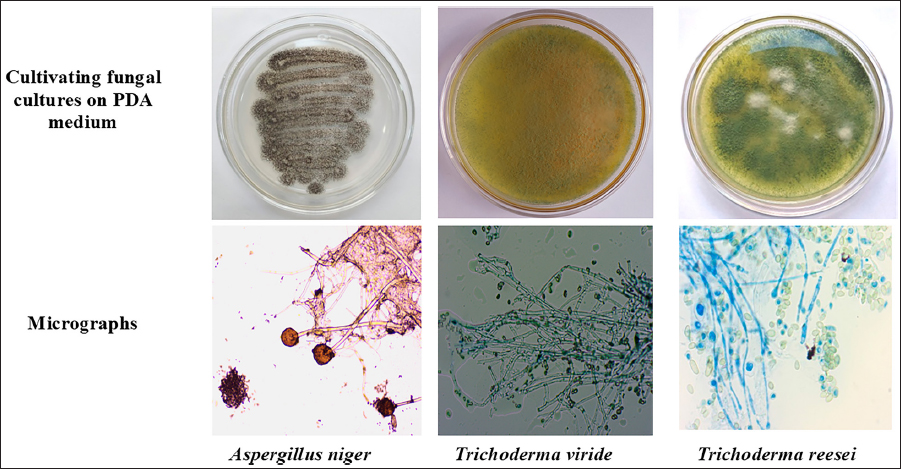
INTRODUCTION
In the current decade, the world has witnessed a notable upswing in waste production, primarily fuelled by the rapid expansion of the human population and the brisk pace of economic growth [1]. This surge has resulted in the generation of millions of metric tons of organic waste, from both municipal and agricultural sectors as a consequence of anthropogenic activities [1]. Within the Indian context, agricultural activities play a substantial role in waste generation, yielding an estimated 550 million tonnes of agricultural residues annually. In the current decade, the rapid increase in waste production, particularly agricultural residues, has become a pressing environmental concern, with millions of metric tons generated annually. Effective utilization of lignocellulosic biomass through enzymatic degradation offers a promising solution for waste management and bioresource utilization. Fungi, with their ability to produce diverse extracellular enzymes, play a pivotal role in breaking down lignocellulosic materials into usable compounds. Key contributors include rice, wheat, sorghum, pearl millet, and maize, which together contribute approximately 236 million tonnes of agricultural residues as straw [1]. These primary materials, including rice straw, corn stover, sugarcane bagasse, and sawdust, comprise substantial quantities of lignocelluloses [2]. Cellulose constitutes the primary component of lignocellulosic materials, accompanied by hemicellulose and lignin. Cellulose, a linear polymer of D-glucose units connected through β-1,4-glycosidic bonds, stands out as a predominant element in plant biomass. Approximately 45% of the total dry weight of wood is composed of this polymer [3,4].
The demand for food drives continuous crop production, often leading farmers to resort to field burning after harvest [5]. In India, particularly in regions like Haryana, rice and wheat sowing predominantly occurs in June-July and November-December, followed by harvesting in October-November and April-May [6]. However, this short timeframe allows farmers only a brief window of three to four weeks to transit to the next growing season. Additionally, there is a lack of cost-effective technologies applicable to small-scale agricultural fields for the collection of residual agricultural biomass [7]. Consequently, farmers often resort to in-situ crop waste burning as a quick, cost-effective, and efficient method for preparing the fields for subsequent crop cycles [8].
The hydrolytic degradation of cellulose is contingent upon the synergistic catalytic activity of three enzymes: endoglucanase, cellobiohydrolase, and β-glucosidase. Cellulases facilitate cellulose hydrolysis, yielding glucose, cellobiose, and cello-oligosaccharides as principal products [9-11]. Subsequently, these agricultural residues are an exceptional carbon substrate for microbial cellulase synthesis. Cellulases can be generated by various species of fungi, bacteria, and yeast and can utilise lignocelluloses as their primary carbon substrate. Nevertheless, fungi emerge as promising microorganisms for synthesising these enzymes because of their ability to produce a diverse array of extracellular enzymes. Among these, A. niger stands out as an optimal candidate due to its robust enzyme production, adaptability to diverse environmental conditions, and ability to thrive on a variety of lignocellulosic substrates. This makes it particularly advantageous for industrial applications compared to other fungi. However, a significant need remains to enhance its cellulolytic activity to levels comparable to extensively studied Trichoderma species. This enables them to efficiently break down complex lignocellulosic substrates into soluble compounds, with their enzyme levels surpassing those observed in yeast and bacteria [12,13]. Cellulases find extensive application in various sectors such as biofuel, food, animal feed, beverages, textiles, pulp and paper, pharmaceuticals, agriculture, and more [12-14]. In agricultural applications, introducing cellulolytic fungi optimizes compost quality through their efficient degradation of agricultural waste substrates [12,15]. Furthermore, utilizing agricultural waste for compost production is a significant alternative to sustainable waste management. In the context of employing enzymes in the pulp and paper industry, alkaliphile enzymes exhibit significant activity at alkaline pH levels. This characteristic presents considerable potential for their application in dewatering and refining steps and bleaching processes without requiring pH adjustments [16].
Soil microbial activity is impacted by agricultural practices, fertilizer input, organic residues, and crop rotation. Soil respiration, a vital indicator of microbial activity, is crucial for understanding biological processes in soil ecosystems. Additionally, soil bioactivity is closely connected to pH, organic matter, texture, climate, moisture, and temperature, influencing enzyme production by fungal species. Therefore, changes observed in the host, such as age and environmental factors like climate and geographical location, can affect the biology of fungi [17-19]. Thus, the present study aimed to address the burgeoning issue of waste management by considering the influence of agricultural practices, soil factors, and climate. The objective was to isolate a local fungal strain that flourishes on the decomposed agricultural wastes in the unique soil conditions of Vadodara, where reports on such strains are currently unavailable. Furthermore, it investigates diverse random mutagenesis approaches to enhance the cellulolytic activity of the tested fungi. These efforts were driven by the need for efficient degradation of lignocellulosic materials, utilizing rice residues as substrate. By bridging the gap between waste management challenges and enzymatic solutions, the present research aims to contribute to the advancement of sustainable practices in agricultural as well as industrial sectors. Thus, the objective of this study is to isolate and enhance the cellulolytic activity of A. niger through mutagenesis techniques, evaluating its potential to produce industrially viable cellulase enzymes using rice straw under solid-state fermentation. This approach aims to address the twin challenges of agricultural waste management and the demand for efficient enzyme production in biofuel and other industrial processes.
