ARTICLE HIGHLIGHTS
In Northern Bangladesh, UTIs were highly prevalent (51.56%)
Male UTI rates were much lower (26.51%) than female UTI rates (64.40%)
Escherichia coli (41.66%), Enterococcus faecalis (23.48%), and Klebsiella pneumoniae (18.93%) were the most common uropathogens found
Antibiotic resistance was alarmingly high, with 88.6% of isolates labeled MDR
Several commonly used antibiotics showed effectiveness against isolated bacteria, but resistance was observed against linezolid, cephradine, azithromycin, nalidixic acid, cefuroxime sodium, co-trimoxazole, cefixime, and ceftriaxone
Patient age, place of residence, marital status, and prior antibiotic use were found to be statistically associated with MDR UTIs.
1. INTRODUCTION
With 150 million cases annually, urinary tract infections (UTIs) are among the major diseases caused by bacteria in hospital and outpatient settings [1]. Globally, the societal costs of these infections, including lost productivity and medical expenses, are about 6 billion US dollars annually [2]. There are many risk factors for cystitis, including feminine gender, recurrent history of UTIs, sexual misconduct, vaginal infection, diabetes, being overweight, and genetic predisposition [1,3]. The three groups most at risk for developing UTIs are young boys, older men, and women of all ages. Urinary tract surgery, indwelling catheters, and obstruction from stones in the urinary tract raise the risk of urosepsis, which has a fatality rate of up to 20% [4,5]. Furthermore, UTIs exacerbate the patient’s condition through frequent recurrences, pyelonephritis with septicemia, kidney impairment in young children, and premature delivery [1].
The most prevalent causes of UTIs in humans are Escherichia coli, Klebsiella pneumoniae, Staphylococcus saprophyticus, Proteus mirabilis, Enterococcus faecalis, group B Streptococcus, Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus epidermidis, Enterobacter spp., Citrobacter spp., Serratia spp., and Candida spp., with changes in their relative incidence [6]. The empirical antibiotic selection is aided by sensitivity data from regional microbiological testing laboratories while treating UTI patients. Due to rare reporting of mild UTI cases to laboratories, these statistics are mainly helpful in diagnosing complex UTIs. Because of this, UTIs are often treated with antibiotics without laboratory confirmation, especially in remote and small-town settings where obtaining a urine culture is challenging and antibiotics are often misused.
However, bacteria associated with UTIs alter their genome structure and other metabolism processes to become resistant to newly introduced antibiotics [7]. Bacterial strains resistant to antibiotics continuously evolve to coincide with evolutionary processes that encourage the emergence of resistant strains by facilitating the transmission of antibiotic resistance traits via genetic elements, including plasmids, transposons, and integrons [8]. The emergence of multidrug-resistant (MDR) bacteria from diverse sources, such as humans [9], poultry [10], cattle [11], and fish [12], has been the central focus of several recent investigations. This has heightened the requirement for regular antibiotic sensitivity testing to identify the preferred medicine and screened for the emergence of MDR strains [13]. The World Health Organization (WHO) has undertaken several approaches to combat antibiotic resistance [14,15]. Some of these include setting criteria for bacteria that may effectively coordinate the monitoring of antibiotic resistance across the most prevalent bacteria, establishing a national response team, and adopting metrics to track and assess the consequences of antimicrobial resistance. However, even though richer countries are using these methods well, many developing countries have few treatment options due to a lack of resources.
Antimicrobial resistance is a universal health concern, yet it affects developing countries more than developed ones [16]. Recent studies have shown that uropathogens are becoming more resistant to commonly prescribed antibiotics in the eastern regions of Bangladesh, India, and Nepal [17]. Because this varies from region to region, it is essential to regularly assess the state of bacteria and their sensitivity patterns in UTIs. It is also likely that the underlying causes of UTIs and the degree of their resistance to the most popular treatments have evolved over the years. To promote the appropriate use of prescribed drugs, it is, therefore, necessary for public health to periodically and continually assess the local incidence of resistant bacteria and their susceptibility profiles to these infections. For this reason, this study was undertaken to isolate and identify the most common bacteria and determine their antibiotic-resistant patterns and potential risk determinants in patients with UTIs in northern Bangladesh.
