1. INTRODUCTION
Analysis of human mitochondrial DNA (mtDNA) is essential for forensic investigations and population genetics research. Understanding human evolution heavily relies on the study of the frequency and pattern of changes in mtDNA sequences, which have a mutation rate that is 10 times higher than that of the nuclear genome [1]. The mtDNA control region, also known as the hypervariable regions, is a crucial mutational hotspot in the entire genome, comprising three hypervariable regions (HV1, HV2, and HV3). This region is unique in forensics as it is inherited solely from the mother and does not undergo recombination, meaning that all maternal relatives will share the same mtDNA haplotype [2-4]. However, this feature limits the power of discrimination, making it challenging to distinguish between closely related individuals or those with the same haplotype. Despite its limitations, mtDNA analysis is still an available choice when biological evidence is damaged or exhibits mixed short tandem repeat profiles. In such situations, mtDNA offers greater precision and reliability when compared to nuclear DNA analysis [4-6].
The putative genetic structure of the population is an essential component to assess mtDNA match comparison with unrelated individuals. Therefore, the study of population and forensic parameters in a given population, such as the number of haplotypes (H), polymorphic sites (S), nucleotide diversity (π), haplotype diversity (Hd), and haplogroup distribution are an important tool in population and forensic genetics [7-11]. Sequencing of either hypervariable regions or the entire mtDNA may be used to study these parameters.
The consistent advancements in sequencing technology, such as Next-Generation Sequencing (NGS), which allows the examination of the entire mtDNA genome, have led to the development of a substantial forensic mtDNA database. For example, MITOMAP and EMPOP databases are used to analyze the vast majority of mtDNA data collected [12-15]. Nonetheless, the reference mitogenomes and/or control region sequences are either unavailable or insufficient for a variety of Indian populations, including Gujarat.
India is known for its diverse population, encompassing differences in social, linguistic, cultural, geographical, ethnic, and genetic aspects. The population of India can be classified based on caste, tribe, religion, region, and language, with four significant linguistic families: Indo-European, Dravidian, Austroasiatic, and Tibeto-Burman. As a geographical region located at the intersection of Africa, Eurasia, and the Pacific, India served as a corridor for the dispersal of modern humans from Africa around 100,000 years ago [16,17]. Several molecular genetic studies conducted in the late 1990s on Indian populations using high-resolution RFLP and sequencing analysis aimed to comprehend complex relationships between different Indian and worldwide sub-populations. These studies reveal that India’s genetic diversity is higher than other comparable global regions, with variations in mtDNA indicating human dispersal throughout the country during the middle Palaeolithic era [18-20]. Moreover, Recent research has uncovered India’s evolutionary history, encompassing ancient settlements and gene flow from West and East Eurasia, achieved through identifying haplogroups and Indian-specific haplogroups. Genetic relationships among castes, tribes, and communities in India have been investigated, although a limited number of studies have included the state of Gujarat [21-25]. For mtDNA analysis to be useful in forensic investigations, it is important to have a large database of mtDNA profiles from different populations. This database can be used as a reference to compare mtDNA samples obtained from crime scenes or from individuals involved in a case. The unavailability of these data for Gujarat and related populations negatively affects mtDNA-based forensic investigations of cases in which people from such populations are involved. Thus, the present study is an effort to create the necessary data set for the Gujarat population.
Gujarat is the fifth-largest state in the Northwest region of India and the ninth-most populous state overall. It is bounded to the west and southwest by the Arabian Sea and to the north by Pakistan. It has a population of sixty million, which represents 4.99% of India’s total population [26-28]. The population is diverse with 11 Major tribes constituting approximately 15% of the total state population with a history dating back to the Harappan Civilization [29]. The numerous migrations and invasions throughout its history have resulted in a complex admixture with high levels of genetic and phenotypic variation, with a variation among the caste population as high as 40%. Several major haplogroups with the following frequency percentages have also been reported from this region: M (44.1%), U7 (12%), N (2.9%), R* (N) (8.8%), and W(N) (5%) [24].
mtDNA analysis has always been used in forensic and population genetic studies. Thus, the purpose of this study was to analyze the HV1 and HV2 mtDNA sequences of the Gujarat population to generate an mtDNA reference dataset. Furthermore, we investigate genetic variation, identify haplogroups, their frequencies, and geographic origins, as well as estimate forensic and population parameters that can be utilized in population genetic studies and forensic mtDNA typing.
2. MATERIALS AND METHODS
2.1. Population Samples
A total of 5–10 mL of whole blood samples from 72 (n1) maternally unrelated consented individuals from north (N), south (S), central (C) and the Saurashtra (T) regions of Gujarat, were selected for sequencing of the entire mtDNA genome. The participants were evenly split between male and female individuals, with half of the samples collected from each gender. The age range of the participants spanned from 20 to 60 years, and the mean age was calculated to be 34 years. All samples were kept at 4°C until further processing. This study was granted ethical approval by the Institutional Ethical Committee. Along with these samples, HV1 and HV2 regions from 104 (n2) unrelated individuals from our earlier work on the Gujarat population (accession numbers; EMPOP EMP00859 and NCBI OM908544-OM908751) were also considered [30]. As a result, a total of 176 (n1 + n2) samples from Gujarat were considered to analyze HV1 and HV2 of mtDNA for this study. Additionally, mtDNA sequence data were collected for the purpose of inter-population comparative analysis. These data were obtained from published sources and were gathered from three different neighboring states of Gujarat. Figure 1 illustrates the overall number of samples that were collected as well as their distribution.
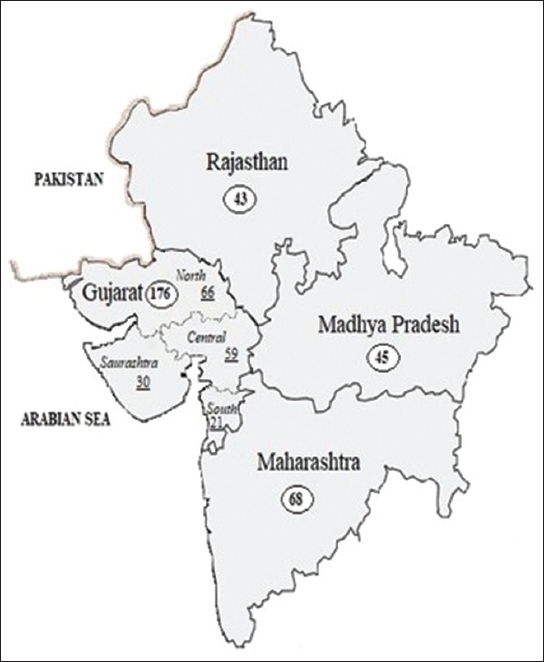 | Figure 1: A schematic map of four states in India displays the total number of samples and their geographic distribution. The number inside the circle represents the total number of samples from the entire state, and the underlined numbers represent number of samples from various regions in Gujarat. [Click here to view] |
2.2. DNA Extraction and Quantitation
Extraction of mtDNA from the 72 samples was carried out immediately after the samples were collected. They were extracted and purified using DNeasy® Blood and Tissue kit (Qiagen, Hilden, Germany) [31]. DNA extractions were carried out in a biosafety chamber (Class II/A2) to avoid contamination of extraneous DNA. Extracted DNA was stored at −20°C until further processing. The eluted DNA samples were quantified using the Quantifiler® Trio DNA Quantification Kit (Applied Biosystems, USA) as per manufacturer’s protocol and analyzed by HID Real-Time PCR Analysis Software V1.2 (Applied Biosystems, USA).
2.3. DNA Amplification and Sequencing
Amplification and sequencing of mtDNA were carried out using kits and reagents provided by Applied Biosystems, USA. The whole mitogenome was sequenced using the Precision ID mtDNA Whole Genome Panel. The panel comprises two pools containing a total of 162 primers and 283 degenerate primers for amplification and sequencing of the entire mtDNA genome. mtDNA library for all samples was prepared by automated workflow on Ion Chef with Precision ID Library kit. The library was quantified using a TaqMan® Quantitation Kit after purification with AMPure™ XP Reagent. Diluted libraries were loaded onto the semiconductor sequencing chip for amplification and sequencing using HID Ion Chef™ and Ion Gene Studio™ S5. Next Generation Sequencing was performed on the Ion Torrent S5™ System as per the manufacturer’s protocol [32]. The NGS data of all samples were analyzed using the Ion Torrent Converge™ v2.1 software (Applied Biosystems, USA). The whole mtDNA genome sequence variants were submitted to the mtDNA population database EMPOP (www.empop.org) as per the guideline [33], for evaluating variations and double-checking designated haplogroups with EMPOP accession number EMP00864 [34]. The FASTA format sequences were submitted to GenBank (accession number OP004728-OP004801).
2.4. Statistical Analysis to Understand the Population Structure of Gujarat
Geneious Prime® 2019.1.2 (Biomatters, USA) was used to align and extract HV1 and HV2 regions from FASTA format sequences. All sequences were assembled by aligning and comparing them to annotated revised Cambridge Reference Sequence (rCRS) [35].
Furthermore, the occurrence of poly-C tracts sequencing errors has previously been demonstrated, where the exact number of cytosine residues is difficult to determine due to variable numbers of cytosines present in these homopolymeric tracts [36-39]. Thus, the number of cytosine residues in these regions was ignored for comparative or population study purposes in accordance with SWGDAM, ISFG, and FBI’s Interpretation Guidelines for mtDNA Sequencing. It was assumed that the number of cytosines in these homopolymeric regions would be the same (as rCRS) across all comparisons [39-42]. Therefore, we reported the pattern and frequency of these tracts in Table S1 of the supplementary material for all samples but omitted them from our statistical population genetics analysis.
Population genetic parameters such as the nucleotide diversity (π), Hd, and the number of haplotypes were computed with Arlequin v3.5.2.2 [43] and DnaSP v.6 [44]. In Arlequin v3.5.2.2, population structure and genetic differentiation were calculated using the analysis of molecular variance (AMOVA) (estimated using 1000 permutations) and pairwise fixation index (Fst). The forensic parameters, including the random match probability and discrimination power, were calculated manually. The random match probability was calculated using the formula (p=ΣX2), where X is the frequency of each observed haplotype [45], while the discrimination power was calculated using the formula (1-ΣX2), where X is the frequency of each observed haplotype [46]. Haplogroups were identified and assigned using EMPOP [34]. The matrilineal relationships within the population, which were determined based on haplogroups are illustrated by constructing a Neighbour Joining tree using the Tamura-Nei model [47] using Geneious Prime® 2019.1.2 software (Biomatters, USA). We used Brinkmann et al. [48] method to manually calculate the maximum and minimum estimates of the probability ratio of obtaining an mtDNA haplotype match within Gujarat and between Gujarat and its other three neighboring states.