2. MATERIALS AND METHODS
2.1 Isolation and Procurement of Cellulase-Producing Fungal Strains
Four distinct agricultural field sites near Parul University, Vadodara, were randomly selected to collect lignocellulosic material, specifically rice straw, with aseptic techniques followed throughout the collection procedure. Rice straw was carefully collected in sterile polythene bags using sterile tools from each site and brought to the laboratory for further processing. Upon arrival at the laboratory, the collected biomass underwent aseptic oven drying at 60°C for 6 h to eliminate moisture and contaminants while preserving material integrity. A designated 6 x 10 feet plot at the Parul University experimental fields adjacent to the Research and Development Cell was chosen for the decomposition study of unseparated straw (excluding seeds). Each rice straw sample from the selected sites was enclosed in perforated nylon bags with a pore size of 100 microns. A total of 12 blocks measuring 30 x 30 cm were marked out in the experimental plot, with four nylon bags (triplicate) containing biomass from each site placed in separate blocks at a depth of 60 cm for 30 days [20].
After the 30-day decomposition period, the decomposed biomass from each block was collected under aseptic conditions. To isolate the mycobiota from each sample, a working solution was prepared following serial dilution plate methods [21] with 10−1, 10−2, 10−3, and 10−4 dilutions for each sample and Potato Dextrose Agar (HiMedia Laboratories) Medium [22] supplemented with 30 ppm Streptomycin, were used as a growth medium. The Petri dishes containing PDA medium were inoculated with specified dilutions and incubated at 27 ± 2°C for 5 days. The resulting fungal strains [Table 1] were subsequently subcultured onto fresh Petri plates containing Potato Dextrose Agar Medium for further examinations and the preparation of axenic cultures. To compare the cellulolytic activity of the isolated strain with superior cellulolytic activity, prominent cellulase producers, namely T. viride (1051) and T. reesei (992) sourced from the National Collection of Industrial Microorganisms (NCIM), Pune (India).
Table 1: Qualitative and quantitative distribution of fungal species isolated aseptically from decomposed wheat straw, rice straw, and bagasse using serial dilution plate method after 30 days of decomposition.
| Fungal Species | Dilutions |
|---|
| 1:100 | 1:1000 | 1:10000 | 1:100000 |
|---|
| TI | PI | TI | PI | TI | PT | TI | PI |
| Aspergillus niger | 21 | 41.2 | 12 | 63.16 | 7 | 50 | 7 | 63.64 |
| Aspergillus luchuensis | 4 | 7.8 | - | - | - | - | - | - |
| Aspergillus terreus | 3 | 5.9 | - | - | - | - | - | - |
| Alternaria citri | 1 | 2.0 | 1 | 5.26 | - | - | - | - |
| Cladosporium sp. | - | - | - | - | 1 | 7.14 | - | - |
| Curvularia lunata | 2 | 3.9 | 2 | 10.53 | - | - | - | - |
| Drechslera sp. | - | - | - | - | 2 | 14.29 | - | - |
| Fusarium incarnatum | 7 | 13.7 | - | - | 4 | 28.57 | - | - |
| Neurospora sp. | - | - | - | - | - | - | 1 | 9.09 |
| Penicillium crysogenum | 9 | 17.6 | - | - | - | - | - | - |
| Penicillium crysogenum | - | - | - | - | - | - | 1 | 9.09 |
| Penicillium digitatum | - | - | 3 | 15.79 | - | - | - | - |
| Penicillium sp. | 4 | 7.8 | - | - | - | - | - | - |
| Trichoderma viride | - | - | 1 | 5.26 | - | - | - | - |
| Verticillium sp. | - | - | - | - | - | - | 2 | 18.18 |
| Number of species | 8 | 5 | 4 | 4 |
| Total Isolates | 51 | 19 | 14 | 11 |
| Simpson’s index (D) | 0.22 | 0.41 | 0.31 | 0.4 |
| Simpson’s index of diversity (1-D) | 0.78 | 0.59 | 0.69 | 0.6 |
| Simpson’s Reciprocal Index (1/D) | 4.51 | 2.44 | 3.25 | 2.5 |
2.2 Preparation of Inoculums
For inoculum preparation, mature fungal colonies exhibiting full sporulation were used, and spores were harvested using aseptic methods and subsequently suspended in 30 ml of distilled water supplemented with 0.2% Tween 20 (Sigma-Aldrich Co. St Louis, MO, USA), followed by dilution to achieve a concentration of 1.0 x 106 cells per mL through enumeration utilizing a Neubauer chamber [23].
2.3 Parameters for Solid State Fermentation (SSF)
The solid rice wastes were collected in an aseptic manner from agricultural areas located in close proximity to Parul University in Vadodara, Gujarat. These wastes were subsequently brought to the laboratory in sterile zip-lock bags. The biomass was classified into distinct nutrient classes and subsequently underwent a drying process in an oven at a temperature of 60°C for a duration of 6 h to attain a reduction in moisture content ranging from 10% to 12%, which is in accordance with the desired moisture content for feed applications, ensuring the absence of contaminations.
Two subsets for SSF were prepared. The first subset consisted of 60 mL of Mendel mineral salt medium [24], cellulose (1.2 g), and 2.4 g of rice in a 750 ml Erlenmeyer flask, while the second subset contained the same components except for the absence of added cellulose. Both sets were adjusted to a pH of 4.5 and then incubated at 27ºC. Additionally, similar sets were prepared for the conditions viz. pH 4.5 at 30ºC, pH 5 at 27ºC, and pH 5 at 30ºC following the existing body of work [25-34]. These modifications enabled investigation of the impact of pH and temperature on the SSF process. For each pH and temperature subset, two triplicate sets without added inoculum served as control and consisted of 0.8 grams of rice straw mixed with 20 mL of Mandel’s mineral salts medium containing cellulose (0.4 g). Additionally, another control group contained 20 mL of Mandel’s medium, and 0.8 grams of rice straw without added cellulose. These subsets were incubated for a period of 5 days. After SSF, the residual fermented mixtures were added with 60 mL of sodium citrate buffer (50 mM, pH 4.5) in each flask, this mixture was agitated in an orbital shaker at 180 rpm and 27ºC for 60 m and samples were intermittently taken, filtered using Whatman no. 1 filter paper, and then subjected to centrifugation at 9000 rpm and 4ºC for 20 m to collect the culture supernatant containing crude enzymes.
2.4 Enzyme Assay(s) and Calculations for the Quantification of Cellulolytic-Enzyme Activity of Test Fungal Strains
The endoglucanase activity (CMCase, Endo-1,4-β-D-glucanase) was evaluated with a 1 mL reaction mixture containing 0.5 mL of culture supernatant (crude enzyme) and 0.5 mL of 1% (w/v) CMC (Sigma-Aldrich Co. St Louis, mo. USA) solution in sodium citrate buffer (50 mM, pH 4.5). The reaction mixture was incubated at 50ºC for 30 m, and reducing sugar was determined using the 3,5-dinitrosalicylic acid (DNS) (HiMedia Laboratories) method at 540 nm spectrophotometrically [35].
The Fpase activity was determined following the method suggested by Wood and Bhat with minor modifications by incubating 0.1 mL culture supernatant (enzyme) with 50 mg of sterile filter paper (Whatman no. 1) in 1.9 mL sodium citrate buffer (50 mM, pH 5). The reaction mixture was incubated at 50ºC for 60 m. The released reducing sugar amount was estimated by the DNS method at 540 nm spectrophotometrically [36].
