1. INTRODUCTION
Healthcare-associated infections are a significant public health issue, leading to increased morbidity, mortality, and imposing a large economic burden on society [1,2]. Acinetobacter baumannii is one of the common causes of healthcare-associated infections [2] and is considered an opportunistic pathogen responsible for serious nosocomial infections, such as pneumonia, sepsis, and meningitis [3,4]. In addition, A. baumannii has the ability to form biofilms and can grow in a wide temperature range of 15–44°C with simple nutritional requirements and high resistance to disinfectants and antiseptics [5-7]. These traits allow for the prolonged survival and spread of A. baumannii in the environment.
Carbapenems are the first-line treatment for infections caused by A. baumannii due to their high efficacy and low toxicity [5,8]. However, the emergence and spread of carbapenem-resistant A. baumannii (CRAB) has become a major concern globally and has been classified as a critical pathogen by the World Health Organization (WHO) [2]. CRAB is resistant to many antibiotics, including carbapenems and third-generation cephalosporins [2]. In cases of CRAB infections, last-line antibiotics such as colistin, polymyxin B, or tigecycline are effective, but their high toxicity limits their usage [5,9]. Several studies have reported the effectiveness of combination therapy using anti-virulence agents (inhibitors active against virulence factors of pathogens) and available antibiotics against A. baumannii [3,10]. Further, information on the resistance of clinical isolates of A. baumannii could contribute to the long-term effectiveness of these treatments.
The susceptibility pattern of A. baumannii to antibiotics varies among hospitals [11]. Can Tho General Hospital is a large medical facility that treats patients from various provinces and cities in the Mekong Delta. This study aimed to reports the clinical factors and the antibiotic resistance profile of A. baumannii strains from Can Tho General Hospital. Thereby, we intended to contribute information to improve the effectiveness of the treatment of A. baumannii infections.
2. MATERIALS AND METHODS
2.1. Bacterial Isolation and Species Identification
This study aimed to comprehensively analyze respiratory specimens collected from patients diagnosed with hospital-acquired pneumonia (HAP), including ventilator-associated pneumonia, at Can Tho General Hospital (Vietnam) from April 2021 to April 2022. The specimens were obtained from various departments within the hospital, including the intensive care unit (ICU), the department of internal medicine (DIM), the department of endocrinology (DOE), and other departments. Only one bacterial strain for one patient was selected for the study despite the possibility of obtaining multiple specimens from each patient. The bacterial isolates were identified using a combination of standard microbiological techniques, including Gram staining, catalase and oxidase tests, motility testing, and automated identification using the Vitek 2 GN ID card with the Vitek-2 Compact system (Biomerieux, France) [12-14]. A. baumannii strains are Gram-negative, twitching motility, oxidase negative, and catalase positive [6].
Our study was conducted after obtaining approval from the Scientific Council and the Medical Council of Can Tho University of Medicine with approval number 21.128-DHYDCT.
2.2. The Minimal Inhibitory Concentration of Testing Antibiotics against A. baumannii Isolates
The antimicrobial susceptibility of the bacterial isolates was assessed using the AST-GN67 card with the Vitek-2 Compact automated microbial identification system. The susceptibility interpretation was based on the Clinical Laboratory Standards Institute guidelines 2014 [15]. The antibiotics tested in this study included penicillin agents (ampicillin/sulbactam – SAM and piperacillin/tazobactam – TXP), cephalosporin agents (cefazolin – CZO, ceftazidime – CAZ, ceftriaxone – CRO, and cefepime – FEP), carbapenem agents (imipenem – IPM, ertapenem – ETP, and meropenem – MEM), aminoglycoside agents (gentamicin – GEN and tobramycin – TOB), fluoroquinolone agents (ciprofloxacin – CIP and levofloxacin – LVX), folate pathway inhibition agent (trimethoprim/sulfamethoxazole – SXT), and polymyxin agent (colistin – COL). The bacterial isolates were classified into three categories based on their resistance patterns: Multidrug-resistant (MDR), extensively drug-resistant (XDR), and pan-drug-resistant (PDR). MDR was defined as non-susceptibility to at least one agent in three or more antimicrobial categories, XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories, and PDR was defined as non-susceptibility to all agents in all antimicrobial categories [16]. The A. baumannii strains were designated as carbapenem-resistant A. baumannii (CRAB) when they were non-susceptible to both imipenem and meropenem [8,15].