3. RESULTS
The majority of studies in the fields of population genetics and forensic science that involve the analysis of mtDNA depend substantially on haplotype and haplogroup analysis. mtDNA haplotypes are the unique combination of variations when aligned to a reference sequence rCRS. The haplogroups are variations in haplotypes that are typically inherited together. Therefore, haplotypes aid in defining haplogroups. And hence, maternally related individuals have similar haplogroups with minimal to no variation in their haplotypes [48-50]. A precise calculation of the Hd, random match probability, discrimination power, haplogroup frequency, and other population and forensic parameters in a particular population can offer significant knowledge, such as the population’s historical background, migration patterns, genetic variation, and can assist with forensic investigations. For instance, lower Hd indicates shared haplotypes among individuals, meaning the more likely it is that two unrelated individuals would share it by chance, rendering a match with this mtDNA type less convincing [6,9,51].
3.1. Intra-population Analysis: Genetic Diversity, Population, and Forensic Parameters
High-quality sequences of mtDNA control region (HV1 and HV2) of 176 individuals were provided to be used as reference data in Gujarat. The mtDNA haplotypes and haplogroups of all individuals are presented in the supplementary material Table S2. Gujarat had a total of 780 polymorphic sites (S), which define 146 unique haplotypes that belonged to 10 distinct haplogroups (M, U, R, N, HV, W, H, T, J, D). A summary of the population’s genetic diversity and forensic parameters of all samples are listed in Table 1.
Table 1: Forensic and population genetic indices (parameters) based on HV1 and HV2 regions for each sub-population samples from Gujarat.
| Parameters | Region | ||||
|---|---|---|---|---|---|
| North (N) | Central (C) | Saurashtra (T) | South (S) | Gujarat (Total) | |
| Sample size | 66 | 59 | 30 | 21 | 176 |
| Number of polymorphic sites (S) | 628 | 770 | 606 | 63 | 780 |
| Nucleotide diversity (π) | 0.0267 | 0.0869 | 0.0484 | 0.0099 | 0.0483 |
| Mean pairwise differences | 26.2340 | 85.4038 | 47.5425 | 9.7952 | 47.4828 |
| Number of haplotypes | 60 | 54 | 29 | 21 | 146 |
| Haplotype diversity (Hd) | 0.9967 | 0.9965 | 0.9977 | 1.0000 | 0.9970 |
| Random match probability | 0.0184 | 0.0204 | 0.0356 | 0.0476 | 0.0056 |
| Discrimination power | 0.9816 | 0.9796 | 0.9644 | 0.9524 | 0.9944 |
The overall nucleotide diversity (π) was 0.0483, indicating a moderate level of genetic diversity throughout the Gujarat region. However, the level of nucleotide diversity differs significantly across the four distinct regions (ranging from 0.0099 to 0.0869), with certain areas exhibiting notably higher levels of diversity compared to others. The Hd was calculated to be 0.99, indicating a high level of genetic variation among the studied subpopulations in Gujarat. In addition, the probability of two randomly selected individuals sharing the same haplotype was assessed and was found to be as low as 0.0184 (N), 0.0204 (C), 0.0356 (T), and 0.0476 (S), while the discrimination power was 0.9816 (N), 0.9796 (C), 0.9644 (T), and 0.9524 (S).
To further evaluate the genetic diversity of the subpopulations, the mean number of pairwise differences (MPD) was calculated. The results indicated that Central Gujarat had the highest MPD (85.403857 ± 37.258705), suggesting that this subpopulation has the highest genetic diversity among all studied subpopulations. In contrast, the southern region of Gujarat exhibited the lowest MPD (9.795238 ± 4.671970), indicating a lower level of genetic diversity compared to the other subpopulations. In addition, demographic parameters such as Fu and Li’s Fs and Tajima’s D were calculated among the four sub-subpopulations in Gujarat. The results indicated a negative value for both Fu and Li’s Fs (−23.9132) and Tajima’s D (−2.1077).
3.2. Haplotypes and Haplogroups Distribution
In the population of Gujarat, the haplogroup with the highest frequency was M (52.27%), followed by U (18.18%) and R (13.64%). The highest number of sub-haplogroups was also found in M, with 43 sub-haplogroups, whereas U contained only 18 sub-haplogroups. The haplogroups D4 and J1b1b were observed only once. Additional information about the frequency of haplogroups and sub-haplogroups in the population is presented in Table 2, while Figure 2 displays a phylogenetic tree (haplogroup tree) depicting matrilineal relationships for the entire population.
Table 2: The detected haplogroups, their frequency, and the geographical origin of the Gujarat population.
| Macro/Sub Haplogroup | Frequency (%) | Macro/Sub | Frequency (%) | Possiblea Origin | Macro/Sub Haplogroup | Frequency (%) | Macro/Sub Haplogroup | Frequency (%) | Possiblea Origin |
|---|---|---|---|---|---|---|---|---|---|
| Haplogroup | |||||||||
| M | 11.364 | Asian | U1a1a | 0.568 | U1a1c1d1 | 0.568 | West Eurasian | ||
| M2a1a | 1.705 | M2b1a | 0.568 | South Asian | U2e1b | 0.568 | U2e2a1a2 | 0.568 | West Eurasian |
| M3a1+204 | 2.273 | M3a1a | 2.244 | South Asian | U4b1a1a1 | 0.568 | U5a1 | 0.568 | West Eurasian |
| M3a1b | 1.136 | M3a2a | 0.568 | South Asian | U5a1b | 0.568 | U5a1b1 | 0.568 | West Eurasian |
| M3d | 1.136 | M3d1 | 1.136 | South Asian | U5a1f1 | 1.136 | U5a2a1 | 0.568 | West Eurasian |
| M4a | 1.705 | M4b | 1.136 | South Asian | U7 | 1.136 | U7a | 5.114 | West Eurasian |
| M5a | 0.568 | M5a1a | 0.568 | South Asian | U7a3b | 1.136 | U7a4a1a | 0.568 | West Eurasian |
| M5a2a | 0.568 | M5a2a1 | 0.568 | South Asian | U2 | 1.136 | U2a | 0.568 | South Asian |
| M5a2a1a | 1.136 | M5a3b | 0.568 | South Asian | U2a1b | 0.568 | U2b2 | 1.705 | South Asian |
| M5a4 | 0.568 | M5b2 | 0.568 | South Asian | Total Freq | 18.182 | |||
| M5b2b | 0.568 | M5c1 | 0.568 | South Asian | |||||
| M6 | 0.568 | M6a1a | 0.568 | South Asian | R | 2.273 | R2 | 1.136 | South Asian |
| M6a1b | 1.136 | M30 | 3.409 | South Asian | R5 | 0.568 | R5a1a | 0.568 | South Asian |
| M30+16234 | 2.273 | M30b | 0.568 | South Asian | R5a2 | 1.136 | R6+16129 | 0.568 | South Asian |
| M30c1 | 0.568 | M30c1a | 0.568 | South Asian | R6a1 | 0.568 | R6a2 | 0.568 | South Asian |
| M30f | 1.705 | M33a1b | 0.568 | South Asian | R6b | 1.136 | R8a1a1a1 | 0.568 | South Asian |
| M33a2 | 0.568 | M33a3 | 0.568 | South Asian | R30a1b | 0.568 | R30a1b1 | 0.568 | South Asian |
| M33b | 0.568 | M37e2 | 0.568 | South Asian | R30b2a | 2.273 | R32 | 1.122 | South Asian |
| M38a | 0.568 | M39 | 1.136 | South Asian | Total Freq | 13.636 | |||
| M39b | 1.136 | M49 | 0.568 | South Asian | |||||
| M52a | 0.568 | M57b | 0.568 | South Asian | N | 3.409 | East Asian | ||
| M57b1 | 1.705 | M65b | 0.568 | South Asian | N1a1b1 | 0.568 | N1a2 | 0.568 | West Eurasian |
| Total Freq | 52.273 | Total Freq | 4.545 | ||||||
| W | 0.568 | West Eurasian | |||||||
| W+194 | 0.568 | W4 | 0.568 | West Eurasian | HV | 2.841 | HV2a | 0.568 | West Eurasian |
| W6 | 0.568 | W6b | 0.568 | West Eurasian | T1a5 | 0.568 | T2b34 | 0.568 | West Eurasian |
| Total Freq | 2.841 | T2d1b | 0.568 | West Eurasian | |||||
| H13a2a1 | 0.568 | H29 | 1.136 | West Eurasian | J1b1b | 0.568 | West Eurasian | ||
| H7b | 0.568 | West Eurasian | D4 | 0.568 | East Asian | ||||
| Total Freq | 2.273 | Total Freq | 6.249 | ||||||
[a] Kyoung, “mtDNA Haplogroup Specific Control Region Mutation Motifs,” Am J Hum Genet, vol. 75, pp. 752–770, 2004. M. van Oven, “PhyloTree Build 17: Growing the human mitochondrial DNA tree,” Forensic Sci. Int. Genet. Suppl. Ser., vol. 5, pp. e392–e394, 2015, doi: https://doi.org/10.1016/j.fsigss. 2015.09.155.