For estimating avicelase activity, a total reaction mixture of 4 mL, containing 1 mL of crude enzyme and 3 mL of 1% Avicel (Sigma-Aldrich Co. St Louis, mo. USA) in 50 mM citric acid buffer (pH 4.8) was prepared. This resulting reaction mixture was incubated at 50ºC for 1 h, with measurements following the same method as adopted for CMCase [37].
As a substrate, the β-Glucosidase Activity was estimated using p-nitrophenyl-β-D-glucopyranoside (pNPG) (Sigma-Aldrich Co. St Louis, mo. USA). The total assay mixture (1 mL) consisting of 0.9 mL of pNPG (1 mM) and 0.1 mL of culture supernatant (crude enzyme) was incubated at 50ºC for 30 m. The developing yellow colour signifying the release of p-nitrophenol (pNP) by enzymatic hydrolysis was estimated at 420 nm spectrophotometrically [35,38].
To determine the concentration of unknown proteins, standard curves were plotted employing glucose (for determining endoglucanase, FPase and avicelase activity) and p-nitrophenol (for determining β-glucosidase activity) standards. The equation gives the relationship between absorbance (O.D.) and concentration:
Y = MX + C
Where:
Y is the absorbance (O.D.),
M is the slope of the standard curve,
C is the intercept (in this case, C is 0),
X is the concentration of glucose or p-nitrophenol.
Calculation of slope for the standard curve:
M = Y2-Y1/X2-X1
Where:
Y2 and Y1 are two succeeding obtained optical densities,
X2 and X1 are two succeeding concentrations
The calculated slope values for glucose and p-nitrophenol standard curve were 0.00226 and 0.046 respectively.
Enzyme Activity Calculation from Concentration:
Unit Activity is defined as the amount of enzyme which produce 1µM of the product per minute and per ml under standard assay conditions [39]. Therefore, the unit activity was calculated following the given equation,

All spectrophotometric estimations were performed using a Shimadzu UV-1900i UV-VIS-NIR Spectrophotometer at the Research and Development cell, Parul University. The enzymatic activities of CMCase, FPase, and Avicelase were measured in micromoles of reducing sugar released per gram (Units/mL). In contrast, beta-D-galactosidase activity was quantified as micromoles of p-nitrophenol released per minute per gram of reaction mixture (Units/mL). All assays were meticulously performed in triplicate, and the graphs and tabulated data reflect the mean values obtained.
2.5 Mutagenesis and Screening of Survivor Fungal Mutant
In the physical mutagenesis procedure, 1 mL of fungal spore suspension of wild-type fungi was evenly dispersed onto CMC-PDA plates containing congo-red dye (0.1% −0.2% w/v), under aseptic conditions. These plates were exposed to UV-radiations using 6W UV lamp (λ= 240 nm), positioned at a distance of 10 cm from the sample. The exposure was carefully conducted using a sequence of irradiation times, viz. 15 sec, 1 min, 5 min, 15 min, 30 min, to 60 min, maintaining consistency across all plates containing the three test fungi in triplicates and subsequently, subjected for a 7-day incubation period at 30°C along with control plates [40]. All UV treatments were conducted in a closed UV exposure chamber with protective barriers to minimize researcher exposure. Appropriate personal protective equipment (PPE), including UV-blocking goggles and gloves, were used during handling.
For chemical mutagenesis, a phosphate buffer solution containing the mutagenic agent Methylnitronitrosoguanidine (NTG or MNNG, HiMedia Laboratories) was prepared. Fully sporulated plates of three wild-type test fungi, aged seven days, were employed for microbial inoculation. Each fungal strain was distributed into six 15 mL centrifuge tubes. 1 mL of the NTG (7mM) containing citrate buffer solution (100 mM, pH 7) was added to each tube, containing 9 mL of fungal suspension [41]. These tubes were incubated for different incubation time intervals i.e. 30 min, 60 min, 90 min, 120 min, 150 min, and 180 min, for each fungal strain having similar compositions. Each tube was centrifuged at 5000 rpm for 10 m, separating fungal cells from the NTG solution. The supernatant, containing excess NTG, was discarded, and fungal cell pellets were collected carefully, without disturbing the tubes. These pellets were inoculated at the center of Carboxymethyl Cellulose-Potato Dextrose Agar (CMC-PDA) plates and incubated at 27°C for 4 days. As suggested by El-Ghonemy et al. [42] the expression of induced mutations in survivor fungal strains was assessed based on the halos formed around the fungal colonies due to cellulolytic-enzyme activity and compared to appropriate control plates. For Ethyl Methane Sulphonate (EMS) (HiMedia Laboratories) based chemical mutagenesis, seven-day-old fully sporulated plates of wild-type strains of test fungi were harvested and suspended in 50 mL of sterile distilled water with 50 µl of Tween 80 and 2.9 g of NaCl (Sigma-Aldrich Co. St Louis, mo. USA) to improve spore dispersion and stability. Six flasks per fungal species were prepared, each containing 50 mL of distilled water with varying concentrations of Ethyl Methane Sulphonate (EMS): Flask 1 (250 µg/mL), Flask 2 (200 µg/mL), Flask 3 (150 µg/mL), Flask 4 (100 µg/mL), Flask 5 (50 µg/mL), and Flask 6 (25 µg/mL), and kept for 24 h of incubation. Subsequently, from each flask, 1 mL of fungal suspension was inoculated onto different CMC-PDA plates and incubated at 27°C for 4 days along with control plates [43].
To ensure safety, NTG and EMS, both classified as potential carcinogens, were handled exclusively within a fume hood. Waste materials were securely contained in sealed containers for proper hazardous waste disposal in compliance with established protocols. Appropriate personal protective equipment (PPE), including nitrile gloves and respiratory masks, was employed throughout the preparation and disposal processes. Contaminated glassware and waste materials were neutralized using a 10% sodium thiosulfate solution before final disposal to mitigate associated risks.
2.6 Estimation of Cellulase Enzyme Production by Survivor Fungal Mutants
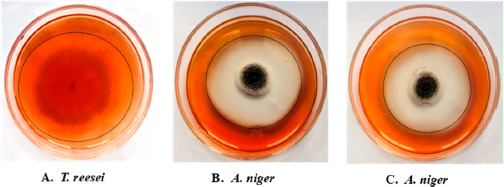
As described by Oetari et al. [44] the CMCase test was carried out to evaluate the fungal enzymatic activity of survivor fungal mutants. Endoglucanase (CMCase) activity was determined using a 5% (w/v) Congo red solution (HiMedia Laboratories). Positive CMCase activity was confirmed by a clear zone around fungal colonies. The Enzymatic Index (EI) was calculated as R/r, where R is the diameter of the clear zone and r is the diameter of the fungal colony, enabling quantitative assessment of enzymatic activity.