2.3. Polymerase Chain Reaction (PCR) Detection Carbapenemase-encoding Genes of CRAB Isolates
The presence of the blaOXA-23, blaOXA-51, and blaIMP genes was determined in all CRAB isolates. Based on the previous research results in Vietnam (data not show), the blaOXA-51 and blaOXA-23 genes represent class D beta-lactamase biosynthetic genes. The blaIPM gene represents the class B beta-lactamase biosynthetic gene. Genomic DNA was extracted from the cells using the Invisorb® Spin Universal kit (Invitek Molecular, Germany). The PCR amplification was carried out using the MyTaq™ Red Mix Kit (Meridian Bioscience, USA) and the C1000 Touch Thermal Cycler system (Bio-Rad, USA), and the reaction mixture included specific primers [Table 1] for each gene described by Woodford et al. [17]. The PCR amplification conditions included an initial denaturation step at 95°C for 3 min, followed by 40 cycles of amplification at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The final extension step was maintained at 72°C for 5 min [18]. The PCR products were analyzed through a 2% agarose gel and visualized using the gel documentation device (Bio-Rad, USA).
Table 1: Primers used to identify carbapenemase-encoding genes in this study [17].
| Gene | Primers sequence (5’→3’) | Tm (°C) | Product (bp) |
|---|---|---|---|
| blaOXA-23 | |||
| Forward primer | GATCGGATTGGAGAACCAGA | 56.72 | 501 |
| Reverse primer | ATTTCTGACCGCATTTCCAT | 55.69 | |
| blaOXA-51 | |||
| Forward primer | TAATGCTTTGATCGGCCTTG | 58.93 | 353 |
| Reverse primer | TGGATTGCACTTCATCTTGG | 56.50 | |
| blaIMP | |||
| Forward primer | GGAATAGAGTGGCTTAAYTCT | 50.37 | 189 |
| Reverse primer | CCAAACYACTASGTTATCTC | 44.39 |
F: Forward primer, R: Reverse primer
2.4. Statistical Analysis
The difference in detection rates of bacterial strains in categorical variables was analyzed using the Chi-square test, which was implemented using the R programming statistical package.
3. RESULTS
3.1. The Epidemiological Characteristics of Isolated A. baumannii Strains
The study aimed to investigate the prevalence of bacterial infections and the specific role of A. baumannii in such infections. The study analyzed 434 patient samples, with a majority of positive cases caused by A. baumannii (31.1%, 135/434 case), followed by Klebsiella pneumoniae (23.2%) and Escherichia coli (20.5%). The study highlights the need for greater attention toward A. baumannii as a potential source of infection.
The researchers also examined the prevalence of A. baumannii infections among different age and gender groups [Figure 1]. The results showed that elderly patients (60+ years) had the highest positive rate for A. baumannii infections, with no infections observed among children aged 0–17 years. In addition, the prevalence of A. baumannii infections was higher among females than males, with 51.85% of all cases occurring in females (P > 0.05).
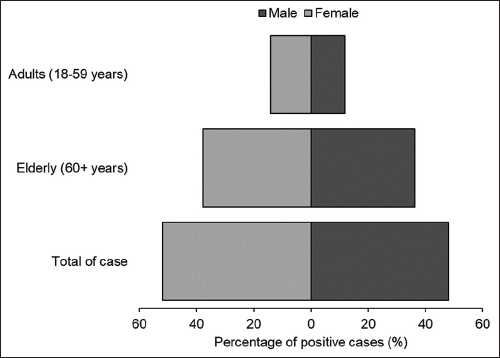 | Figure 1: Positive percentage of Acinetobacter baumannii infections by gender and age. [Click here to view] |
Furthermore, the study explored the distribution of A. baumannii infections in different medical departments, with the ICU showing the highest positive rate (62.9%), followed by the DIM (12.5%) and the DOE (13.3%). This emphasizes the importance of identifying departments with high-positive rates to facilitate targeted infection control measures [Figure 2].