2. MATERIALS AND METHODS
2.1. Study Area and Sampling Size Determination
Between November 2020 and December 2021, a cross-sectional investigation was conducted at the Department of Veterinary and Animal Sciences, the University of Rajshahi, in collaboration with a private diagnostic facility in the districts of Bogra and Rangpur, Bangladesh, to evaluate the risk factors and antibiogram profiling of bacteria among individuals suffering from UTIs [Figure 1]. The calculation of the size of the samples was done by the formula of the survey [18]: n = z2p (1-p)/d2, where z (95% confidence interval) = 1.96; P (prevalence) = 20.73%, and d = acceptable error (5%). The data utilized in the calculation of sample size was collected from the Northern International Medical College Hospital in Dhaka, Bangladesh, which reported a prevalence of 20.73% of patients with UTIs [19]. Using an arbitrary questionnaire for symptoms, 256 samples were obtained.
 | Figure 1: Geographical location of the study areas of urinary tract infections in the northern region of Bangladesh. (a) study location of bogra and rangpur district in bangladesh map, (b) twelve upazilas of bogra district, and (c) eight upazilas of rangpur district. [Click here to view] |
2.2. Collection and Processing of Samples and Data Collection
The research methods and guidelines employed in this study were approved by the Institutional Animal, Medical Ethics, Biosafety, and Biosecurity Committee of the Institute of Biological Science at the University of Rajshahi (Memo No. 56/321/IAMEBBC/IBSc). Each patient who took part in the trial gave their informed consent. Midstream urine samples were taken from each suspected patient using the standard protocol and placed in sterile, comprehensive glass containers. The samples were then sent to the Rajshahi University Department of Veterinary and Animal Sciences within an hour while they were still sterile and in a cold chain for microbiological investigation. All patients were given instructions on how to collect samples aseptically. Sociodemographic information about each patient was noted, including gender, age, place of residence, degree of education, and marital status. Other information recorded were urine color and appearance, pH, blood or purulent material presence, pregnancy status, whether the UTIs originated in a hospital or as outpatients, and prior antibiotic use.
2.3. Isolation, Identification and Characterization of Uropathogens
HiChrome UTI agar medium (HiMedia, Mumbai, India) was used to screen uropathogens such as E. coli, K. pneumoniae, P. aeruginosa, P. mirabilis, E. faecalis, and S. aureus based on enzyme-substrate reactions, leading to the distinctive colony and color production [Figure 2] [20]. Briefly, the uncentrifuged, evenly mixed urine samples were inoculated on UTI agar plates and incubated at 37°C aerobically for 18–24 h. Following incubations, the cultures were subcultured on various media, such as Mannitol salt agar, Eosin Methylene Blue, MacConkey agar, and Sheep Blood Agar, and examined and documented. A UTI diagnosis was made when at least 105 colony-forming units (CFU) of bacteria per milliliter of urine were found to be present. The isolates on the selective media were stored in glycerol at 40% at −80°C. Then, gram staining, string tests, sugar fermentation tests, methyl red tests, Voges-Proskauer tests, catalase tests, coagulase tests, the reaction in triple sugar iron (TSI) agar tests, and indole tests were used to study the shape and biochemistry of the colonies [21].
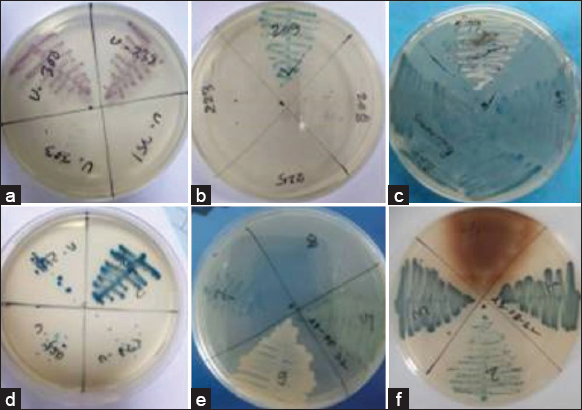 | Figure 2: Detection of various types of bacteria from human urine using chromogenic media. (a) pink color for Escherichia coli, (b) blue, small color for Enterococcus faecalis, (c) colorless (greenish pigment) for Pseudomonas aeruginosa, (d) blue to purple, mucoid color for Klebsiella pneumonie, (e) golden yellow color for Staphylococcus aureus, and (f) Light brown color for Proteus mirabilis. [Click here to view] |
2.4. Antibiotic Susceptibility Testing