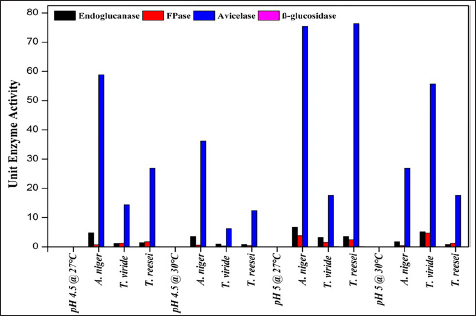
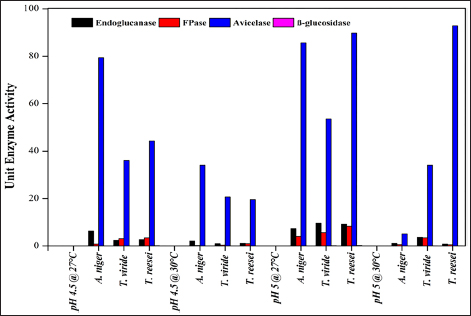
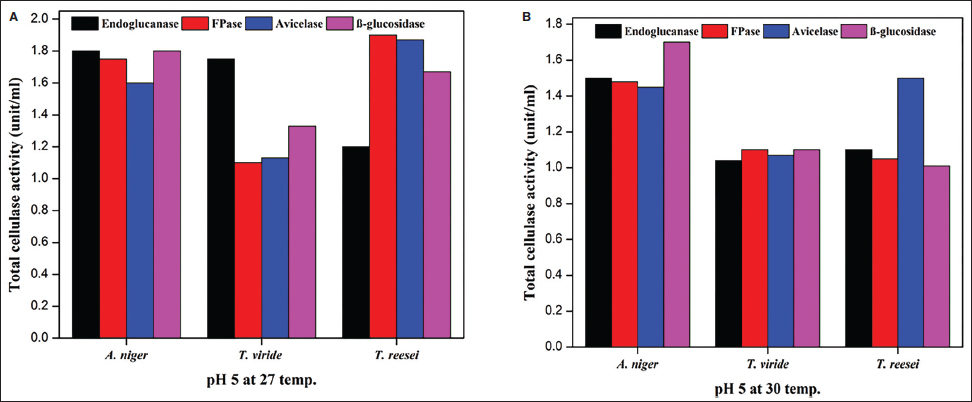
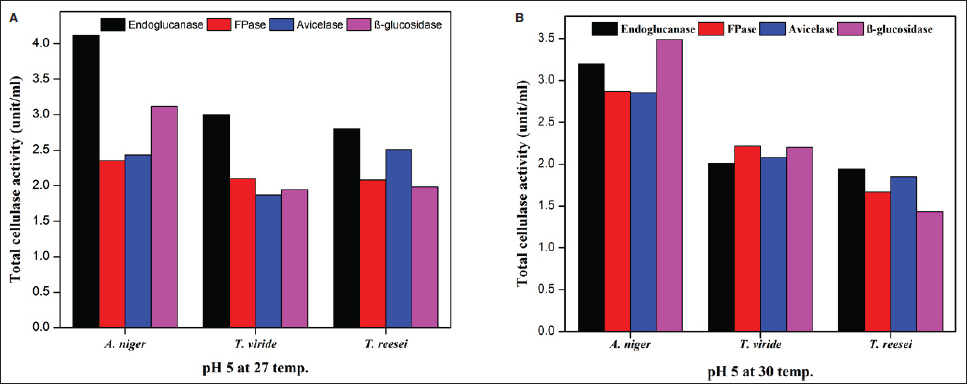
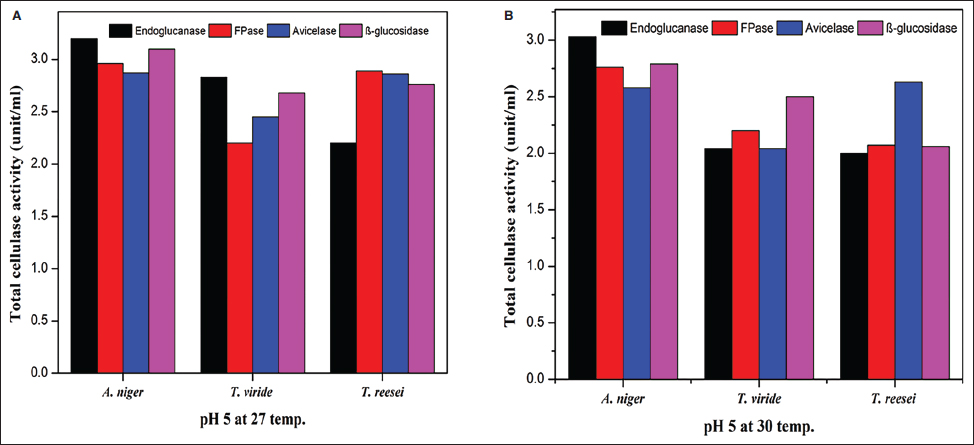
Cellulase-producing mutant fungi were then evaluated for their ability to degrade agricultural residues under various conditions, including pH 4.5 and 27°C, pH 5 and 30°C, and pH 5 and 27°C and compared with wild-type strains. Similar to wild-type strains, extracellular enzyme assays were conducted for endoglucanase, FPase, Avicelase, and β-Glucosidase activities for the screening of superior mutants.
2.7 Statistical Analysis
All experiments were conducted in triplicate, and the data were presented as the mean ± standard deviation in accordance with scientific protocol. To assess significant differences (p < 0.05) between pairs of group means for the variable, a Post hoc analysis using Tukey’s HSD (Honest Significant Difference) test was employed following a single-factor ANOVA.
4. DISCUSSION
In the present investigation, a diverse range of fungal isolates were obtained following isolation procedures and A. niger was selected based on its frequency in the culture plates. The T. viride, and T. reesei strains were chosen for their documented proficiency in cellulase production and compared with A. niger strain for the cellulolytic activities. Extensive literature has highlighted the industrial significance of lignocellulolytic enzymes, particularly cellulase, and both Aspergillus and Trichoderma species are recognized as potential producers of cellulase [50]. Among these, T. viride and T. reesei have been extensively studied for their cellulolytic potential [51]. Nathan et al. emphasized T. viride’s wide distribution and its capability as a cellulolytic organism, alongside its ability to produce biocontrol agents and growth-promoting factors [52]. Similarly, T. reesei, a rapidly growing filamentous fungus commonly found in soil environments, has gained tremendous attention for its prolific secretion of cellulolytic enzymes [53]. Consequently, these fungi were chosen for comprehensive evaluation alongside A. niger to assess their cellulolytic prowess.
The present study exhibited a noteworthy enhancement in enzyme production across different fungal strains compared to the strains studied in previous studies [Table 8]. For endoglucanase activity, A. niger shows significant productivity of 30.36 ± 9.73 Units/mL using rice straw at pH 5 and 27°C. This is substantially higher than previous findings such as 2.37 Units/mL on wheat straw and waste paper deinking at pH 8 and 45°C [54] and 3.92 Units/mL on M3 medium at pH 6 and 40°C [55]. Similarly, T. reesei exhibited an FPase activity of 17.32 ± 2.3 Units/mL using rice straw at pH 5 and 27°C, surpassing the FPase activity obtained in previous studies like 1.95 Units/mL from Trichoderma sp. on municipal waste at pH 6.5 and 45°C [56]. Additionally, A. niger exhibits FPase activity of 12.00 ± 2.52 Units/mL , superior to the 1.19 Units/mL reported in a previous study using municipal waste as substrate at pH 6-7 and 40°C [56].