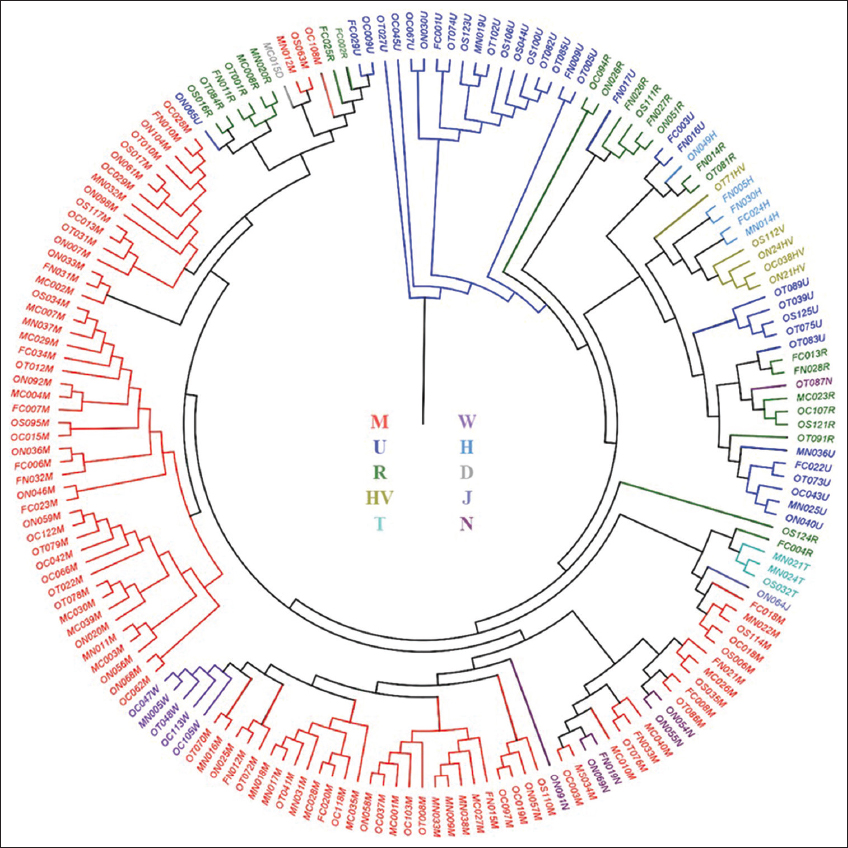 | Figure 2: Phylogenetic relationship of the four geographic regions (Central, North, South and Saurashtra) based on the major mtDNA haplogroups. Different colors represent major haplogroups according to the following: M (red), U (blue), R (green), N (purple), HV (yellow), W (dark violet), H (sky blue), T (cyan), J (violet), D (grey). The second letter of the sample ID at each tip node represent the geographical location in Gujarat: C–Central, N–North, S–South and T–Saurashtra. [Click here to view] |
We observed that the majority of mtDNA lineages in the Gujarat population belong to either the South Asian (Indian) haplogroup M (52.27%) and R (13.64%) or the Western-Eurasian haplogroups H (2.27%), HV (3.41%), T (1.70%), J (0.57%), U (18.18%), and W (2.84%). There was only one individual who belonged to D4 (0.57%), an East Asian haplogroup.
3.3. Inter-population Analysis: Genetic Variation and Population Structure
A comparative analysis of the genetic variation and differentiation was conducted between our population samples, and those from Maharashtra, Rajasthan, and Madhya Pradesh. Figure 1 shows the number of sample population data from the three states obtained from published literature [52]. The sequences from selected regions were downloaded from GenBank (accession numbers: FJ 383814 to FJ 383174). The AMOVA as well as F-statistics (Fst) were calculated from the haplotype frequencies using the Arlequin software. Our findings indicate that genetic variation within populations accounted for 97.57%, while only 2.43% of the variation was observed between populations, as illustrated in Table 3. In addition, the pairwise Fst values, as indicated in Table 4, were both statistically significant and comparable. Gujrat was compared with the three neighboring states and the highest variation in population structure was observed between Gujarat and Rajasthan (Fst 0.02689). The least variation was observed between Gujarat and Madhya Pradesh (Fst 0.0145).
Table 3: Analysis of molecular variance (AMOVA) of four different populations in India.
| Source of variation | Degree of freedom | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among populations | 3 | 4.078 | 0.01223 Va* | 2.43 |
| Within populations | 328 | 160.835 | 0.49035 Vb† | 97.57 |
| Total | 331 | 164.913 | 0.50258 | |
| Fixation Index (Fst) ‡ = 0.02434/P-value=0.000/number of permutations :1023 | ||||
Variance:
* Va: Variance for population among groups,
† Vb: Variance for haplotypes within a population within a group, Fst‡: Permuting haplotypes among populations within groups
Table 4: Analysis of molecular variance; pairwise Fst and probability values for four different populations in India.
| State | Fst value | Gujarat | Madhya Pradesh | Maharashtra | Rajasthan | P-value |
|---|---|---|---|---|---|---|
| Gujarat | 0.00000±0.0000 | 0.00000±0.0000 | 0.00000±0.0000 | |||
| Madhya Pradesh | 0.0145 | 0.00000±0.0000 | 0.00000±0.0000 | |||
| Maharashtra | 0.01952 | 0.03236 | 0.00000±0.0000 | |||
| Rajasthan | 0.02689 | 0.04019 | 0.04509 |
In forensics, it is important to consider matching probability rather than genetic distances [48,53]. Thus, mtDNA sequences from Gujarat were compared to those from its three neighboring states to examine if there were any regional differences that would affect the possibility of finding sequence matches by chance. Table 5 represents the likelihood of finding a match within Gujarat rather than between populations. The maximum probability of finding two distinct haplotypes is 99.97% when sampling from Gujarat and Maharashtra, 99.95% when sampling from Gujarat and Madhya Pradesh, and 99.64% when sampling from Gujarat and Rajasthan. To rephrase, the probability of finding a match within Gujarat is approximately 26.3 times higher than between Gujarat and Maharashtra, 15.8 times higher than between Gujarat and Madhya Pradesh, and 2.2 times higher than between Gujarat and Rajasthan. The lower estimates of mwmin/mbmin for Gujarat- Maharashtra, Gujarat - Madhya Pradesh, and Gujarat - Rajasthan are 7.7 times, 4.6 times, and 0.6 times, respectively.
Table 5: HV1 and HV2 sequence matching probabilities within Gujarat and between Gujarat and neighbouring populations.
| Gujarat (G) | Maharashtra (M) | Madhya Pradesh (MP) | Rajasthan (R) | |
|---|---|---|---|---|
| Na | 176 | 68 | 45 | 43 |
| dwminb | 0.9920 | 0.9485 | 0.9511 | 0.9248 |
| mw maxc | 0.0079 | 0.0515 | 0.0489 | 0.0752 |
| mwmind | 0.0023 | 0.0373 | 0.0273 | 0.0532 |
| mbmine | - | G-M: 0.0003 | G-MP: 0.0005 | G-R: 0.0036 |
| mwmax/mbminf | - | G-M: 26.3 | G-MP: 15.8 | G-R: 2.2 |
| mwmin/mbming | - | G-M: 7.7 | G-MP: 4.6 | G-R: 0.6 |
a Number of samples,
b Minimum diversity within the population (defined as h by Nei 1987)
c Maximum matching probability within the population
d Minimum matching probability within the population
e Minimum matching probability between two populations
f Maximum estimate to find a match within a population than between two populations
g Minimum estimate to find a match within a population than between two populations c-gcalculated as Brinkmann et al. (1999) [b] Nei M, “Molecular evolutionary genetics.” Columbia University Press, New York, P 178, 1987
4. DISCUSSION
Gujarat has a remarkable level of mtDNA diversity, implying that the genetic makeup of the population has been changed over time by a complex interplay of numerous influences. The history of human migration and settlement is thought to be a major driver of genetic variety in the region. Gujarat has been populated for thousands of years and has been a major center of trade and commerce for much of its history, resulting in a mix of cultural and genetic influences from neighboring countries such as West Asia, Central Asia, and East Africa [28,54].
The high Hd observed in the studied subpopulations indicates the presence of relatively few identical or shared haplotypes, with low random match probability and high discrimination power. The limited recent exchange of genes across linguistic and caste boundaries is suggested by the small number of shared haplotypes between the subpopulations [21,55]. Furthermore, this is of significant forensic importance, as it suggests that chance matches may occur in one in a hundred individuals in the North, two in a hundred in the Central, three in a hundred in the Saurashtra, and four in a hundred in the South. Central Gujarat had the highest MPD, which can be attributed to the presence of three major cities: Ahmedabad, Vadodara, and Anand. These cities have been commercial hubs and have attracted immigrants from other states, resulting in higher genetic diversity. Conversely, the southern region of Gujarat had the lowest MPD due to its small size, with the Arabian Sea and the Western Ghats on either side restricting gene flow.
The overall negative values of the demographic parameters (Fu and Li’s Fs and Tajima’s D) observed in all four sub-subpopulations are indicative of recent population expansion or selection. Our study also revealed a high level of Hd and low nucleotide diversity (π). It is possible that a period of fast population growth contributed to the increased stability of rare mutations, as has been suggested in previous studies [56,57].
More than half of Gujarat’s population belongs to the haplogroup M, which accounts for 52.27% of the population. Prior research conducted by Quintana-Murci and colleagues reported that the frequency of this haplogroup in Gujarat was 44.1% [58]. This increase in frequency could be the result of population growth in a larger geographic area.
The haplogroup M, originating from L3, exhibited 14 (13, if M should not be considered) distinct subclades. M30 (motifs;195A, 16223T) and M3 (motif;16126C) superclades were shown to be the most common, accounting for about 33.70% of the M haplogroup. These haplogroups were defined by fast mutations “speedy mutation” at their motif’s sites, and their phylogenetically status has consequently been challenged [58,59]. The M 30 sub-clade has a more recent expansion time at 33,042 YBP [60]. Four samples of M30 with a specific mutation at 16234 branched out, forming M30+16234, previously reported in the Shin population in Pakistan [61]. The second most frequent super-clade M3 was seen more frequently in the North region and the founder age for this haplogroup is less than 25,000 years [52]. M37, M38, M49, and M52 were the least frequent subclades.
The haplogroup U can be considered among the initial maternal founders in Southwest Asia and Europe having subclades older than 30 thousand years [62]. The clade originated from R with the following motifs 11467G, 12308G, and 12372A [63]. Being the second most frequent lineage in India and Europe, it is geographically distributed through North Africa and Central Asia as well [21,58,64]. Similarly, it was also found to be the second most frequent in the population of Gujarat with a frequency of 18.18. The subclade U7 (motifs;152C, 16318T) was found to be the most predominant with a frequency of 7.96 (U7a being the most frequent) which was found previously in Iran, India and Pakistan [24]. This subclade is comparatively recent (16–19 thousand years) with a wide geographical range across Europe, Near East, and South Asia [62]. It is also highly likely to have emanated from Near East [65]. The subclade U2 (motif;16051G) and U5 (motif;16270T) followed behind closely at 5.11 and 3.41 frequency, respectively, with no apparent geographical variation between the four regions. U4 (motifs; 16356C, 195C) subclade was the least frequent in the studied population.