The antibiotic susceptibility test (AST) for the uropathogens that had been isolated was done with the Kirby-Bauer disk diffusion method [22]. Twenty two commonly used antibiotics under ten classes were employed: aminoglycosides (gentamicin-10 μg, amikacin-30 μg, and netilmicin-30 μg), carbapenems (imipenem-10 μg and meropenem-10 μg), cephalosporins (cephradine-30 μg, cefuroxime sodium-30 μg, ceftriaxone-30 μg, ceftazidime-30 μg, cefepime-30 μg, and cefixime-5 μg), fluoroquinolones (ciprofloxacin-5 μg, moxifloxacin-5 μg, levofloxacin-5 μg, and nalidixic acid-30 μg), monobactams (Aztreonam-30 μg), penicillins + b-lactamase inhibitors (amoxicillin 20 μg-clavulanic acid-10 μg), tetracyclines (doxycycline-30 μg), macrolides (azithromycin-30 μg), folate pathway inhibitors (co-trimoxazole = sulfamethoxazole-23.75 μg and trimethoprim-1.25 μg), oxazolidinones (Linezolid-30 μg), and nitrofuran (nitrofurantoin-300 μg). The AST was done by freshly spreading each isolated bacterial growth culture with an equal concentration of 0.5 McFarland solutions on Mueller-Hinton agar (HiMedia, India) plates. Clinical and Laboratory Standards Institute guidance was followed to classify the results as sensitive or resistant [23]. According to the preceding study [24], MDR isolates were classified. In addition, the multiple antibiotic resistance (MAR) index was calculated using the following formula: MAR = a/b, where “a” represents the number of drugs that were resistant to a specific isolate and “b” denotes the average number of tested antibiotics [25].
2.5. Statistical Analysis
All data from the lab investigation and the questionnaire survey were entered into a Microsoft Excel spreadsheet. The coded data were processed using IBM’s statistics is a statistical software suite version 24 for processing and analysis (Armonk, NY, USA). To summarize the data, descriptive statistics were utilized. The proportions of participants who identified as positive or negative about culture were compared using the Chi-square test. An estimate of the prevalence of UTIs was calculated. The risk factors linked to UTIs were measured using binary logistic regression analysis. Statistics were judged significant at P < 0.05.
3. RESULTS
3.1. Societal and Demographic Characteristics
Of 256 patients with suspected UTIs, 164 (64.07%) and 92(35.93%) were female and male, respectively. The average age of the patients was 37.05 ± 10.5 years, with 27 (10.54%) of them being under the age of 15 years or under, 57 years (22.27%) being between the ages of 16 and 30 years, 56 (21.88%) being between the ages of 31 and 45 years, and 46 and 60 years, 59 (23.04%) being beyond the age of 60 years. One hundred and seventy-two (67.19%) patients lived in rural areas and 206 (80.47%) of the study participants had completed elementary school, secondary school, or tertiary education [Table S1].
3.2. Frequency of UTIs and uropathogens with Associated Factors
Of the 256 urine samples tested bacteriologically, 132 (51.56%) had significant bacteriuria (105 CFU/mL), and females had a higher (64.40%; 85/132) prevalence of UTIs than males (35.60%; 47/132). The age group with the highest rate of UTIs was 61 years or older (26.51%; 35/59), followed by 16–30 years (21.21%; 28/57), 46–60 years (21.21%; 28/57), 31–45 years (18.94%; 25/56), and 15 years or under (12.13%; 16/27). Interestingly, males aged 61 or older and 15 or under showed a higher prevalence of UTIs (16.67% and 6.06%, respectively) than females (9.84% and 5.30%, respectively). Whereas, in the age groups of 16–30 (21.21%), 31–45 (5.15%), and 46–60 (12.87%), females were more prone than males (0.75%, 6.06%, and 6.06%, respectively).
One hundred thirty-two bacterial isolates were detected from buoyant urine sample collections, including 94 (71.21%) Gram-negative and 38 (28.79%) Gram-positive bacteria. We further grouped these into two Gram-positive and four Gram-negative bacteria. We found the majority of isolated bacteria were Gram-negative, including E. coli (41.66%; 55/132), followed by E. faecalis (23.48%; 31/132) and K. pneumoniae (18.93%; 25/132) [Table 1 and Figure 2]. However, we did not find any significant correlation between gender and the types of bacterial infections found (P = 0.141).