Table 8: Comparative analysis of enzyme productivity between current study and previous research across various fungal strains, substrates, and environmental conditions.
| Enzyme | Microbial Strain | Productivity | Substrate/Medium | Conditions | Ref. |
|---|
| pH | Temp. (°C) |
|---|
| Endoglucanase | A. niger | 30.36 ± 9.73 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. viride | 29.18 ± 6.6 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. reesei | 25.58 ± 7.1 U ml−1 | Rice straw | 5 | 27 | Present study |
| A. niger | 2.37 IU ml−1 | Wheat straw and waste paper deinking | 8 | 45 | [54] |
| A. brasiliensis | 40.50 U g−1 | Theobroma grandiflorum residue | - | - | [71] |
| A. niger | 3.92 IU ml−1 | M3 medium | 6 | 40 | [55] |
| A. niger | 0.153 µmol/ml/min | Corn starch, sodium nitrate | 3 | 28 | [72] |
| A. niger | 4.165 U ml−1 | Prickly pear | 3-8 | 29.56-31.22 | [73] |
| A. nidulans | 68.58 ± 0.55 U ml−1 | Wheat straw | 6 | 30 | [74] |
| A. niger | 1.19 U ml−1 | Municipal waste | 6-7 | 40 | [75] |
| Rhizopus sp. | 7.859 U ml−1 | Prickly pear | 3-8 | 30.41-27.86 | [73] |
| Trichoderma sp. | 1.95 U ml−1 | Municipal waste | 6.5 | 45 | [75] |
| F. oxysporum | 0.445 IU ml−1 | PDA-CMC | 4−9 | 50 | [56] |
| Penicillium oxalicum | 9.2 U g−1 | Sorghum waste | 4 | 40 | [57] |
| Aspergillus sp. IN5 | 3.83.80 ± 0.30 U g−1 | Soybean meal | 6 | 35 | [76] |
| Pleurotus ostreatus | 0.868 U ml−1 | Bean straw | 4 | 30 | [60] |
| P. ostreatus | 0.444 U ml−1 | Wheat straw | 4 | 30 | [60] |
| FPase | A. niger | 12.00 ± 2.52 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. viride | 12.48 ± 3.15 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. reesei | 17.32 ± 2.3 U ml−1 | Rice straw | 5 | 27 | Present study |
| A. nidulans | 12.0 ± 0.06 U ml−1 | Wheat straw | 6 | 30 | [74] |
| A. niger | 1.504 U ml−1 | Municipal waste | 6-7 | 40 | [75] |
| Trichoderma sp. | 1.77 U ml−1 | Municipal waste | 6.5 | 45 | [75] |
| F. oxysporum | 9.25 IFPU ml−1 | PDA-CMC | 4−9 | 50 | [56] |
| Aspergillus sp. IN5 | 0.24 ± 0.00 U g−1 | Soybean meal | 7 | 35 | [76] |
| A. fumigatus SB12 | 0.358 ± 0.05 IU/mL | PDA-CMC | - | 40 | [77] |
| Pleurotus ostreatus | 0.191 U ml−1 | Bean straw | 4 | 30 | [60] |
| P. ostreatus | 0.216 U ml−1 | Wheat straw | 4 | 30 | [60] |
| Penicillium oxalicum | 4.2 U g−1 | Sorghum waste | 4 | 40 | [57] |
| A. niger | 245.73 ± 14.9 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. viride | 131.42 ± 12.2 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. reesei | 225.27 ± 16.5 U ml−1 | Rice straw | 5 | 27 | Present study |
| Avicelase | Penicillium oxalicum | 8.4 U g−1 | Sorghum waste | 4 | 40 | [57] |
| Aspergillus flavus | 799.23 U g−1 | Clover Husk | 6 | 50 | [78] |
| A. terreus RWY | 10.3 ± 0.66 U g−1 | Bagasse | - | 45 | [58] |
| A. niger | 67 U g−1 | Bagasse | - | - | [59] |
| β-Glucosidase | A. niger | 0.19 ± 0.03 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. viride | 0.43 ± 0.08 U ml−1 | Rice straw | 5 | 27 | Present study |
| T. reesei | 0.56 ± 0.03 U ml−1 | Rice straw | 5 | 27 | Present study |
| A. nidulans | 1.89 ± 0.037 U ml−1 | Wheat straw | 6 | 30 | [74] |
| Aspergillus sp. IN5 | 176.94 ± 7.72 Ug−1 | Soybean meal | 5 | 35 | [76] |
| Pleurotus ostreatus | 0.389 U ml−1 | Bean straw | 4 | 30 | [60] |
| P. ostreatus | 0.245 U ml−1 | Wheat straw | 4 | 30 | [60] |
For Avicelase activity, A. niger displayed a remarkable productivity of 245.73 ± 14.9 Units/mL using rice straw at pH 5 and 27°C, significantly higher than other studies [57-59]. In the present study on β-Glucosidase activity, T. reesei demonstrated 0.56 ± 0.03 Units/mLenzyme productivity using rice straw at pH 5 and 27°C, improving over other reported activities by Pleurotus ostreatus using substrates such as bean straw and wheat straw having activity of 0.389 Units/mL and 0.245 Units/mL respectively [60]. Therefore, the results from this study exhibited higher enzymatic activities across different enzymes under study and fungal strains compared to several studies, under optimized conditions using rice straw as the substrate. These findings highlight the potential of these fungal strains in sustainable industrial applications and more efficient biomass conversion processes. Enhanced cellulase production from NTG-treated A. niger strains can improve the enzymatic hydrolysis of lignocellulosic biomass, boosting biofuel production. These strains can also aid waste management by accelerating organic waste degradation, reducing landfill use, and supporting composting.
A thorough analysis of the activity of specific enzymes in several fungal species under various environmental conditions yields essential insights into the regulation of enzyme production. This information is crucial for industrial uses, especially in producing biofuels. All tested fungal strains exhibit considerably increased enzyme activity when cellulose is added. The findings of the present study strongly align with those of Bhardwaj et al. [61], who noted that commercial cellulose derivatives accelerate the rate of cellulase enzyme production. Initially, microorganisms do not need to contend with recalcitrant lignin materials and can directly begin producing cellulases to digest added cellulose. Therefore, we strongly recommend the inclusion of some cellulose or cellulose-like carbon source in the medium for the enhanced production of cellulolytic enzymes for industrial and agricultural practices.