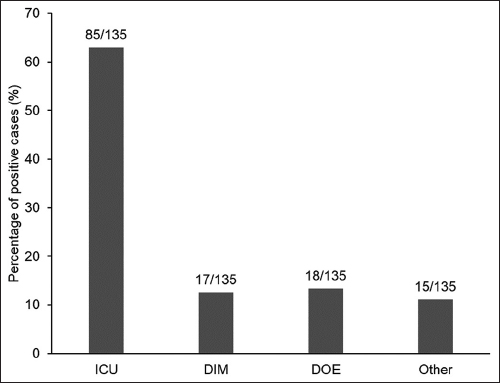 | Figure 2: Prevalence of Acinetobacter baumannii infections by department. [Click here to view] |
3.2. Profile of Antibiotic Resistance Phenotypes of A. baumannii Strains
This study investigated the resistance of A. baumannii strains to various classes of antibiotics. A total of 135 A. baumannii strains were analyzed, and they were classified based on their susceptibility to different antibiotics [Figure 3]. The results showed that a high proportion of strains were resistant to most of the antibiotics tested, including carbapenems, penicillins, cephalosporins, aminoglycosides, and fluoroquinolones. In the carbapenems class, more than 86% of the strains were resistant to imipenem, ertapenem, and meropenem. Similarly, in the penicillins class, more than 85% of the strains were resistant to ampicillin/sulbactam and piperacillin/tazobactam. All strains in the cephalosporin class were resistant to cefazolin, while almost 90% were resistant to ceftazidime, ceftriaxone, and cefepime. A high proportion of strains were also resistant to aminoglycosides and fluoroquinolones. Only colistin and trimethoprim/sulfamethoxazole showed some efficacy, but even so, some strains were resistant to them.
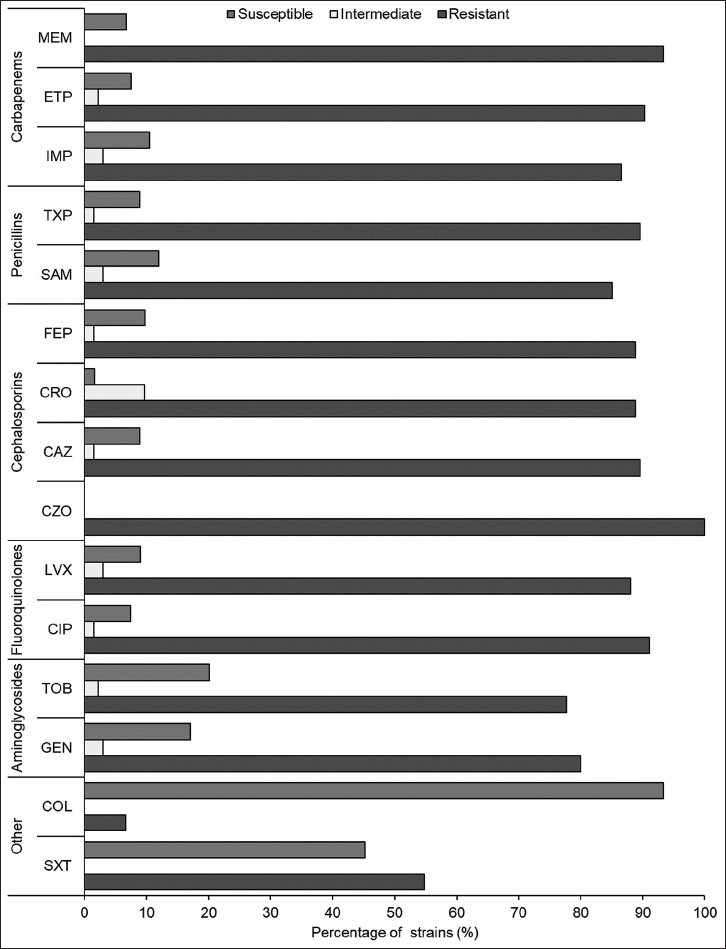 | Figure 3: Antibiotic susceptibility profiles of bacterial phenotypes (%). [Click here to view] |
The distribution of antibiotic-resistant strains was presented, and among the 135 clinical isolates, 77.04% were found to be XDR, while 9.63% were MDR. Nine isolates were pan-drug-resistant (PDR), and the remaining nine were non-resistant. The high prevalence of XDR strains suggests that A. baumannii infections may be difficult to treat, and alternative treatment options are needed. This study highlights the urgent need for the development of new antibiotics to combat antibiotic-resistant A. baumannii strains. Figure 3 shows the susceptibility of A. baumannii strains to various antibiotics, and Figure 4 shows the distribution of antibiotic-resistant strains.
