. Crit Rev Biotechnol 2018;38:1277-96. [https://doi.org/10.1080/07388551.2018.1472551]
7. Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP. Disease resistance mechanisms in plants. Genes (Basel) 2018;9:339. [https://doi.org/10.3390/genes9070339]
8. Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions:Shaping the evolution of the plant immune response. Cell 2006;124:803-14. [https://doi.org/10.1016/j.cell.2006.02.008]
9. Jia Y. Marker assisted selection for the control of rice blast disease. Pestic Outlook 2003;14:150-2. [https://doi.org/10.1039/b308503c]
10. BundóM, Coca M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol J 2015;14:1357-67. [https://doi.org/10.1111/pbi.12500]
11. Liu D, Chen X, Liu J, Ye J, Guo Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 2012;63:3899-911. [https://doi.org/10.1093/jxb/ers079]
12. Weeks DP, Spalding MH, Yang B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J 2016;14:483-95. [https://doi.org/10.1111/pbi.12448]
13. Siddiq EA, Vemireddy LR. Advances in genetics and breeding of rice:An overview. In:Rice Improvement. Cham:Springer;2021. [https://doi.org/10.1007/978-3-030-66530-2_1]
14. Ikeda T, Tanaka W, Mikami M, Endo M, Hirano HY. Generation of artificial drooping leaf mutants by CRISPR-Cas9 technology in rice. Genes Genet Syst 2016;90:231-5. [https://doi.org/10.1266/ggs.15-00030]
15. Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, et al. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 2016;11:e0154027. [https://doi.org/10.1371/journal.pone.0154027]
16. Nieves-Cordones M, Mohamed S, Tanoi K, Kobayashi NI, Takagi K, Vernet A, et al. Production of low-Cs(+) rice plants by inactivation of the K(+) transporter OsHAK1 with the CRISPR-Cas system. Plant J 2017;92:43-56. [https://doi.org/10.1111/tpj.13632]
17. Shao G, Xie L, Jiao G, Wei X, Sheng Z, Tang S, et al. CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in rice. Chin J Rice Sci 2017;31:216-22.
18. Zhou H, He M, Li J, Chen L, Huang Z, Zheng S, et al. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci Rep 2016;6:37395. [https://doi.org/10.1038/srep37395]
19. Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 2013;6:1975-83. [https://doi.org/10.1093/mp/sst119]
20. Zegeye WA, Tsegaw M, Zhang Y, Cao L. CRISPR-based genome editing:Advancements and opportunities for rice improvement. Int J Mol Sci 2022;23:4454. [https://doi.org/10.3390/ijms23084454]
21. Zhang A, Liu Y, Wang F, Li T, Chen Z, Kong D, et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol Breed 2019;39:47. [https://doi.org/10.1007/s11032-019-0954-y]
22. Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, et al. Arobust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 2015;8:1274-84. [https://doi.org/10.1016/j.molp.2015.04.007]
23. Xia H, Huang W, Xiong J, Yan S, Tao T, Li J, et al. Differentially methylated epiloci generated from numerous genotypes of contrasting tolerances are associated with osmotic-tolerance in rice seedlings. Front Plant Sci 2017;8:11. [https://doi.org/10.3389/fpls.2017.00011]
24. Ricroch A, Clairand P, Harwood W. Use of CRISPR systems in plant genome editing:Toward new opportunities in agriculture. Emerg Top Life Sci 2017;1:169-82. [https://doi.org/10.1042/ETLS20170085]
25. Ahmad S, Wei X, Sheng Z, Hu P, Tang S. CRISPR/Cas9 for development of disease resistance in plants:Recent progress, limitations and future prospects. Brief Funct Genomics 2020;19:26-39. [https://doi.org/10.1093/bfgp/elz041]
26. Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 2019;70:667-97. [https://doi.org/10.1146/annurev-arplant-050718-100049]
27. Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, et al. Methodologies for improving HDR efficiency. Front Genet 2019;9:691. [https://doi.org/10.3389/fgene.2018.00691]
28. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 2019;20:490-507. [https://doi.org/10.1038/s41580-019-0131-5]
29. Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, et al. ACRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 2015;169:971-85. [https://doi.org/10.1104/pp.15.00636]
30. Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 2014;12:797-807. [https://doi.org/10.1111/pbi.12200]
31. Ashokkumar S, Jaganathan D, Ramanathan V, Rahman H, Palaniswamy R, Kambale R, et al. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS One 2020;15:e0237018. [https://doi.org/10.1371/journal.pone.0237018]
32. Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 2015;33:41-52. [https://doi.org/10.1016/j.biotechadv.2014.12.006]
33. Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G. CRISPR for crop improvement:An update review. Front Plant Sci 2018;9:985. [https://doi.org/10.3389/fpls.2018.00985