Avicelase activity in A. niger was consistently strong in all settings, indicating a specific capacity to break down cellulose. Conversely, under the studied conditions, β-glucosidase appeared to have a less substantial role in the fast breakdown of cellulose as evidenced by its limited fluctuation and decreased activity. The results highlight the importance of adjusting fermentation conditions to the specific fungal strains and targeting enzymes to maximize enzyme outputs. The notable increase in enzyme activity under particular conditions suggests that new cost-effective and efficient methods of producing critical enzymes may be developed. A. niger shows promise in its capacity to degrade cellulose, a valuable property for both industrial and agricultural uses.
Developing a thorough understanding of the complex relationship between the synthesis of enzymes and the combined influence of internal and external stimuli can produce significant benefits for genetic engineering methods targeted at enhancing enzyme yields. This can be accomplished by adjusting enzyme stability and activity in response to changes in pH and temperature or by adjusting regulatory components that react to cellulose. This has significant implications for biotechnological applications and can direct future investigations into the molecular mechanisms underlying these responses and the potential industrial uses of these fungi.
Investigating the genetic and molecular mechanisms that control these activities in fungi is crucial to building on the understanding gained from environmental factors regulating enzyme activity. The synthesis and secretion of enzymes that break down cell walls is essential for fungi that feed on plant materials. Together, these enzymes break down the cell wall and release sugar monomers necessary for energy production. The synthesis of enzymes involved in breaking down lignocellulosic biomass is critically controlled at the transcriptional level and is responsive to the type of substrate present [62,63]. Critical transcriptional regulators, such as XYR1, the primary regulator of xylanases and cellulases, and CRE1, which governs carbon catabolite repression, have been identified. Additional factors influencing these regulatory pathways include negatively acting regulators ACE1, RCE1, and RXE1, as well as positively acting factors like ACE2 and the CCAAT binding complex HAP2/3/5. Notably, XYR1 is recognized as the central transcriptional activator for a wide array of genes, including those for hemicellulases and cellulases, underscoring its pivotal role in enzyme production [64]. Therefore, it becomes significantly important to investigate these genes with a larger sample size to have a clearer picture of the degradation processes by these fungal species.
The present investigation provided a clearer picture on how UV, NTG, and EMS mutagenesis affect enzyme activities in A. niger, T. viride, and T. reesei. The enzymatic breakdown of CMC served as a prerequisite for the subsequent clearance of Congo red. This dye clearance was contingent upon both substrate presence and the attainment of adequate cellulase activity by the microorganisms [49]. The research highlights both the potential and challenges of using mutagenic treatments to boost fungal enzyme production, emphasizing the need for further genetic and molecular research to refine mutagenesis strategies for more efficient fungal engineering and enhanced enzymatic capabilities. Random mutagenesis is a simple method for modifying an organism’s genetics and functions. Using successive stepwise mutagenesis-selection methods and mutagens with different modes of action has been shown to increase product yield [65]. Kumar et al. [66] discovered that UV-induced mutation significantly enhances the production of endoglucanase and β-glucosidase by microorganisms. Present work on UV- and EMS-induced mutations showed variable results, with UV mutagenesis being more effective under specific conditions than EMS. Also, studies on UV and EMS mutagenesis have shown variable outcomes. In a UV-mutagenesis study of A. niger FCBP-02, a 1.5- to 2.0-fold enhancement in cellulase activity was observed [67], while in another study, a maximum cellulase production of 330 µM/min/mg with 10 minutes of UV treatment was exhibited [68]. These findings indicate that while UV mutagenesis enhances enzyme activity, the improvements are less pronounced than NTG mutagenesis. Furthermore, the present study demonstrates that NTG mutagenesis significantly enhances cellulase production in A. niger. This finding aligns with previous research indicating the efficacy of NTG in boosting cellulase activity. For instance, in a recent report, a 4.5-fold increase in endoglucanase B activity in A. niger with NTG treatment was recorded, achieving 428.5 µM/mL/min compared to 94 µM/mL/min in the native enzyme [69]. Our results corroborate these findings, with NTG-treated mutants exhibiting a 4.12-fold increase in endoglucanase activity, reaching 30.36 Units/mL under optimal conditions of pH 5 and 27°C. No reports have been encountered stating the role of NTG mutagenesis in demonstrating an increase in avicelase activity. To the best of our knowledge, the present work is the first-ever report suggesting enhanced avicelase activity in A. niger following NTG mutagenesis.These treatments have the potential to increase the rate of enzyme production and could aid in managing lignocellulosic waste materials in situ through SSF. Current study also highlights the advantages of solid-state fermentation (SSF) over submerged fermentation (SmF) for industrial applications. SSF offers benefits such as higher product yields, lower energy requirements, and reduced wastewater production. This aligns with findings from a study conducted by Ahmed et al., which optimized cellulase and xylanase production by A. niger using brewery-spent grain in SSF, achieving significant enzyme activities [70]. Using rice straw as a substrate in SSF further underscores the potential of this method for sustainable industrial enzyme production. The results of the present study support this notion, as mutant strains of A. niger exhibited improved endoglucanase and β-glucosidase activity. This enhancement could be valuable in addressing the ongoing issue of indiscriminate burning, which contributes to significant environmental pollution. Ultimately, the presented work confirms that NTG mutagenesis is a potent method for enhancing cellulase production in A. niger, with improvements surpassing those achieved through UV or EMS treatments. The application of SSF using agricultural residues like rice straw presents a viable strategy for large-scale enzyme production, contributing to both waste valorization and industrial biotechnology advancements.
REFERENCES
1. Kumar A, Agrawal A. Recent trends in solid waste management status, challenges, and potential for the future Indian cities – A review. Curr Res Environ Sustain. 2020;2.
2. Badhan AK, Chadha BS, Kaur J, Saini HS, Bhat MK. Production of multiple xylanolytic and cellulolytic enzymes by thermophilic fungus Myceliophthora sp. IMI 387099. Bioresour Technol. 2007;98:504–10.
3. Sohail M, Siddiqi R, Ahmad A, Khan SA. Cellulase production from Aspergillus niger MS82: effect of temperature and pH. N Biotechnol. 2009;25:437–41.
4. Pérez J, Muñoz-Dorado J, De La Rubia T, Martínez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overviewPérez. Int Microbiol. 2002;5:53–63.
5. Grover D, Chaudhry S. Ambient air quality changes after stubble burning in rice–wheat system in an agricultural state of India. Environ Sci Pollut Res. 2019;26:20550–9.
6. Gadde B, Bonnet S, Menke C, Garivait S. Air pollutant emissions from rice straw open field burning in India, Thailand and the Philippines. Environ Pollut. 2009;157:1554–8.
7. He Q, Zhao X, Lu J, Zhou G, Yang H, Gao W, et al. Impacts of biomass-burning on aerosol properties of a severe haze event over Shanghai. Particuology. 2015;20:52–60.
8. Venkataraman C, Habib G, Kadamba D, Shrivastava M, Leon J.-F, Crouzille B, et al. Emissions from open biomass burning in India: Integrating the inventory approach with high-resolution Moderate Resolution Imaging Spectroradiometer (MODIS) active-fire and land cover data. Glob Biogeochem Cycles. 2006;20.