Table 1: Prevalence of uropathogens among genders in the study population.
| Uropathogens | Male (n=47) | Female (n=85) | Total | Pearson Chi-square | P-value |
|---|---|---|---|---|---|
| Gram-negative | |||||
| Escherichia coli | 20 (43.47%) | 35 (44.30%) | 55 (41.66%) | 8.298, df=5 | 0.141 |
| Klebsiella spp. | 8 (17.39%) | 17 (21.51) | 25 (18.93%) | ||
| Pseudomonas spp. | 4 (8.69%) | 7 (8.86%) | 11 (8.33%) | ||
| Proteus spp. | 3 (6.5%) | 0% | 3 (2.27%) | ||
| Gram-positive | |||||
| Enterococcus spp. | 8 (17.39%) | 23 (29.11%) | 31 (23.48%) | ||
| Staphylococcus spp. | 4 (8.69%) | 3 (3.79%) | 7 (5.30%) |
The age group of 15 years and younger had the highest concentration of E. coli (60%), followed by that of 46–60 years (52%), 31–45 years (39.29%), 16–30 years (37.93%), and ≥61 years (31.42%). K. pneumoniae (28.58%), P. aeruginosa (20%), E. faecalis (8.58%), and P. mirabilis (2.86%) were the most common organisms among people aged ≥61 years. For 46–60 years, E. faecalis (24%), K. pneumoniae (20%), and P. aeruginosa (4%) were the most common organisms. E. faecalis (32.14%) was the second most frequent organism for 31–45 years, followed by K. pneumoniae (21.42%), P. aeruginosa (3.57%), and S. aureus (3.57%). S. aureus (10.34%), K. pneumoniae (6.90%), and P. aeruginosa (3.44%), followed by E. faecalis (41.38%) as the second most prevalent organism over 16–30 years. In addition, for ≤15 years, K. pneumoniae (13.33%) and P. mirabilis (13.33%) were the second most common organisms, followed by P. aeruginosa (6.67%) and E. faecalis (6.67%). Regarding the bacterial isolate, no obvious correlation was found within the age group [Table 2].
Table 2: Prevalence of uropathogens among the age groups of the study population.
| Uropathogens | ≤15 years (n=15) | 16–30 years (n=29) | 31–45 years (n=28) | 46–60 years (n=25) | ≥61 years (0=35) | Chi-square |
|---|---|---|---|---|---|---|
| Gram-negative | ||||||
| Escherichia coli | 9 (60%) | 11 (37.93%) | 11 (39.29%) | 13 (52%) | 11 (31.42%) | 36.2, df=20 |
| Klebsiella spp. | 2 (13.33%) | 2 (6.90%) | 6 (21.42%) | 5 (20%) | 10 (28.58%) | |
| Pseudomonas spp. | 1 (6.67%) | 1 (3.44%) | 1 (3.57%) | 1 (4%) | 7 (20%) | |
| Proteus spp. | 2 (13.33%) | 0 | 0 | 0 | 1 (2.86%) | |
| Gram-positive | ||||||
| Enterococcus spp. | 1 (6.67%) | 12 (41.38%) | 9 (32.14%) | 6 (24%) | 3 (8.58%) | |
| Staphylococcus spp. | 0 | 3 (10.34%) | 1 (3.57%) | 0 | 3 (8.58%) |
Of the 132 bacterial isolates, 88 (66.67%) came from people who lived in rural areas, and the remaining 44 (33.33%) came from people who lived in cities. UTIs are significantly related to pregnancies and prior antibiotic usage (P < 0.05). Among the studied participants, outpatients were more (81.81%; 108/132) than hospital patients (18.19%, 24/132). Approximately 58.33% (77/132) of the patients were married, compared to 30.30% (40/132) of single and 11.37% (15/132) of widowed. Most of the study participants had completed their education, with 22.72% (30/132) having attended primary school, 25% having attended secondary school, and 28.79% having attended university, but just 23.49% (31/132) were illiterate [Table 3].
Table 3: Association of urinary tract infections with sociodemographic features of research participants in Bangladesh’s Northern region (n=256).