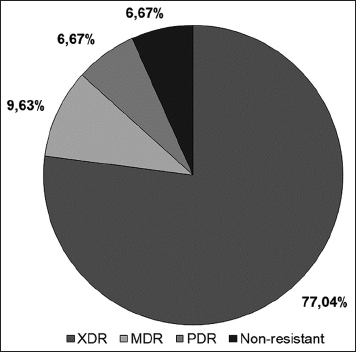 | Figure 4: Antibiotic resistance polymorphism (%) in clinical isolates. The percentage of each category is presented, including extensively drug-resistant, multidrug-resistant, pandrug-resistant, and non-resistant isolates. [Click here to view] |
3.3. Antibiotic Resistance Phenotyping and Carbapenemase-encoding Genes Profile of CRAB Strains
Carbapenem-resistant A. baumannii (CRAB) infections pose a major challenge due to antibiotic resistance. In this study, 117 CRAB strains were genotyped for resistance to imipenem and meropenem antibiotics, with 86.7% (n = 117) found to be resistant to both antibiotics. Analysis of antibiotic resistance polymorphism in four different hospital departments showed that the ICU department had the highest prevalence of XDR strains (63.2%), while the DOE department had the highest prevalence of MDR strains (3.41%). Overall, the findings indicate the need for increased surveillance and improved infection control measures in the ICU department to prevent the spread of antibiotic-resistant CRAB strains. In addition, the results suggest that tailored treatment options are necessary for patients infected with CRAB strains in different hospital departments, based on observed patterns of antibiotic resistance polymorphism [Figure 5].
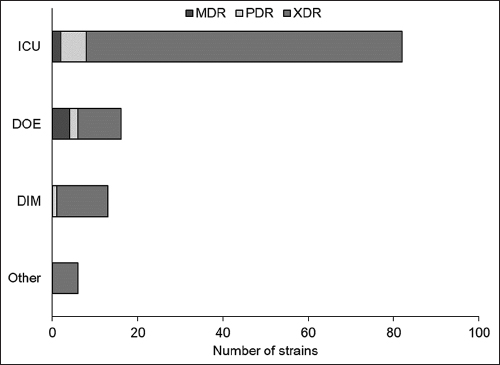 | Figure 5: Antibiotic resistance polymorphism of carbapenem-resistant Acinetobacter baumannii strains by department. The three categories of antibiotic resistance are multidrug-resistant, pandrug resistant, and extensively drug resistant (XDR). The figure reveals that the intensive care unit department has the highest number of XDR strains, followed by the department of internal medicine and the department of endocrinology the departments. The other department has only six strains, and all of them fall under the XDR category. [Click here to view] |
Further, analysis of the CRAB strains revealed that 76.9% carried at least one of the tested resistance genes, with blaOXA-23 and blaOXA-51 genes being the most prevalent. The combination of blaOXA-23 and blaOXA-51 genes was detected in 83.76% of the strains. XDR strains were found to have the most common profile of the presence of blaOXA-23 and blaOXA-51 genes, followed by the blaOXA-23/blaOXA-51 combination, and the blaOXA-23/blaOXA-51/blaIMP combination. Among the MDR strains, only the blaOXA-23/blaOXA-51/blaIMP combination was detected in one strain, and among the PDR strains, the most common profile was the blaOXA-23/blaOXA-51 combination, followed by the blaOXA-23/blaOXA-51/blaIMP combination [Figure 6].