The Western-Eurasian-specific haplogroups H, HV, J, T, N1 and W shows low frequency in the population. These low-frequency haplogroups and their respective lineages are probably quite useful in providing information on the divergence that took place along the route from Eurasia to South Asia [66,67]. The South Asian M and Western-Eurasian U haplogroups account for the vast majority of the population (71.35%), and their distribution is nearly uniform across Gujarat.
The comparative analysis of the genetic variation and differentiation between our population samples and those from Maharashtra, Rajasthan, and Madhya Pradesh, revealed that the genetic variation within populations was higher than between populations. To determine the effect of geographical substructure on forensic investigations, it is desirable to have a cluster with low within-population variation and high between-population variation [52,68]. Our results suggesting that there was no significant genetic divergence among populations. The differences between them are caused by only 2.43% of total variants, indicating substantial gene flow between them. Although the populations exhibited a high degree of genetic similarity (as evidenced by relatively small and similar Fst values), the pairwise Fst values indicated the existence of some genetic differences among the populations. Notably, the highest variation in population structure was observed between Gujarat and Rajasthan, while the least variation was observed between Gujarat and Madhya Pradesh. The substantial genetic differences observed between the populations of Gujarat and Rajasthan can be due to the historical migration patterns into India, which probably occurred through Rajasthan and Gujarat. Considering Rajasthan’s location at the intersection of Africa, Western Eurasia, and Eastern Eurasia, it is probable that the region served as a critical terrestrial pathway for the migration of human populations, leading to substantial genetic diversity [69,70].
The analysis of the forensic parameter, match probability, between Gujarat and the three neighboring states revealed a notable ethnic disparity. The results indicate that it is more likely to find a sequence match within the population of Gujarat than between Gujarat and the other three neighboring populations. This finding underscores the importance of employing micro-geographic sampling in forensic applications to accurately identify individuals based on their DNA profiles. By sampling individuals from smaller geographic regions, the likelihood of finding a match within the same population increases, thereby improving the reliability of DNA evidence in forensic investigations [48,71]. Considering the current status of the mtDNA data on Indian populations and related genetic parameters, the present study provides some advantages and advancements in the current knowledge. One of the major outcomes is the estimation of various population genetics parameters for the mtDNA and to investigate potential relationships between the sub-populations of Gujarat using phylogenetic analyses. Second, we estimated and compared the population genetics structure between Gujarat and the neighboring states for forensic and population genetic analyses. Third, by incorporating population parameters, forensic scientists can ensure that the criminal justice system operates with accuracy and fairness. Finally, our contribution to the global DNA database (EMPOP) provides accessible forensic mtDNA data references for Gujarat, thereby enhancing the accuracy and efficiency of forensic investigations in the region. In addition, this dataset can have implications for other fields like evolutionary biology, anthropology, and medicine. The study was limited in its ability to determine the precise ancestral migration patterns of the haplogroups studied due to a lack of detailed maternal lineage information for the collected samples. The forensic analysis relies on large amounts of high-quality data, thus it is crucial that further research be carried out with rigorous database sample collection and analysis to encompass the other populations of India.
5. CONCLUSION
The results from the current study demonstrated that sequencing hypervariable regions (HV1 and HV2) can reveal a significant amount of information for tracing maternal lineages and distinguishing between unrelated individuals. To the best of our knowledge, few mtDNA data have been released from Gujarat, hence expanding and improving mtDNA sequence databases is crucial for forensic investigation. We have produced a high-quality database, which may be used as a reference for forensic investigations as well as for population genetics research. Our results show a high Hd with a low random match probability which helps in exploring maternal lineage and forensic analysis. The majority of the maternal lineages that we detected in our sample belonged to haplogroup M, which is a haplogroup that is exclusively present in South Asia (India). West Eurasian haplogroups were also observed in the population indicating genetic continuity with the West Eurasian region during the emergence of these haplogroups. The significant negative neutrality test values show that the population had an excess of rare mutations leading to an increase in diversity.
6. ACCESSION NUMBERS
The nucleotide sequences have been submitted to NCBI GenBank® under accession numbers OP004728-OP004801. The dataset generated is accessible in the EMPOP database under accession number EMP00864
7. SUPPORTING INFORMATION
Supplementary data [Tables S1 and S2] associated with this article can be found in the online version.
8. ACKNOWLEDGEMENTS
The authors greatly appreciate the generosity and kind support of Walther Parson. Thank you to our lab mates Blessy Baby, and Kudzanai Joanna Mushavatu.
9. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
10. FUNDING
This work was financially supported by the regular academic grant from National Forensic Sciences University, Gujarat, India. Mohammed H. M Alqaisi would like to acknowledge the Indian Council for Cultural Relations (ICCR) for their financial support for this work.
11. DECLARATION OF COMPETING INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
12. COMPLIANCE WITH ETHICAL STANDARDS
This study was approved by the Ethical Committee of National Forensic Sciences University wide letter no. NFSU/SDSR/IEC/Certificate/73/21 Date: June 03, 2021. All samples were collected with detailed informed consent.
13. DATA AVAILABILITY
The mtDNA sequences are available on EMPOP database with accession number EMP00864. The GenBank accession number for the submitted sequences are from OP004728-OP004801.
14. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Cann RL, Wilson AC. Length mutations in human mitochondrial DNA. Genetics 1983;104:699-711. [CrossRef]
2. Case JT, Wallace DC. Maternal inheritance of mitochondrial DNA polymorphisms in cultured human fibroblasts. Somatic Cell Genet 1981;7:103-8. [CrossRef]
3. Brown WM, Prager EM, Wang A, Wilson AC. Mitochondrial DNA sequences of primates:Tempo and mode of evolution. J Mol Evol 1982;18:225-39. [CrossRef]
4. Budowle B, Allard MW, Wilson MR, Chakraborty R. Forensics and mitochondrial DNA:Applications, debates, and foundations. Annu Rev Genomics Hum Genet 2003;4:119-41. [CrossRef]
5. Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A 1994;91:?-46. [CrossRef]
6. Holland MM, Parsons TJ. Mitochondrial DNA sequence analysis-validation and use for forensic casework. Forensic Sci Rev 1999;11:21-50.
7. Weir BS. Population genetics in the forensic DNA debate. Proc Natl Acad Sci U S A 1992;89:11654-9. [CrossRef]
8. Balding DJ, Nichols RA. DNA profile match probability calculation:How to allow for population stratification, relatedness, database selection and single bands. Forensic Sci Int 1994;64:125-40. [CrossRef]
9. Verscheure S, Backeljau T, Desmyter S. Reviewing population studies for forensic purposes:Dog mitochondrial DNA. Zookeys 2013;365:381-411. [CrossRef]
10. Sultana GN, Tuli JF, Begum R, Tamang R. Mitochondrial DNA control region variation from Bangladesh:Sequence analysis for the establishment of a forensic database. Forensic Med Anat Res 2014;2:95-100. [CrossRef]
11. Hong SB, Kim KC, Kim W. Population and forensic genetic analyses of mitochondrial DNA control region variation from six major provinces in the Korean population. Forensic Sci Int Genet 2015;17:99-103. [CrossRef]
12. Parson W, Strobl C, Huber G, Zimmermann B, Gomes SM, Souto L, et al. Evaluation of next generation mtGenome sequencing using the Ion Torrent Personal Genome Machine (PGM). Forensic Sci Int Genet 2013;7:632-9. [CrossRef]
13. Court DS. Mitochondrial DNA in forensic use. Emerg Top Life Sci 2021;5:415-26. [CrossRef]
14. Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP:A human mitochondrial genome database. Nucleic Acids Res 1996;24:177-9. [CrossRef]
15. Prieto L, Zimmermann B, Goios A, Rodriguez-Monge A, Paneto GG, Alves C, et al. The GHEP-EMPOP collaboration on mtDNA population data--a new resource for forensic casework. Forensic Sci Int Genet 2011;5:146-51. [CrossRef]
16. Cann RL. Genetic clues to dispersal in human populations:Retracing the past from the present. Science 2001;291:1742-8. [CrossRef]
17. Majumder PP. People of India:Biological diversity and affinities. Evol Anthropol 1998;6:100-10. [CrossRef]
18. Bhasin MK, Khanna A. Study of behavioural traits among nine population groups of Jammu and Kashmir, India. J Hum Ecol 1994;5:131-4. [CrossRef]
19. Papiha SS. Genetic variation in India. Hum Biol 1996;68:607-28.
20. Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, et al. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 1999;9:1331-4. [CrossRef]
21. Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 2003;72:313-32. [CrossRef]
22. Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, et al. Genetic evidence on the origins of Indian caste populations. Genome Res 2001;11:994-1004. [CrossRef]
23. Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, et al. Ethnic India:A genomic view, with special reference to peopling and structure. Genome Res 2003;13:2277-90. [CrossRef]
24. Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, et al. Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet 2004;5:26. [CrossRef]
25. Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, et al. Population genetic structure in Indian Austroasiatic speakers:The role of landscape barriers and sex-specific admixture. Mol Biol Evol 2011;28:1013-24. [CrossRef]
26. Government of Gujarat. Gujarat State Portal;2020. Available from:https://gujaratindia.gov.in/state-profile/demography.htm [Last accessed on 2023 Jun 22].
27. Census of India 2011:Provisional Population Totals;2011. Available from:https://censusindia.gov.in/nada/index.php/catalog/1428 [Last accessed on 2023 Jun 22].
28. Patel AB. Traditional bamboo uses by the tribes of Gujarat. Indian J Tradit Knowl 2005;4:179-84.
29. Herman CF. “Harappan“Gujarat?:The archaeology-chronology connection. Paléorient 1996;22:77-112. [CrossRef]
30. Alqaisi MH, Ekka MM, Patel BC. Forensic evaluation of mitochondrial DNA heteroplasmy in Gujarat population. India. Ann Hum Biol 2022;49:332-41. [CrossRef]
31. Qiagen. DNeasy Blood and Tissue Handbook. Germany:Qiagen;2020. 1-62.
32. Fisher Scientific. Precision ID mtDNA Panels with the HID Ion S5 ™/HID Ion GeneStudio™ S5 System:Manual Library Preparation. Hampton:Fisher Scientific;2021.