9. Deswal D, Khasa YP, Kuhad RC. Optimization of cellulase production by a brown rot fungus Fomitopsis sp. RCK2010 under solid state fermentation. Bioresour Technol. 2011;102:6065–72.
10. Gao J, Weng H, Zhu D, Yuan M, Guan F, Xi Y. Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid-state cultivation of corn stover. Bioresour Technol. 2008;99:7623–9.
11. Singhania RR, Sukumaran RK, Patel AK, Larroche C, Pandey A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb Technol. 2010;46:541–9.
12. Sari SLA, Setyaningsih R, Wibowo NFA. Isolation and screening of cellulolytic fungi from Salacca edulis leaf litter. Biodiversitas. 2017;18:1282–8.
13. Wen Z, Liao W, Chen S. Production of cellulase/β-glucosidase by the mixed fungi culture Trichoderma reesei and Aspergillus phoenicis on dairy manure. Process Biochem. 2005;40:3087–94.
14. Angela S, Oziniel R, Ignatious N, Thembekile N. Diversity of cellulase- and xylanase-producing filamentous fungi from termite mounds. J Yeast Fungal Res. 2019;10:15–29.
15. Sakpetch P, H-Kittikun A, Chandumpai A. Isolation and screening of potential lignocellulolytic microorganisms from rubber bark and other agricultural residues. Walailak J Sci Technol. 2017;14:953–67.
16. Muthezhilan R, Ashok R, Jayalakshmi S. Production and optimization of thermostable alkaline xylanase by Penicillium oxalicum in solid state fermentation. J Microbiol. 2007:20–8.
17. Friedel JK, Munch JC, Fischer WR. Soil microbial properties and the assessment of available soil organic matter in a haplic luvisol after several years of different cultivation and crop rotation. Soil Biol Biochem. 1996;28:479–88.
18. Emmerling C, Udelhoven T, Schröder D. Response of soil microbial biomass and activity to agricultural de-intensification over a 10 year period. Soil Biol Biochem. 2001;33:2105–14.
19. Sunitha VH, Devi DN, Srinivas C. Extracellular Enzymatic activity of endophytic fungal strains isolated from medicinal plants. World J Agric Sci. 2013;9:1–9.
20. Yan C, Yan SS, Jia TY, Dong SK, Ma CM, Gong ZP. Decomposition characteristics of rice straw returned to the soil in northeast China. Nutr Cycl Agroecosyst. 2019;114:211–24.
21. Waksman SA. A method for counting the number of fungi in the soil. J Bacteriol. 1922;7:339–41.
22. Dodge BO, Raper KB, Thom C. A manual of the Penicillia. Bull Torrey Bot Club. 1950;77:515.
23. Petrikkou E, Rodríguez-Tudela JL, Cuenca-Estrella M, Gómez A, Molleja A, Mellado E. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J Clin Microbiol. 2001;39:1345–7.
24. Jafari N, Jafarizadeh-Malmiri H, Hamzeh-Mivehroud M, Adibpour M. Optimization of UV irradiation mutation conditions for cellulase production by mutant fungal strains of Aspergillus niger through solid state fermentation. Green Process Synth. 2017;6:333–40.
25. Buragohain P, Sharma N, Pathania S. Use of wheat straw for extracellular cellulase production from Aspergillus niger F 8 isolated from compost. J Biofuels. 2016;7:37.
26. Acharya PB, Acharya DK, Modi HA. Optimization for cellulase production by Aspergillus niger using saw dust as substrate. African J Biotechnol. 2008;7:4147–52.
27. Legodi LM, La Grange D, Van Rensburg ELJ, Ncube I. Isolation of Cellulose degrading fungi from decaying banana pseudostem and Strelitzia alba. Enzyme Res. 2019;2019.
28. Archer DB, Peberdy JF. The molecular biology of secreted enzyme production by fungi. Crit Rev Biotechnol. 1997;17:273–306.
29. Juwaied AA, Abdulamier A, Al-Amiery H, Abdumuniem Z, Anaam U. Optimization of cellulase production by Aspergillus niger and Tricoderma viride using sugar cane waste. J Yeast Fungal Res. 2011;2:19–23.
30. Mrudula S, Murugammal R. Production of cellulase by Aspergillus niger under submerged and solid state fermentation using coir waste as a substrate. Brazilian J Microbiol. 2011;42:1119–27.
31. Kumari S, Prasad D, Swayamprabha S. Production and optimisation of cellulases on pre-treated groundnut shell by Aspergillus niger. 2010;3:21–37.
32. Singh RKRRS, Singh T, Pandey A, et al. RNA Metabolism. Bioresour Technol. 2020;8:393–435.
33. Okada G. Purification and properties of a cellulase from aspergillus niger. Agric Biol Chem. 1985;49:1257–65.
34. Tong CC, Rajendra K. Effect of Carbon and nitrogen sources on the growth and production of cellulase enzymes of a newly isolated Aspergillus sp. Pertanika. 1992;15:45–50.
35. Gokhale DV, Deobagkar DN. Differential Expression of xylanases and endoglucanases in the hybrid derived from intergeneric protoplast fusion between a Cellulomonas sp. and Bacillus subtilis. Appl Environ Microbiol 1989;55:2675–80.
36. Wood TM, Bhat KM. Methods for measuring cellulase activities. Methods Enzymol. 1988;160:87–112.
37. Grajek W, Technology P. Influence of water activity on the enzyme biosynthesis and enzyme activities produced by Trichoderma viride TS in solid-state fermentation. Enzyme Microbial Technol. 1987;9:658–62.
38. Zhang L, Fu Q, Li W, Wang B, Yin X, Liu S, et al. Identification and characterization of a novel β-glucosidase via metagenomic analysis of Bursaphelenchus xylophilus and its microbial flora. Sci Rep. 2017;7:1–11.
39. Adney, B and Baker J. Measurement of cellulase activities: Laboratory analytical procedure (LAP). Natl Renew Energy Lab. 1996:8.
40. Jafari N, Jafarizadeh-Malmiri H, Hamzeh-Mivehroud M, Adibpour M. Optimization of UV irradiation mutation conditions for cellulase production by mutant fungal strains of Aspergillus niger through solid state fermentation. Green Process Synth. 2017;6:333–40.
41. Foster PL. In vivo mutagenesis. Methods Enzymol. 1991;204:114–25.
42. El-Ghonemy DH, Ali TH, El-Bondkly AM, Moharam MES, Talkhan FN. Improvement of Aspergillus oryzae NRRL 3484 by mutagenesis and optimization of culture conditions in solid-state fermentation for the hyper-production of extracellular cellulase. Antonie van Leeuwenhoek. 2014;106:853–64.
43. Isaac GS, Abu-Tahon MA. Enhanced alkaline cellulases production by the thermohalophilic aspergillus terreus AUMC 10138 mutated by physical and chemical mutagens using corn stover as substrate. Brazilian J Microbiol. 2015;46:1269–77.
44. Oetari A, Susetyo-Salim T, Sjamsuridzal W, Suherman EA, Suherman M, Suherman R, et al. Occurrence of fungi on deteriorated old dluwang manuscripts from Indonesia. Int Biodeterior Biodegrad. 2016;114:94–103.