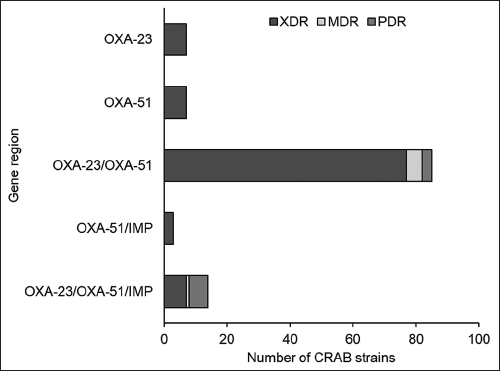 | Figure 6: Antibiotic resistance polymorphism of carbapenem-resistant Acinetobacter baumannii strains by gene region. OXA-23 is blaOXA-23, OXA-51 is blaOXA-51, and IMP is blaIMP. [Click here to view] |
4. DISCUSSION
In this study, respiratory specimens were positive for A. baumannii, Pseudomonas sp., and Enterobacteriaceae. These groups of bacteria are reported by the WHO to be in the critical group in hospital-associated infections [2]. ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter species) are bacteria involved in nosocomial infections of concern worldwide [19]. Among them, carbapenem resistance was most commonly reported in A. baumannii [20]. The carbapenem resistance phenotype is very common for MDR and XDR A. baumannii strains [21]. The immune system in the elderly is not responding as strongly as in young adults as determined by impaired B- and T-cell production in the bone marrow and thymus and impaired function of mature lymphocytes in secondary lymphoid tissues [22]. Most of the patients over 60 years of age in the study were hospitalized inpatients. Weakened immune system, underlying medical conditions, and hospital stay increase the risk of HAP in patients [23]. The intensive care unit was reported to have a high prevalence of this bacterium [24,25]. Patients admitted to the ICU are in poor health, often require prolonged hospital stays, undergo invasive procedures, and are treated with broad-spectrum antibiotics [11,25]. In addition, the PDR A. baumannii strains were isolated from the patient treated for SARS-CoV-2 in the ICU. Cases of secondary infection with SARS-CoV-2 were reported in several studies. A. baumannii was reported to account for a high proportion of secondary Gram-negative bacteremia [26]. A. baumannii carrying the blaOXA-23 gene has caused outbreaks in patients positive for SARS-CoV-2 in multiple intensive care units in Japan [26].
Carbapenems (especially meropenem and imipenem) are considered the first-line treatment for A. baumannii infections. However, the non-guided use of antibiotics led to the emergence of CRAB. CRAB has spread to Asia-Pacific, the Indian continent, North America, Latin America, and Europe [27]. The mechanism of antibiotic resistance in A. baumannii is complex, and coexist multiple virulence factors [7]. Resistance of A. baumannii can be influenced by different virulence factors, including – but not limited to, beta-lactam-degrading enzymes, and/or non-enzymes [25]. Genes encoding Group D carbapenemases could be found on chromosomes and plasmids but not on integrons [7]. Group B carbapenemases are a group of metal-bound enzymes. These enzymes were encoded by mobile genetic elements (plasmid and integron) [26]. Genes encoding class D beta-lactamases are common in A. baumannii, including blaOXA-23, ISAb1-blaOXA-51, blaOXA-58, blaOXA-40, and blaOXA-143 considered as the mechanism responsible for carbapenem resistance in A. baumannii [20,28].
Non-enzymatic mechanisms are primarily related to changes in membrane permeability, and/or to the action of membrane protein pumps/channels. The activity of reducing membrane permeability, and the presence of capsular polysaccharides, lipopolysaccharides, phospholipases, and outer membrane vesicles, were reported to influence the multidrug resistance of A. baumannii [24,25]. Reduction in the number of porin channels in the outer membrane, overexpression of MDR pumps, and changes in the characteristics of penicillin-binding proteins, increased multidrug resistance in A. baumannii [7,24].
In this study, A. baumannii strains showed a high rate of resistance to testing antimicrobial agents. The isolates were resistant to most antibiotics belonging to the groups of penicillins, cephalosporins, carbapenems, aminoglycosides, and fluoroquinolones. Carbapenem resistance in A. baumannii is increasing in many areas. Ababneh reported 90.6% of A. baumannii isolates in Jordanian hospitals as CRAB, of which 10.4% were MDR and 84.4% were XDR. The percentage of strains carrying the blaOXA-51 gene was highest (89,5%), followed by blaOXA-23 (88.3%) and New Delhi Metallo-β-lactamase (blaNDM-1) (10.4%) [29]. In other studies, the blaOXA-23 gene was reported to be the most common and dominant gene in CRAB [21,28]. In particular, the spread of the blaOXA-23 gene is reported to gradually replace the prevalence of blaOXA-58 in A. baumannii in the ICU in several regions of Italy [28]. Most of the isolates carried the blaOXA-23 gene. Pulsed-field gel electrophoresis analysis showed cross-contamination of A. baumannii in patients and the environment. In Vietnam, a marked increase in carbapenem-resistant and MDR A. baumannii has been recorded in many different hospitals. Drug resistance of clinical A. baumannii strains is mainly due to the action of the blaOXA-23 gene [30-32]. In addition, the factor ISaba1 was reported to increase the expression of blaOXA genes in A. baumannii strains [32,33]. MDR in A. baumannii, especially in Vietnam, represents a concern in controlling this pathogen. Therefore, an assessment of antibiotic resistance patterns in each medical facility is necessary.