33. Parson W, Gusmão L, Hares DR, Irwin JA, Mayr WR, Morling N, et al. DNA Commission of the International Society for Forensic Genetics:Revised and extended guidelines for mitochondrial DNA typing. Forensic Sci Int Genet 2014;13:134-42. [CrossRef]
34. Parson W, Dür A. EMPOP--a forensic mtDNA database. Forensic Sci Int Genet 2007;1:88-92. [CrossRef]
35. Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999;23:147. [CrossRef]
36. Bendall KE, Sykes BC. Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet 1995;57:248-56.
37. Ballard D, Winkler-Galicki J, Weso?y J. Massive parallel sequencing in forensics:Advantages, issues, technicalities, and prospects. Int J Legal Med 2020;134:1291-303. [CrossRef]
38. Imaizumi K, Parsons TJ, Yoshino M, Holland MM. A new database of mitochondrial DNA hypervariable regions I and II sequences from 162 Japanese individuals. Int J Legal Med 2002;116:68-73. [CrossRef]
39. Connell JR, Benton MC, Lea RA, Sutherland HG, Haupt LM, Wright KM, et al. Pedigree derived mutation rate across the entire mitochondrial genome of the Norfolk Island population. Sci Rep 2022;12:6827. [CrossRef]
40. Budowle B, Dizinno JA, Wilson MR. Interpretation guidelines for mitochondrial dna sequencing. Proceedings of the tenth international symposium on human identification. Madison, WI:Promega Corporation, 1999:1-9
41. Methods A. Scientific Working Group on DNA Analysis Methods. In:Interpretation Guidelines for Mitochondrial DNA Analysis by Forensic DNA Testing Laboratories;2013. p. 1-26.
42. Connell JR, Benton MC, Lea RA, Sutherland HG, Haupt LM, Wright KM, et al. Evaluating the suitability of current mitochondrial DNA interpretation guidelines for multigenerational whole mitochondrial genome comparisons. J Forensic Sci 2022;67:1766-75. [CrossRef]
43. Excoffier L, Lischer HE. Arlequin suite ver 3.5:A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010;10:564-7. [CrossRef]
44. Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6:DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 2017;34:3299-302. [CrossRef]
45. Stoneking M, Hedgecock D, Higuchi RG, Vigilant L, Erlich HA. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet 1991;48:370-82.
46. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989;123:585-95. [CrossRef]
47. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512-26.
48. Brinkmann C, Forster P, Schürenkamp M, Horst J, Brinkmann B, Rolf B. Human Y-chromosomal STR haplotypes in a Kurdish population sample. Int J Legal Med 1999;112:181-3. [CrossRef]
49. Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 2004;303:223-6. [CrossRef]
50. García-Olivares V, Muñoz-Barrera A, Lorenzo-Salazar JM, Zaragoza-Trello C, Rubio-Rodríguez LA, Díaz-de Usera A, et al. Abenchmarking of human mitochondrial DNA haplogroup classifiers from whole-genome and whole-exome sequence data. Sci Rep 2021;11:20510. [CrossRef]
51. Arora D, Singh A, Sharma V, Bhaduria HS, Patel RB. HgsDb:Haplogroups Database to understand migration and molecular risk assessment. Bioinformation 2015;11:272-5. [CrossRef]
52. Chandrasekar A, Kumar S, Sreenath J, Sarkar BN, Urade BP, Mallick S, et al. Updating phylogeny of mitochondrial DNA macrohaplogroup m in India:Dispersal of modern human in South Asian corridor. PLoS One 2009;4:e7447. [CrossRef]
53. Palo JU, Hedman M, Ulmanen I, Lukka M, Sajantila A. High degree of Y-chromosomal divergence within Finland--forensic aspects. Forensic Sci Int Genet 2007;1:120-4. [CrossRef]
54. Ali M, Liu X, Pillai EN, Chen P, Khor CC, Ong RT, et al. Characterizing the genetic differences between two distinct migrant groups from Indo-European and Dravidian speaking populations in India. BMC Genet 2014;15:86. [CrossRef]
55. Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Rani MV, et al. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet 2001;109:339-50. [CrossRef]
56. Brandstätter A, Peterson CT, Irwin JA, Mpoke S, Koech DK, Parson W, et al. Mitochondrial DNA control region sequences from Nairobi (Kenya):Inferring phylogenetic parameters for the establishment of a forensic database. Int J Legal Med 2004;118:294-306. [CrossRef]
57. Bowen BW, Grant WS. Phylogeography of the sardines (Sardinops spp.):Assessing biogeographic models and population histories in temperate upwelling zones. Evolution 1997;51:1601-10. [CrossRef]
58. Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, et al. Where west meets east:The complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 2004;74:827-45. [CrossRef]
59. Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 2002;71:1150-60. [CrossRef]
60. Rajkumar R, Banerjee J, Gunturi HB, Trivedi R, Kashyap VK. Phylogeny and antiquity of M macrohaplogroup inferred from complete mt DNA sequence of Indian specific lineages. BMC Evol Biol 2005;5:26. [CrossRef]
61. Khan MU, Sabar MF, Baig AA, Naqvi AU, Ghani MU. Forensic and genetic characterization of mtDNA lineages of Shin, a unique ethnic group in Pakistan. Pak J Zool 2021;53:133-41. [CrossRef]
62. Sahakyan H, Kashani BH, Tamang R, Kushniarevich A, Francis A, Costa MD, et al. Origin and spread of human mitochondrial DNA haplogroup U7. Sci Rep 2017;7:46044. [CrossRef]
63. Van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009;30:E386-94. [CrossRef]
64. Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 2000;67:1251-76. [CrossRef]
65. Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R. The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the old world. In:Genomic Diversity. Springer:Boston, MA;1999. 135-52. [CrossRef]
66. Bhatti S, Abbas S, Aslamkhan M, Attimonelli M, Trinidad MS, Aydin HH, et al. Genetic perspective of uniparental mitochondrial DNA landscape on the Punjabi population, Pakistan. Mitochondrial DNA A DNA Mapp Seq Anal 2018;29:714-26. [CrossRef]
67. Li ZY, Wu XJ, Zhou LP, Liu W, Gao X, Nian XM, et al. Late Pleistocene archaic human crania from Xuchang, China. Science 2017;355:969-72. [CrossRef]
68. Roewer L, Croucher PJ, Willuweit S, Lu TT, Kayser M, Lessig R, et al. Signature of recent historical events in the European Y-chromosomal STR haplotype distribution. Hum Genet 2005;116:279-91. [CrossRef]
69. Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin SM, Stoneking M. Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet 2003;11:253-64. [CrossRef]
70. Dada R, Saraswathy KN, Meitei KS, Mondal PR, Kaur H, Kucheria K, et al. Genetic sketch of the six population groups of Rajasthan:A study based on 12 autosomal loci. Anthropol Sci 2011;119:259-64. [CrossRef]
71. Pfeiffer H, Brinkmann B, Hühne J, Rolf B, Morris AA, Steighner R, et al. Expanding the forensic German mitochondrial DNA control region database:Genetic diversity as a function of sample size and microgeography. Int J Legal Med 1999;112:291-8. [CrossRef]
Table S1: Pattern and frequency of poly-C tracts in HV1 and HV2 based on sequencing 72 samples using NGS (Ion Torrent) and 104 samples previously sequenced by the Sanger sequencing method.
| Location | Position | Pattern | No. of C | NGS method (72 samples) | Sanger method (104 samples) | Both methods (Total 176 samples) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||||
| HV1 | 16182, 16183 and 16189 | 16182-(A > C) (A > C) 5C (T > C) 4C A-16194 | 12 | 1 | 1.39 | 0 | 0 | 1 | 0.57 | |
| 16183 and 16189 | 16182-A (A > C) 5C (T > C) 4C A-16194 | 11 | 2 | 2.78 | 1 | 0.96 | 3 | 1.7 | ||
| 16189 | 16182-AA 5C (T > C) 4C A-16194 | 10 | 5 | 6.94 | 2 | 1.92 | 7 | 3.98 | ||
| Total | 11 | 6.25 | ||||||||
| HV2 | 309 | 302-A 7C (ins 1C) T-310 | 7 | 32 | 44.44 | 44 | 42.3 | 76 | 43.18 | |
| 302-A 7C (ins 2C) T-310 | 8 | 2 | 2.78 | 1 | 0.96 | 3 | 1.7 | |||
| Total | 79 | 44.88 | ||||||||
| *315 | 302-A 7C T 5C (ins 1C) G-316 | 13 | 72 | 100 | 104 | 100 | 176 | 100 | ||
N: number of individuals,
* 315.1C is not included in frequency calculations due to its exceptionally high prevalence in the population
Table S2: Mitochondrial DNA HV1 and HV2 sequence haplotypes and haplogroups of Gujarat population.