| Factors | Bacteriological growth (%) | Total (%) | χ2 | P-value | |
|---|---|---|---|---|---|
| Positive=132 (51.56) | Negative=124 (48.43) | ||||
| Age (years) | |||||
| ≤15 | 16 (12.13) | 11 (8.88) | 27 (10.54) | 3.408 | 0.492 |
| 16–30 | 28 (21.21) | 29 (23.38) | 57 (22.27) | ||
| 31–45 | 25 (18.94) | 31 (25) | 56 (21.88) | ||
| 46–60 | 28 (21.21) | 29 (23.38) | 57 (22.27) | ||
| ≥61 | 35 (26.51) | 24 (19.36) | 59 (23.04) | ||
| Sex | |||||
| Male | 47 (35.60) | 45 (36.30) | 92 (35.93) | 0.013 | 0.909 |
| Female | 85 (64.40) | 79 (63.70) | 164 (64.07) | ||
| Residence | |||||
| Rural | 88 (66.67) | 84 (67.74) | 172 (67.19) | 0.307 | 0.579 |
| Urban | 44 (33.33) | 40 (32.26) | 84 (32.81) | ||
| Disease acquired from | |||||
| Outpatient | 108 (81.81) | 108 (87.10) | 216 (84.38) | 1.351 | 0.245 |
| Inpatient | 24 (18.19) | 16 (12.90) | 40 (15.62) | ||
| Marital status | |||||
| Single | 40 (30.30) | 33 (26.61) | 73 (28.51) | 1.095 | 0.578 |
| Married | 77 (58.33) | 80 (64.51) | 157 (61.33) | ||
| Widowed | 15 (11.37) | 11 (8.88) | 26 (10.16) | ||
| Prior antibiotic use | |||||
| Yes | 101 (76.51) | 30 (24.20) | 131 (51.18) | 70.05 | 0 |
| No | 31 (23.49) | 94 (75.80) | 125 (48.82) | ||
| Educational status | |||||
| Illiterate | 31 (23.49) | 19 (15.32) | 50 (19.53) | 3.953 | 0.267 |
| Primary school | 30 (22.72) | 29 (23.39) | 59 (23.05) | ||
| Secondary school | 33 (25) | 42 (33.88) | 75 (29.30) | ||
| University | 38 (28.79) | 34 (27.41) | 72 (28.12) | ||
| Pregnancy | |||||
| Yes | 12 (9.10) | 3 (2.41) | 15 (5.86) | 5.159 | 0.023 |
| No | 120 (90.90) | 121 (97.59) | 241 (94.14) | ||
3.3. Antimicrobial Susceptibility Profile of Bacterial Uropathogens
Antimicrobial susceptibility testing revealed that Gram-negative and Gram-positive bacterial isolates had various sensitivity patterns to single and multiple antimicrobials often used to treat infections [Tables 4 and 5]. The majority of the Gram-negative isolates were sensitive to imipenem (98.93%), meropenem (97.88%), amikacin (93.61%), netilmicin (93.61%), nitrofurantoin (75.82%), gentamicin (74.47%), moxifloxacin (73.40%), levofloxacin (73.40%), ceftazidime (65.96%), aztreonam (61.70%), ciprofloxacin (59.58%), ceftriaxone (58.18%), amoxicillin + clavulanic acid (57.44%), doxycycline (55.31%), and cefepime (54.25%). In contrast, Gram-negative bacteria were highly resistant to linezolid (77.66%), cephradine (73.40%), nalidixic acid (70.32%), cefuroxime sodium (63.82%), co-trimoxazole (60.63%), cefixime (54.26%), and azithromycin (54.26%). Over 55% of E. coli isolates tested positive for resistance to nalidixic acid, linezolid, cephradine, cefuroxime sodium, azithromycin, and ceftriaxone. Low resistance rates were found for meropenem (1.81%), nitrofurantoin (5.46%), netilmicin (7.28%), and amikacin (9.10%). Over 55% of K. pneumoniae was resistant to linezolid, cephradine, cefixime, cefuroxime sodium, and co-trimoxazole. However, K. pneumoniae showed resistance to imipenem, meropenem, amikacin, and netilmicin at a 4%–8% range. In addition, roughly 55% of P. aeruginosa isolates exhibited resistance to co-trimoxazole, linezolid, nitrofurantoin, cefuroxime sodium, amoxicillin + clavulanic acid, doxycycline, nalidixic acid, and cephradine, but gentamicin resistance rates were generally low. Each isolate of P. aeruginosa was 100% effective against amikacin, netilmicin, imipenem, meropenem, and aztreonam. P. mirabilis was completely resistant against cephradine, cefuroxime sodium, moxifloxacin, doxycycline, azithromycin, co-trimoxazole, and linezolid, whereas 66.67% of it was only resistant against gentamicin, cefepime, ciprofloxacin, levofloxacin, and aztreonam. The sensitivity of P. mirabilis to amikacin, netilmicin, imipenem, meropenem, and ceftriaxone was 100%.