45. Gilman JC. A Manual of Soil Fungi. Vol II. Biotech Books; 1957.
46. Subramanian CV. Hyphomycetes, an account of Indian Species, except Cercosporae. 1974.
47. Barnett H, Hunter B. Illustrated genera of imperfect fungi. 1972.
48. Nagamani A, Kunwar CM. Handbook of Soil Fungi. 2006.
49. Meddeb-Mouelhi F, Moisan JK, Beauregard M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme Microb Technol. 2014;66:16–9.
50. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–77.
51. Domingues FC, Queiroz JA, Cabral JMS, Fonseca LP. The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme Microb Technol. 2000;26:394–401.
52. Nathan VK, Rani ME, Rathinasamy G, Dhiraviam KN, Jayavel S. Process optimization and production kinetics for cellulase production by Trichoderma viride VKF3. Springerplus. 2014:3.
53. Mandels M, Reese ET. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol. 1957;73:269–78.
54. Dixit M, Shukla P. Multi-efficient endoglucanase from Aspergillus niger MPS25 and its potential applications in saccharification of wheat straw and waste paper deinking. Chemosphere. 2023;313:137298.
55. Abdullah R, Chudhary SN, Kaleem A, Iqtedar M, Nisar K, Iftikhar T, et al. Enhanced production of β-glucosidase by locally isolated fungal strain employing submerged fermentation. Biosci J. 2019;35:1552–9.
56. Dutta S deb, Tarafder M, Islam R, Datta B. Characterization of cellulolytic enzymes of Fusarium soil Isolates. Biocatal Agric Biotechnol. 2018;14:279–85.
57. Bomtempo FVS, Santin FMM, Pimenta RS, de Oliveira DP, Guarda EA. Production of cellulases by Penicillium oxalicum through solid state fermentation using agroindustrial substrates. Acta Sci Biol Sci. 2017;39:321–9.
58. Sharma R, Kocher GS, Bhogal RS, Oberoi HS. Cellulolytic and xylanolytic enzymes from thermophilic Aspergillus terreus RWY. J Basic Microbiol. 2014;54:1367–77.
59. Hanif A, Yasmeen A, Rajoka MI. Induction, production, repression, and de-repression of exoglucanase synthesis in Aspergillus niger. Bioresour Technol 2004;94:311–9.
60. Diro BB, Daba T, Genemo TG. Production and characterization of cellulase from mushroom (Pleurotus ostreatus) for effective degradation of cellulose. Int J Biol Pharm Sci Arch. 2021;2:135–50.
61. Bhardwaj N, Kumar B, Agrawal K, Verma P. Current perspective on production and applications of microbial cellulases: a review. Bioresour Bioprocess. 2021;8:1–34.
62. de Souza WR. Microbial degradation of lignocellulosic biomass. In: Sustainable Degradation of Lignocellulosic Biomass - Techniques, Applications and Commercialization. InTech; 2013:207.
63. Olsson L, Christensen TMIE, Hansen KP, Palmqvist EA. Influence of the carbon source on production of cellulases, hemicellulases and pectinases by Trichoderma reesei Rut C-30. Enzyme Microb Technol. 2003;33:612–9.
64. Luo Y, Valkonen M, Jackson RE, Palmer JM, Bhalla A, Nikolaev I, et al. Modification of transcriptional factor ACE3 enhances protein production in Trichoderma reesei in the absence of cellulase gene inducer. Biotechnol Biofuels. 2020;13:137.
65. Chandra M, Kalra A, Sangwan NS, Gaurav SS, Darokar MP, Sangwan RS. Development of a mutant of Trichoderma citrinoviride for enhanced production of cellulases. Bioresour Technol. 2009;100:1659–62.
66. Kumar AK. UV mutagenesis treatment for improved production of endoglucanase and β-glucosidase from newly isolated thermotolerant actinomycetes, Streptomyces griseoaurantiacus. Bioresour Bioprocess. 2015;2:1–10.
67. Shafique S, Bajwa R, Shafique S. Mutagenesis and genotypic characterization of Aspergillus niger FCBP-02 for improvement in cellulolytic potential. Nat Prod Commun. 2009;4:557–62.
68. Daud Q ul ain, Hamid Z, Sadiq T, et al. Enhanced production of cellulase by escherichia coli Engineered with UV-mutated cellulase gene from Aspergillus niger UVMT-I. BioResources. 2019;14:9054–63.
69. Morán-Aguilar MG, Calderón-Santoyo M, Domínguez JM, Aguilar-Uscanga MG. Optimization of cellulase and xylanase production by Aspergillus niger CECT 2700 using brewery spent grain based on Taguchi design. Biomass Convers Biorefinery. 2023;13:7983–91.
70. Ahmad W, Zafar M, Anwar Z. Heterologous expression and characterization of mutant cellulase from indigenous strain of Aspergillus niger. PLoS One. 2024;19:e0298716.
71. de Souza Falcão L, Santiago do Amaral T, Bittencourt Brasil G, Melchionna Albuquerque P. Improvement of endoglucanase production by Aspergillus brasiliensis in solid-state fermentation using cupuaçu (Theobroma grandiflorum) residue as substrate. J Appl Microbiol. 2022;132:2859–69.
72. Patel KB, Patel SS, Patel BK, Chauhan HC, Rajgor M, Kala JK. Production and optimization of endoglucanase by Aspergillus sp., Trichoderma sp. and Penicillium sp. Int J Curr Microbiol Appl Sci. 2017;6:1318–25.
73. dos Santos TC, Filho GA, De Brito AR, Pires AJV, Bonomo RCF, Franco M. Production and characterization of cellulolytic enzymes by Aspergillus niger and Rhizopus sp. By solid state fermentation of prickly pear. Rev Caatinga. 2016;29:222–33.
74. Naitam MG, Tomar GS, Kaushik R. Optimization and production of holocellulosic enzyme cocktail from fungi Aspergillus nidulans under solid-state fermentation for the production of poly(3-hydroxybutyrate). Fungal Biol Biotechnol. 2022;9.
75. Gautam SP, Bundela PS, Pandey AK, Khan J, Awasthi MK, Sarsaiya S. Optimization for the production of cellulase enzyme from municipal solid waste residue by two novel cellulolytic fungi. Biotechnol Res Int. 2011;2011:1–8.
76. Boondaeng A, Keabpimai J, Trakunjae C, Vaithanomsat P, Srichola P, Niyomvong N. Cellulase production under solid-state fermentation by Aspergillus sp. IN5: parameter optimization and application. Heliyon. 2024;10:e26601.
77. Barapatre S, Rastogi M, Savita, Nandal M. Isolation of fungi and optimization of ph and temperature for cellulase production. Nat Environ Pollut Technol. 2020;19:1729–35.
78. Selim M, Awad S, Abdel-Fattah GM. Using clover husk for avicelase production from Aspergillus flavus via solid-state fermetnation. J Agric Chem Biotechnol. 2020;11:319–26.