| Sample ID | Region | Haplogroup | Haplotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FC001U | C | U7a3b | 73G | 151T | 152C | 153G | 263G | 309.1C | 315.1C | 16092C | 16189C |
| FC002R | C | R6a1 | 18T | 73G | 150T | 152C | 228A | 263G | 315.1C | 16129A | 16319A |
| FC003U | C | U1a1c1d1 | 73G | 263G | 285T | 309.1C | 309.2C | 315.1C | 16182C | 16183C | 16189C |
| FC004R | C | R5a1a | 73G | 93G | 200G | 263G | 309.1C | 315.1C | 16145A | 16304C | 16519C |
| FN005H | N | H29 | 93G | 263G | 309.1C | 315.1C | 16319A | 16519C | |||
| FC006M | C | M5a2a | 73G | 146C | 263G | 309.1C | 315.1C | 16129A | 16223T | 16519C | |
| FC007M | C | M6a1a | 73G | 146C | 263G | 315.1C | 16189C | 16209C | 16223T | 16231C | 16311C |
| FC008M | C | M65b | 73G | 241G | 263G | 309.1C | 315.1C | 372.1T | 16223T | 16311C | 16519C |
| FN009M | N | M | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16051G | 16319A | 16519C |
| FN010M | N | M5b2b | 73G | 263G | 315.1C | 16048A | 16129A | 16223T | 16519C | ||
| FN011R | N | R6a2 | 73G | 263G | 315.1C | 16129A | 16213A | 16362C | 16519C | ||
| FN012M | N | M2a1a | 73G | 195C | 204C | 263G | 309.1C | 315.1C | 16223T | 16270T | 16319A |
| FC013R | C | R32 | 73G | 152C | 263G | 315.1C | 16145A | 16185T | 16239T | 16325C | |
| FN014R | N | R8a1a1a1 | 73G | 195C | 243G | 315.1C | 16519C | ||||
| FN015M | N | M3a1a | 73G | 263G | 315.1C | 16126C | 16150T | 16223T | 16519C | ||
| FN016U | N | U1a1a | 73G | 195C | 263G | 285T | 309.1C | 315.1C | 385G | 16183C | 16186T |
| FN017U | N | U2a1b | 73G | 195C | 215G | 263G | 309.1C | 309.2C | 315.1C | 16051G | 16206C |
| FC018M | C | M4b | 73G | 263G | 315.1C | 16086C | 16145A | 16189C | 16223T | 16261T | 16311C |
| FN019N | N | N1a1b1 | 73G | 143A | 199C | 204C | 250C | 263G | 297G | 315.1C | 16223T |
| FC020M | C | M3a1+204 | 73G | 204C | 263G | 309.1C | 315.1C | 16126C | 16223T | 16519C | |
| FN021M | N | M5a2a1 | 73G | 263G | 315.1C | 16170G | 16192T | 16223T | 16301T | 16519C | |
| FC022U | C | U5a1f1 | 73G | 195C | 200G | 263G | 315.1C | 16192T | 16256T | 16270T | 16311C |
| FC023M | C | M30c1a | 73G | 146C | 195A | 263G | 309.1C | 315.1C | 16166del | 16223T | 16519C |
| FC024H | C | H7b | 263G | 315.1C | 16519C | ||||||
| FC025R | C | R30a1b | 73G | 152C | 263G | 309.1C | 315.1C | 16126C | 16181G | 16209C | 16362C |
| FN026R | N | R30b2a | 73G | 152C | 215G | 263G | 309.1C | 315.1C | 373G | 16129A | 16311C |
| FN027R | N | R30b2a | 73G | 263G | 309.1C | 315.1C | 373G | 16292T | 16497G | 16519C | |
| FN028R | N | R32 | 73G | 152C | 263G | 315.1C | 16145A | 16185T | 16239T | 16325C | |
| FC029U | C | U2e2a1a2 | 73G | 152C | 217C | 263G | 315.1C | 16051G | 16092C | 16129C | 16168T |
| FN030H | N | H29 | 93G | 263G | 309.1C | 315.1C | 16319A | 16519C | |||
| FN031M | N | M5a | 73G | 152C | 189G | 195C | 225T | 315.1C | 16129A | 16209C | 16223T |
| FN032M | N | M33a3 | 73G | 146C | 152C | 207A | 263G | 315.1C | 16129A | 16223T | 16271C |
| FN033M | N | M38a | 73G | 246C | 309.1C | 315.1C | 16111T | 16223T | 16239T | 16266T | 16390A |
| FC034M | C | M57a | 73G | 146C | 152C | 263G | 309.1C | 315.1C | 16051G | 16223T | 16311C |
| MC001M | C | M3a2a | 73G | 263G | 309.1C | 315.1C | 16126C | 16169T | 16223T | 16519C | |
| MC002M | C | M2b1a | 73G | 152C | 182T | 195C | 263G | 309.1C | 315.1C | 16169.1C | 16183C |
| MC003M | C | M30f | 73G | 195A | 263G | 309.1C | 315.1C | 16223T | 16368C | 16519C | |
| MC004M | C | M6a1b | 73G | 146C | 263G | 309.1C | 315.1C | 16188T | 16223T | 16231C | 16362C |
| MN005W | N | W6b | 73G | 143A | 189G | 194T | 195C | 204C | 207A | 263G | 309.1C |
| MC007M | C | M57b1 | 73G | 146C | 189G | 263G | 315.1C | 16223T | 16311C | 16519C | |
| MC008R | C | R | 73G | 153G | 189G | 195C | 263G | 315.1C | 16129A | 16362C | 16519C |
| MN009M | N | M3d1 | 73G | 263G | 315.1C | 16126C | 16223T | 16344T | 16519C | ||
| MC010M | C | M33a1b | 73G | 152C | 199C | 263G | 315.1C | 16223T | 16519C | ||
| MN011M | N | M30f | 73G | 195A | 263G | 309.1C | 315.1C | 16223T | 16368C | ||
| MN012M | N | M6 | 73G | 152C | 214G | 263G | 315.1C | 16223T | 16362C | ||
| MN014H | N | H13a2a1 | 263G | 309.1C | 315.1C | 16519C | |||||
| MC015D | C | D4 | 73G | 263G | 315.1C | 16223T | 16362C | ||||
| MN016M | N | M3a1a | 73G | 194T | 195C | 204C | 263G | 315.1C | 16126C | 16192T | 16223T |
| MN017M | N | M3a1+204 | 73G | 150T | 204C | 217C | 263G | 315.1C | 16126C | 16223T | 16519C |
| MN018M | N | M3a1b | 73G | 204C | 217C | 263G | 309del | 315.1C | 16126C | 16223T | 16295T |
| MN019U | N | U7a4a1a | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16309G | 16318C | 16519C |
| MN020R | N | R | 73G | 153G | 189G | 195C | 263G | 315.1C | 16129A | 16362C | 16519C |
| MN021T | N | T2b34 | 41T | 61T | 73G | 263G | 309.1C | 315.1C | 319C | 16126C | 16294T |
| MN022M | N | M4b | 73G | 146C | 263G | 315.1C | 16145A | 16223T | 16234T | 16261T | 16311C |
| MC023R | C | R5 | 64T | 73G | 263G | 309.1C | 315.1C | 16304C | 16524G | 16526A | |
| MN024T | N | T1a5 | 73G | 200G | 263G | 309.1C | 315.1C | 16126C | 16163G | 16186T | 16189C |
| MN025U | N | U5a1b | 73G | 263G | 309.1C | 315.1C | 16192T | 16256T | 16270T | 16399G | |
| MC026M | C | M30b | 73G | 152C | 195A | 263G | 309.1C | 315.1C | 16192T | 16223T | 16278T |
| MC027M | C | M3d | 73G | 263G | 315.1C | 16126C | 16223T | 16311C | 16344T | 16519C | |
| MC028M | C | M3a1a | 73G | 204C | 263G | 315.1C | 16126C | 16223T | 16519C | ||
| MC029M | C | M57b1 | 73G | 146C | 189G | 263G | 315.1C | 16209C | 16223T | 16311C | 16519C |
| MC030M | C | M30 | 73G | 195A | 263G | 315.1C | 16223T | 16519C | |||
| MN031M | N | M3a1b | 73G | 204C | 263G | 315.1C | 16126C | 16223T | 16311C | 16519C | |
| MN032M | N | M5a3b | 73G | 194T | 263G | 309.1C | 315.1C | 16129A | 16223T | 16295T | 16519C |
| MN033M | N | M3d | 73G | 263G | 315.1C | 16126C | 16223T | 16344T | 16519C | ||
| MS034M | S | M | 73G | 199C | 263G | 315.1C | 16093C | 16223T | 16239T | 16304C | 16519C |
| MC035M | C | M3a1a | 73G | 204C | 263G | 315.1C | 16126C | 16223T | 16497G | ||
| MN036U | N | U5a2a1 | 73G | 263G | 309.1C | 315.1C | 16114A | 16192T | 16256T | 16270T | 16294T |
| MN037M | N | M57b1 | 73G | 146C | 189G | 263G | 315.1C | 16223T | 16311C | 16519C | |
| MN038M | N | M3d1 | 73G | 263G | 315.1C | 16126C | 16223T | 16344T | 16519C | ||
| MC039M | C | M30 | 73G | 195A | 263G | 315.1C | 16223T | 16519C | |||
| MC040M | C | M37e2 | 73G | 263G | 309.1C | 315.1C | 16093C | 16111T | 16189C | 16223T | 16224C |
| OT001R | T | R6+16129 | 73G | 263G | 309.1C | 315.1C | 16129A | 16213A | 16362C | 16519C | |
| OC003M | C | M | 73G | 199C | 263G | 315.1C | 16093Y | 16223T | 16239T | 16304C | 16519C |
| OT005U | T | U2 | 73G | 152C | 263G | 309.1C | 315.1C | 16051G | 16207G | 16227G | 16519C |
| OS006M | S | M4a | 73G | 152C | 263G | 315.1C | 16145A | 16176T | 16223T | 16261T | 16311C |
| ON007M | N | M | 73G | 263G | 315.1C | 16093C | 16129A | 16223T | 16362C | 16519C | 16527T |
| OT008M | T | M33a2 | 73G | 263G | 315.1C | 16169T | 16172C | 16223T | 16519C | ||
| OC009U | C | U2e1b | 73G | 152C | 217C | 263G | 315.1C | 315.2C | 340T | 16051G | 16082T |
| OT010M | T | M | 73G | 263G | 315.1C | 16129A | 16223T | 16519C | |||
| OT012M | T | M | 73G | 146C | 263G | 309.1C | 315.1C | 16093C | 16129A | 16223T | 16311C |
| OC013M | C | M | 73G | 263G | 315.1C | 16129A | 16209C | 16223T | 16362C | 16519C | |
| OC015M | C | M52a | 73G | 146C | 263G | 309.1C | 315.1C | 16126C | 16218T | 16223T | 16275G |
| OS016R | S | R6b | 73G | 195C | 246C | 263G | 315.1C | 16145A | 16179T | 16227G | 16245T |
| OS017M | S | M5a2a1a | 73G | 263G | 315.1C | 16129A | 16223T | 16265C | 16519C | ||