5. CONCLUSIONS
The study showed that A. baumannii is the leading cause of infections, particularly in patients over 60 years old in both genders, indicating the need for effective infection control measures, especially in ICUs. Colistin, a member of the polymyxin class of antibiotics, was found to be effective against A. baumannii, but resistance to various classes of antibiotics, including carbapenems, was also observed. The prevalence of carbapenem-resistant A. baumannii (CRAB) strains was highest in the ICU, with a significant proportion of XDR strains. The blaOXA-23 and blaOXA-51 genes were found to be the most prevalent resistance genes in CRAB strains, with the combination of blaOXA-23 and blaOXA-51 genes being the most common. This study emphasizes the importance of developing tailored treatment options for CRAB strains based on observed patterns of antibiotic resistance polymorphism in different hospital departments.
6. ACKNOWLEDGMENTS
We would like to express our sincere gratitude to Can Tho University of Medicine and Pharmacy and Can Tho General Hospital for their invaluable support throughout the research project. Their assistance included providing us with access to essential samples and testing facilities, which were crucial for the successful completion of this study. We would also like to extend our heartfelt appreciation to all individuals who contributed to the project and offered their assistance in various research endeavors. Their valuable contributions have greatly enriched our study.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. FUNDING
This research project was funded by Can Tho University of Medicine and Pharmacy and its advisory members (Project code: 840/QD-DHYDCT).
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Son HJ, Cho EB, Bae M, Lee SC, Sung H, Kim MN, et al. Clinical and microbiological analysis of risk factors for mortality in patients with carbapenem-resistant Acinetobacter baumannii Bacteremia. Open Forum Infect Dis 2020;7:ofaa378. [CrossRef]
2. World Health Organization. WHO Publishes List of Bacteria for which New Antibiotics are Urgently Needed. Geneva:World Health Organization;2017.
3. Law SK, Tan HS. The role of quorum sensing, biofilm formation, and iron acquisition as key virulence mechanisms in Acinetobacter baumannii and the corresponding anti-virulence strategies. Microbiol Res 2022;260:127032. [CrossRef]
4. Cha MH, Kim SH, Kim S, Lee W, Kwak HS, Chi YM, et al. Antimicrobial resistance profile of Acinetobacter spp. Isolates from retail meat samples under Campylobacter-selective conditions. J Microbiol Biotechnol 2021;31:733-9. [CrossRef]
5. Ayenew Z, Tigabu E, Syoum E, Ebrahim S, Assefa D, Tsige E. Multidrug resistance pattern of Acinetobacter species isolated from clinical specimens referred to the Ethiopian Public Health Institute:2014 to 2018 trend anaylsis. PLoS One 2021;16:e0250896. [CrossRef]
6. Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, De Vos P, et al. Bergey's Manual®of Systematic Bacteriology. 2nd ed. Boston, MA:Springer New York;2005. [CrossRef]
7. Garnacho-Montero J, Amaya-Villar R. Multiresistant Acinetobacter baumannii infections:Epidemiology and management. Curr Opin Infect Dis 2010;23:332-9. [CrossRef]
8. Chang Y, Luan G, Xu Y, Wang Y, Shen M, Zhang C, et al. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol 2015;6:910. [CrossRef]
9. Labarca JA, Salles MJ, Seas C, Guzmán-Blanco M. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit Rev Microbiol 2014;42:276-92. [CrossRef]
10. Lim J, Hong J, Jung Y, Ha J, Kim H, Myung H, et al. Bactericidal effect of cecropin a fused endolysin on drug-resistant gram-negative pathogens. J Microbiol Biotechnol 2022;32:816-23. [CrossRef]
11. Gupta N, Gandham N, Jadhav S, Mishra R. Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J Nat Sci Biol Med 2015;6:159-62. [CrossRef]
12. Jakovac S, Goic-Barisic I, Pirija M, Kovacic A, Hrenovic J, Petrovic T, et al. molecular characterization and survival of carbapenem-resistant Acinetobacter baumannii isolated from hospitalized patients in Mostar, Bosnia and Herzegovina. Microb Drug Resist 2021;27:383-90. [CrossRef]
13. Yin D, Guo Y, Li M, Wu W, Tang J, Liu Y, et al. Performance of VITEK 2, E-test, Kirby-Bauer disk diffusion, and modified Kirby-Bauer disk diffusion compared to reference broth microdilution for testing tigecycline susceptibility of carbapenem-resistant K. pneumoniae and A. baumannii in a multicenter study in China. Eur J Clin Microbiol Infect Dis 2021;40:1149-54. [CrossRef]
14. Lee CM, Kim YJ, Jung SI, Kim SE, Park WB, Choe PG, et al. Different clinical characteristics and impact of carbapenem-resistance on outcomes between Acinetobacter baumannii and Pseudomonas aeruginosa bacteraemia:A prospective observational study. Sci Rep 2022;12:8527. [CrossRef]
15. CLSI. Performance Standards for Antimicrobial Susceptibility Testing:Twenty-forth Informational Supplement. Wayne, PA:Clinical and Laboratory Standards Institute;2014.
16. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria:An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18:268-81. [CrossRef]
17. Woodford N, Ellington M, Coelho J, Turton J, Ward M, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006;27:351-3. [CrossRef]
18. Le TS, Nguyen PT, Nguyen-Ho SH, Nguyen TP, Nguyen TT, Thai MN, et al. Expression of genes involved in exopolysaccharide synthesis in Lactiplantibacillus plantarum VAL6 under environmental stresses. Arch Microbiol 2021;203:4941-50. [CrossRef]
19. De Oliveira DM, Forde BM, Kidd TJ, Harris PN, Schembri MA, Beatson SA, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 2020;33:e00181-19. [CrossRef]
20. Hu S, Niu L, Zhao F, Yan L, Nong J, Wang C, et al. Identification of Acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci Rep 2019;9:17888. [CrossRef]
21. Ramadan RA, Gebriel MG, Kadry HM, Mosallem A. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa:Characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect Drug Resist 2018;11:1261-9. [CrossRef]
22. Montecino-Rodriguez E, Berent-Maoz B, Dorshkind K. Causes, consequences, and reversal of immune system aging. J Clin Investig 2013;123:958-65. [CrossRef]
23. Lynch JP 3rd. Hospital-acquired pneumonia:Risk factors, microbiology, and treatment. Chest 2001;119:373S-84. [CrossRef]
24. Jia W, Li C, Zhang H, Li G, Liu X, Wei J. Prevalence of genes of OXA-23 carbapenemase and AdeABC efflux pump associated with multidrug resistance of Acinetobacter baumannii isolates in the ICU of a comprehensive hospital of Northwestern China. Int J Environ Res Public Health 2015;12:10079-92. [CrossRef]
25. Wieland K, Chhatwal P, Vonberg RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa:Results of a systematic review. Am J Infect Control 2018;46:643-8. [CrossRef]
26. Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 2021;10:373. [CrossRef]
27. Liu C, Chen K, Wu Y, Huang L, Fang Y, Lu J, et al. Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg Microbes Infect 2022;11:1730-41. [CrossRef]
28. Zarrilli R, Bagattini M, Migliaccio A. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in Italy. Ann Ig Med Prev Comunità2021;33:401-9.
29. Ababneh Q, Aldaken N, Jaradat Z, Al Sbei S, Alawneh D, Al-Zoubi E, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolated from three major hospitals in Jordan. Int J Clin Pract 2021;75:e14998. [CrossRef]
30. Quoc CH, Phuong TN, Duc HN, Le TT, Thu HT, Tuan SN, et al. Carbapenemase genes and multidrug resistance of Acinetobacter baumannii:A cross sectional study of patients with Pneumonia in Southern Vietnam. Antibiotics (Basel) 2019;8:148. [CrossRef]
31. Nhu NT, Lan NP, Campbell JI, Parry CM, Thompson C, Tuyen HT, et al. Emergence of carbapenem-resistant Acinetobacter baumannii as the major cause of ventilator-associated pneumonia in intensive care unit patients at an infectious disease hospital in Southern Vietnam. J Med Microbiol 2014;63:1386-94. [CrossRef]
32. Anh NT, Nga TV, Tuan HM, Tuan NS, Minh YD, Chau NV, et al. Molecular epidemiology and antimicrobial resistance phenotypes of Acinetobacter baumannii isolated from patients in three hospitals in Southern Vietnam. J Med Microbiol 2017;66:46-53. [CrossRef]
33. Tran QK, Tran DH, Pham HV, Nguyen VT, Tran XB, Larsson M, et al. Study on the co-infection of children with severe community-acquired pneumonia. Pediatrics International 2022;64:e14853. [CrossRef]