| OC018M | C | M4a | 73G | 263G | 309.1C | 315.1C | 16111T | 16145A | 16176T | 16223T | 16261T |
| OC019M | C | M39b | 73G | 153G | 263G | 315.1C | 16075C | 16223T | 16304C | 55.1T | 59del |
| ON020M | N | M30 | 73G | 195A | 263G | 315.1C | 16223T | 16519C | |||
| ON021H | N | HV | 263G | 315.1C | 16356C | 16519C | |||||
| OT022M | T | M30 | 73G | 195A | 225A | 263G | 309.1C | 315.1C | 16223T | 16362C | 16519C |
| OS024H | N | HV | 263G | 315.1C | 16356C | 16519C | |||||
| ON025M | N | M2a1a | 73G | 195C | 204C | 263G | 315.1C | 16223T | 16270T | 16319A | 16352C |
| ON026R | N | R2 | 73G | 152C | 263G | 309.1C | 315.1C | 16071T | 16093C | 16519C | |
| OT027U | T | U7 | 73G | 152C | 200G | 263G | 309.1C | 315.1C | 16093C | 16209C | 16309G |
| OC028M | C | M5b2 | 73G | 263G | 315.1C | 16048A | 16129A | 16223T | 16519C | ||
| OC029M | C | M5a1a | 73G | 263G | 315.1C | 334C | 16129A | 16189C | 16223T | 16265G | 16291T |
| ON030U | N | U7a | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16318T | 16519C | |
| OT031M | T | M | 73G | 263G | 315.1C | 16129A | 16209C | 16223T | 16519C | ||
| OS032T | S | T2d1b | 73G | 150T | 194T | 200G | 263G | 309.1C | 315.1C | 16126C | 16294T |
| ON033M | N | N | 73G | 152C | 195C | 225T | 263G | 315.1C | 16093C | 16129A | 16209C |
| OS034M | S | M | 73G | 146C | 189G | 263G | 309.1C | 315.1C | 16148T | 16223T | 16242T |
| OS035M | S | M | 73G | 152C | 263G | 279C | 309.1C | 315.1C | 16192T | 16223T | 16311C |
| ON036M | N | M5a4 | 73G | 146C | 263G | 315.1C | 16129A | 16223T | 16224C | 16519C | |
| OT037M | C | M | 73G | 263G | 315.1C | 16126C | 16169T | 16183del | 16223T | 16519C | |
| OC038H | C | HV | 263G | 315.1C | 16356C | 16519C | |||||
| OT039U | T | U2b2 | 73G | 146C | 234G | 263G | 315.1C | 16051G | 16093C | 16239T | 16288C |
| ON040U | N | U5a1b1 | 73G | 263G | 315.1C | 16192T | 16256T | 16270T | 16291T | 16399G | |
| OT041M | T | M3a1+204 | 73G | 204C | 217C | 263G | 315.1C | 16126C | 16223T | 16311C | 16519C |
| OC042M | C | M30+16234 | 73G | 195A | 263G | 309.1C | 315.1C | 16223T | 16234T | 16274A | 16519C |
| OC043U | C | U5a1 | 73G | 263G | 315.1C | 16129A | 16192T | 16256T | 16270T | 16399G | |
| OS044U | S | U7a | 73G | 151T | 152C | 263G | 315.1C | 16309G | 16318T | 16519C | 16527T |
| OC045U | C | U7 | 73G | 152C | 263G | 309.1C | 315.1C | 16093C | 16309G | 16318T | 16519C |
| ON046M | N | M30c1 | 73G | 146C | 195A | 263G | 309.1C | 315.1C | 16093C | 16166del | 16223T |
| OC047W | C | W4 | 73G | 143A | 189G | 194T | 195C | 196C | 204C | 207A | 263G |
| OT048W | T | W6 | 73G | 189G | 194T | 195C | 204C | 207A | 263G | 309.1C | 315.1C |
| ON049H | N | HV2a | 72C | 73G | 195C | 263G | 309.1C | 315.1C | 16217C | 16286G | |
| ON051R | N | R30b2a | 73G | 150T | 263G | 309.1C | 315.1C | 373G | 16292T | 16311C | 16497G |
| ON054N | N | N | 73G | 207A | 263G | 309.1C | 315.1C | 16223T | 16256T | 16266T | 16311C |
| ON055N | N | N | 73G | 207A | 263G | 309.1C | 315.1C | 16223T | 16256T | 16266T | 16311C |
| ON056M | N | M30f | 73G | 195A | 200G | 263G | 309.1C | 315.1C | 16126C | 16223T | 16368C |
| ON057M | N | M39 | 55.1T | 59del | 60del | 65.1T | 66T | 73G | 153G | 207A | 263G |
| ON058M | N | M | 73G | 263G | 309.1C | 315.1C | 16126C | 16169T | 16223T | 16519C | |
| ON059M | N | M30+16234 | 73G | 195A | 263G | 309.1C | 309.2C | 315.1C | 16223T | 16234T | 16519C |
| ON061M | N | M5a2a1a | 73G | 263G | 315.1C | 16129A | 16223T | 16265C | 16519C | ||
| OC062M | C | M | 73G | 195A | 263G | 309.1C | 315.1C | 16145A | 16223T | 16271C | 16519C |
| OS063M | S | M | 73G | 152C | 214G | 263G | 315.1C | 16223T | 16327T | 16362C | |
| ON064J | N | J1b1b | 73G | 263G | 271T | 295T | 315.1C | 16069T | 16126C | 16145A | 16261T |
| ON065U | N | U4b1a1a1 | 73G | 195C | 263G | 315.1C | 16356C | 16362C | 16519C | ||
| OC066M | C | M30+16234 | 73G | 152C | 195A | 263G | 315.1C | 16092C | 16223T | 16234T | 16353A |
| OC067U | C | U7a | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16069T | 16274A | 16318T |
| ON068M | N | M30 | 73G | 195A | 263G | 309.1C | 315.1C | 16145A | 16223T | 16311C | 16519C |
| ON069N | N | N1a2 | 73G | 199C | 204C | 263G | 315.1C | 16111T | 16223T | 16291T | 16301T |
| OT070M | T | M | 73G | 194T | 195C | 204C | 263G | 315.1C | 16126C | 16192T | 16223T |
| OT071H | T | HV | 263G | 315.1C | 16354T | ||||||
| OT072M | T | M2a1a | 73G | 195C | 204C | 263G | 315.1C | 16223T | 16270T | 16319A | 16352C |
| OT073U | T | U5a1f1 | 73G | 195C | 200G | 263G | 315.1C | 16192T | 16256T | 16270T | 16311C |
| OT074U | T | U7a3b | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16092C | 16207G | 16256T |
| OT075U | T | U2b2 | 73G | 146C | 152C | 234R | 263G | 309.1C | 315.1C | 16051G | 16209C |
| OT076M | T | M | 73G | 152Y | 246C | 263G | 315.1C | 16111T | 16223T | 16368C | 16519C |
| OT078M | T | M30 | 73G | 195A | 263G | 309.1C | 315.1C | 16179del | 16223T | 16519C | |
| OT079M | T | M30+16234 | 73G | 195A | 263G | 309.1C | 315.1C | 16223T | 16234T | 16519C | |
| OT081R | T | R | 73G | 195C | 263G | 309.1C | 315.1C | 16519C | |||
| OT082U | T | U7a | 73G | 151T | 152C | 263G | 315.1C | 16309G | 16318T | 16519C | |
| OT083U | T | U2a | 73G | 150T | 152C | 194T | 263G | 315.1C | 16051G | 16145A | 16172C |
| OT084R | T | R6b | 73G | 195C | 246C | 263G | 315.1C | 16093C | 16179T | 16227G | 16245T |
| OT085U | T | U7a | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16309G | 16318T | 16519C |
| OT086M | T | M | 73G | 263G | 309.1C | 315.1C | 16223T | 16234T | 16295G | 16311C | 16519C |
| OT087N | T | N | 73G | 152Y | 263G | 309.1C | 315.1C | 16037G | 16111T | 16352C | 16526A |
| OT089U | T | U2 | 73G | 146C | 263G | 315.1C | 16051G | 16086C | 16129A | 16353T | 16519C |
| OT091R | T | R30a1b1 | 73G | 263G | 315.1C | 16209C | 16256T | ||||
| ON092M | N | M6a1b | 73G | 146C | 263G | 309.1C | 315.1C | 16188T | 16223T | 16231C | 16362C |
| OC094R | C | R2 | 73G | 152C | 195C | 249G | 263G | 279C | 315.1C | 16071T | 16519C |
| OS095M | S | M | 73G | 146C | 178G | 263G | 315.1C | 16126C | 16223T | 16519C | |
| ON091R | N | N | 73G | 263G | 309.1C | 315.1C | 16223T | 16327T | 16398A | 16519C | |
| OC097M | C | M39b | 55.1T | 59del | 60del | 65.1T | 66T | 73G | 153G | 263G | 315.1C |
| ON098M | N | M5c1 | 73G | 150T | 263G | 315.1C | 16129A | 16145A | 16223T | 16519C | |
| OS100U | S | U7a | 73G | 151T | 152C | 263G | 315.1C | 16176T | 16309G | 16318T | 16519C |
| OT102U | T | U7a | 73G | 151T | 152C | 263G | 309.1C | 315.1C | 16309G | 16318C | 16519C |
| OC103M | C | M | 73G | 263G | 315.1C | 16126C | 16169T | 16223T | 16519C | ||
| ON104M | N | M | 73G | 263G | 315.1C | 16129A | 16223T | 16519C | |||
| OC105W | C | W | 73G | 189G | 195C | 204C | 207A | 263G | 309.1C | 315.1C | 16223T |
| OS106U | S | U7a | 73G | 151T | 152C | 263G | 315.1C | 16309R | 16318T | 16319A | 16519C |
| OC107R | C | R5a2 | 73G | 146C | 152C | 263G | 315.1C | 16266T | 16304C | 16311C | 16356C |
| OC108M | C | M33b | 73G | 152C | 263G | 315.1C | 16223T | 16324C | 16362C | 16519C | |
| OS110M | S | M39 | 55.1T | 59del | 60del | 65.1T | 73G | 263G | 315.1C | 16223T | 16325C |
| OS111R | S | R30b2a | 73G | 152C | 263G | 309.1C | 315.1C | 373G | 16258C | 16292T | 16497G |
| OS112H | S | HV | 263G | 315.1C | 16217C | 16356C | 16519C | ||||
| OC113W | C | W+194 | 73G | 189G | 194T | 195C | 204C | 207A | 263G | 315.1C | 16223T |
| OS114M | S | M4a | 73G | 146C | 263G | 315.1C | 16145A | 16176T | 16223T | 16234T | 16261T |
| OS117M | S | N | 73G | 263G | 315.1C | 16129A | 16209C | 16223T | 16362C | 16519C | |
| OC118M | C | M3a1+204 | 73G | 204C | 263G | 309.1C | 315.1C | 16126C | 16223T | 16519C | |
| OS121R | S | R5a2 | 73G | 152C | 263G | 309.1C | 315.1C | 16266T | 16304C | 16325C | 16356C |
| OC122M | C | M49 | 73G | 195C | 263G | 315.1C | 16223T | 16234T | 16519C | ||
| OS123U | S | U7a | 73G | 151T | 152C | 263G | 315.1C | 16140C | 16207R | 16242T | 16309G |
| OS124R | S | R | 73G | 263G | 309.1C | 315.1C | 16519C | ||||
| OS125U | S | U2b2 | 73G | 146C | 152C | 234G | 263G | 309.1C | 315.1C | 16051G | 16184T |
| Sample ID | Region | Haplogroup | Haplotype | ||||||||
| FC001U | C | U7a3b | 16207G | 16309G | 16318C | 16519C | |||||
| FC002R | C | R6a1 | 16320T | 16362C | 16393T | 16519C | |||||
| FC003U | C | U1a1c1d1 | 16249C | 16311C | 16519C | 16527T | |||||
| FC004R | C | R5a1a | 16524G | ||||||||
| FN005H | N | H29 | |||||||||
| FC006M | C | M5a2a | |||||||||
| FC007M | C | M6a1a | 16356C | 16362C | 16519C | ||||||
| FC008M | C | M65b | |||||||||
| FN009M | N | M | |||||||||
| FN010M | N | M5b2b | |||||||||
| FN011R | N | R6a2 | |||||||||
| FN012M | N | M2a1a | 16352C | 16519C | |||||||
| FC013R | C | R32 | |||||||||
| FN014R | N | R8a1a1a1 | |||||||||
| FN015M | N | M3a1a | |||||||||
| FN016U | N | U1a1a | 16189C | 16249C | |||||||
| FN017U | N | U2a1b | 16215G | 16230G | 16304C | 16311C | 16519C | ||||
| FC018M | C | M4b | 16519C | ||||||||
| FN019N | N | N1a1b1 | 16311C | 16391A | 16519C | ||||||
| FC020M | C | M3a1+204 | |||||||||
| FN021M | N | M5a2a1 | |||||||||
| FC022U | C | U5a1f1 | 16399G | ||||||||
| FC023M | C | M30c1a | |||||||||
| FC024H | C | H7b | |||||||||
| FC025R | C | R30a1b | 16519C | ||||||||
| FN026R | N | R30b2a | 16497G | 16519C | |||||||
| FN027R | N | R30b2a | |||||||||
| FN028R | N | R32 | |||||||||
| FC029U | C | U2e2a1a2 | 16183C | 16189C | 16362C | 16519C | |||||
| FN030H | N | H29 | |||||||||
| FN031M | N | M5a | 16261T | 16319A | 16355T | 16519C | 16527T | ||||
| FN032M | N | M33a3 | 16399G | 16519C | |||||||
| FN033M | N | M38a | 16519C | ||||||||
| FC034M | C | M57a | 16519C | ||||||||
| MC001M | C | M3a2a | |||||||||
| MC002M | C | M2b1a | 16189C | 16223T | 16274A | 16319A | 16320T | 16399G | 16519C | ||
| MC003M | C | M30f | |||||||||
| MC004M | C | M6a1b | 16519C | ||||||||
| MN005W | N | W6b | 315.1C | 16189C | 16223T | 16292T | 16325C | 16355T | 16519C | ||
| MC007M | C | M57b1 | |||||||||
| MC008R | C | R | |||||||||
| MN009M | N | M3d1 | |||||||||
| MC010M | C | M33a1b | |||||||||
| MN011M | N | M30f | |||||||||
| MN012M | N | M6 | |||||||||
| MN014H | N | H13a2a1 | |||||||||
| MC015D | C | D4 | |||||||||
| MN016M | N | M3a1a | 16312G | 16519C | |||||||
| MN017M | N | M3a1+204 | |||||||||
| MN018M | N | M3a1b | 16519C | ||||||||
| MN019U | N | U7a4a1a | |||||||||
| MN020R | N | R | |||||||||
| MN021T | N | T2b34 | 16296T | 16304C | 16519C | ||||||
| MN022M | N | M4b | 16519C | ||||||||
| MC023R | C | R5 | |||||||||
| MN024T | N | T1a5 | 16294T | 16519C | |||||||
| MN025U | N | U5a1b | |||||||||
| MC026M | C | M30b | 16519C | ||||||||
| MC027M | C | M3d | |||||||||
| MC028M | C | M3a1a | |||||||||
| MC029M | C | M57b1 | |||||||||
| MC030M | C | M30 | |||||||||
| MN031M | N | M3a1b | |||||||||
| MN032M | N | M5a3b | |||||||||
| MN033M | N | M3d | |||||||||
| MS034M | S | M | |||||||||
| MC035M | C | M3a1a | |||||||||
| MN036U | N | U5a2a1 | 16526A | ||||||||
| MN037M | N | M57b1 | |||||||||
| MN038M | N | M3d1 | |||||||||
| MC039M | C | M30 | |||||||||
| MC040M | C | M37e2 | 16295T | 16519C | |||||||
| OT001R | T | R6+16129 | |||||||||
| OC003M | C | M | |||||||||
| OT005U | T | U2 | |||||||||
| OS006M | S | M4a | 16519C | ||||||||
| ON007M | N | M | |||||||||
| OT008M | T | M33a2 | |||||||||
| OC009U | C | U2e1b | 16126C | 16129C | 16183C | 16189C | 16256T | 16298C | 16362C | 16519C | |
| OT010M | T | M | |||||||||
| OT012M | T | M | 16390A | 16519C | |||||||
| OC013M | C | M | |||||||||
| OC015M | C | M52a | 16291T | 16356C | 16390A | 16391A | 16519C | ||||
| OS016R | S | R6b | 16266T | 16278T | 16362C | 16519C | |||||
| OS017M | S | M5a2a1a | |||||||||
| OC018M | C | M4a | 16266T | 16291T | 16311C | 16519C | |||||
| OC019M | C | M39b | 60del | 65.1T | 66T | ||||||
| ON020M | N | M30 | |||||||||
| ON021H | N | HV | |||||||||
| OT022M | T | M30 | |||||||||
| OS024H | N | HV | |||||||||
| ON025M | N | M2a1a | 16519C | ||||||||
| ON026R | N | R2 | |||||||||
| OT027U | T | U7 | 16318T | 16519C | |||||||
| OC028M | C | M5b2 | |||||||||
| OC029M | C | M5a1a | 16519C | ||||||||
| ON030U | N | U7a | |||||||||
| OT031M | T | M | |||||||||
| OS032T | S | T2d1b | 16519C | ||||||||
| ON033M | N | N | 16223T | 16261T | 16319A | 16355T | 16519C | ||||
| OS034M | S | M | 16311C | 16519C | 16527T | ||||||
| OS035M | S | M | |||||||||
| ON036M | N | M5a4 | |||||||||
| OT037M | C | M | |||||||||
| OC038H | C | HV | |||||||||
| OT039U | T | U2b2 | 16352C | 16353T | |||||||
| ON040U | N | U5a1b1 | |||||||||
| OT041M | T | M3a1+204 | |||||||||
| OC042M | C | M30+16234 | |||||||||
| OC043U | C | U5a1 | |||||||||
| OS044U | S | U7a | |||||||||
| OC045U | C | U7 | |||||||||
| ON046M | N | M30c1 | 16519C | ||||||||
| OC047W | C | W4 | 309.1C | 315.1C | 16145A | 16189C | 16223T | 16292T | 16320T | 16519C | |
| OT048W | T | W6 | 16192T | 16223T | 16266T | 16292T | 16325C | 16519C | |||
| ON049H | N | HV2a | |||||||||
| ON051R | N | R30b2a | 16519C | ||||||||
| ON054N | N | N | 16519C | ||||||||
| ON055N | N | N | 16519C | ||||||||
| ON056M | N | M30f | 16519C | ||||||||
| ON057M | N | M39 | 309.1C | 315.1C | 16093Y | 16223T | 16304C | ||||
| ON058M | N | M | |||||||||
| ON059M | N | M30+16234 | |||||||||
| ON061M | N | M5a2a1a | |||||||||
| OC062M | C | M | |||||||||
| OS063M | S | M | |||||||||
| ON064J | N | J1b1b | 16357C | 16519C | |||||||
| ON065U | N | U4b1a1a1 | |||||||||
| OC066M | C | M30+16234 | 16362C | 16519C | |||||||
| OC067U | C | U7a | 16519C | ||||||||
| ON068M | N | M30 | |||||||||
| ON069N | N | N1a2 | 16356C | 16519C | |||||||
| OT070M | T | M | 16312G | 16519C | |||||||
| OT071H | T | HV | |||||||||
| OT072M | T | M2a1a | |||||||||
| OT073U | T | U5a1f1 | 16399G | ||||||||
| OT074U | T | U7a3b | 16318T | 16519C | |||||||
| OT075U | T | U2b2 | 16239T | 16244A | 16274A | 16352C | 16353T | ||||
| OT076M | T | M | |||||||||
| OT078M | T | M30 | |||||||||
| OT079M | T | M30+16234 | |||||||||
| OT081R | T | R | |||||||||
| OT082U | T | U7a | |||||||||
| OT083U | T | U2a | 16206C | 16256T | |||||||
| OT084R | T | R6b | 16266T | 16278T | 16362C | 16519C | 64T | ||||
| OT085U | T | U7a | |||||||||
| OT086M | T | M | |||||||||
| OT087N | T | N | |||||||||
| OT089U | T | U2 | |||||||||
| OT091R | T | R30a1b1 | |||||||||
| ON092M | N | M6a1b | 16519C | ||||||||
| OC094R | C | R2 | |||||||||
| OS095M | S | M | |||||||||
| ON091R | N | N | |||||||||
| OC097M | C | M39b | 16075C | 16223T | 16304C | ||||||
| ON098M | N | M5c1 | |||||||||
| OS100U | S | U7a | |||||||||
| OT102U | T | U7a | |||||||||
| OC103M | C | M | |||||||||
| ON104M | N | M | |||||||||
| OC105W | C | W | 16519C | ||||||||
| OS106U | S | U7a | |||||||||
| OC107R | C | R5a2 | 16524G | ||||||||
| OC108M | C | M33b | |||||||||
| OS110M | S | M39 | |||||||||
| OS111R | S | R30b2a | 16519C | ||||||||
| OS112H | S | HV | |||||||||
| OC113W | C | W+194 | 16292T | 16519C | |||||||
| OS114M | S | M4a | 16311C | 16519C | |||||||
| OS117M | S | N | |||||||||
| OC118M | C | M3a1+204 | |||||||||
| OS121R | S | R5a2 | |||||||||
| OC122M | C | M49 | |||||||||
| OS123U | S | U7a | 16318T | 16362C | 16519C | ||||||
| OS124R | S | R | |||||||||
| OS125U | S | U2b2 | 16209C | 16239T | 16352C | 16353T | |||||
Haplogroup and haplotype of 176 maternally unrelated individuals (104 from our previous study starting with the letter O in sample ID and 72 from the current study) from different regions in Gujarat (N: North Gujarat; T: Saurashtra; C: Central Gujarat; S: South Gujarat)