Table 4: Antibiotics susceptibility patterns of Gram-negative bacterial uropathogens isolated from patients with urinary tract infections (%).
| Antibiotic classes | Bacteria | Gram-negative bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli (n=55) | Klebsiella pneumoniae (n=25) | Pseudomonas aeruginosa (n=11) | Proteus mirabilis (n=3) | Total (n=94) | |||||||
| Antibiotics | R | S | R | S | R | S | R | S | R | S | |
| Aminoglycosides | GM | 14 (25.46) | 41 (74.54) | 7 (28) | 18 (72) | 1 (9.10) | 10 (90.90) | 2 (66.67) | 1 (33.33) | 24 (25.53) | 70 (74.47) |
| AK | 5 (9.10) | 50 (90.90) | 1 (4) | 24 (96) | 0 (00) | 11 (100) | 0 (00) | 3 (100) | 6 (6.39) | 88 (93.61) | |
| NET | 4 (7.28) | 51 (92.72) | 2 (8) | 23 (92) | 0 (00) | 11 (100) | 0 (00) | 3 (100) | 6 (6.39) | 88 (93.61) | |
| Carbapenems | IMI | 0 (00) | 55 (100) | 1 (4) | 24 (96) | 0 (00) | 11 (100) | 0 (00) | 3 (100) | 1 (1.07) | 93 (98.93) |
| MEM | 1 (1.81) | 54 (98.18) | 1 (4) | 24 (96) | 0 (00) | 11 (100) | 0 (00) | 3 (100) | 2 (2.12) | 92 (97.88) | |
| Cephalosporins | CE | 39 (70.90) | 16 (29.09) | 20 (80) | 5 (20) | 7 (63.63) | 4 (36.37) | 3 (100) | 0 (00) | 69 (73.40) | 25 (26.60) |
| CXM | 35 (63.63) | 20 (36.37) | 14 (56) | 11 (44) | 8 (72.72) | 3 (27.28) | 3 (100) | 0 (00) | 60 (63.82) | 34 (36.18) | |
| CRO | 32 (58.18) | 23 (41.81) | 12 (48) | 13 (52) | 2 (18.19) | 9 (81.81) | 0 (00) | 3 (100) | 46 (48.93) | 48 (51.07) | |
| CAZ | 22 (40) | 33 (60) | 7 (28) | 18 (72) | 2 (18.19) | 9 (81.81) | 1 (33.33) | 2 (66.67) | 32 (34.04) | 62 (65.96) | |
| CPM | 30 (54.54) | 25 (45.45) | 10 (40) | 15 (60) | 1 (9.10) | 10 (90.90) | 2 (66.67) | 1 (33.33) | 43 (45.74) | 51 (54.25) | |
| CFM | 29 (52.72) | 26 (47.28) | 16 (64) | 9 (36) | 5 (45.46) | 6 (54.54) | 1 (33.33) | 2 (66.67) | 51 (54.26) | 43 (45.74) | |
| Fluoroquinolones | CIP | 23 (41.81) | 32 (58.19) | 10 (40) | 15 (60) | 3 (27.28) | 8 (72.72) | 2 (66.67) | 1 (33.33) | 38 (40.42) | 56 (59.58) |
| MFX | 13 (23.63) | 42 (76.37) | 6 (24) | 19 (76) | 3 (27.28) | 8 (72.72) | 3 (100) | 0 (00) | 25 (26.60) | 69 (73.40) | |
| LEV | 15 (27.28) | 40 (72.72) | 6 (24) | 19 (76) | 2 (18.19) | 9 (81.81) | 2 (66.67) | 1 (33.33) | 25 (26.60) | 69 (73.40) | |
| NA | 44 (80) | 11 (20) | 13 (52) | 12 (48) | 7 (63.63) | 4 (36.37) | ND | 64 (70.32) | 27 (29.68) | ||
| Monobactams | ATM | 24 (43.63) | 31 (56.37) | 10 (40) | 15 (60) | 0 | 11 (100) | 2 (66.67) | 1 (33.33) | 36 (38.30) | 58 (61.70) |
| Penicillins + b-lactamase inhibitors | AUG | 21 (38.19) | 34 (61.81) | 11 (44) | 14 (56) | 7 (63.63) | 4 (36.37) | 1 (33.33) | 2 (66.67) | 40 (42.56) | 54 (57.44) |
| Tetracyclines | DXT | 22 (40) | 33 (60) | 10 (40) | 15 (60) | 7 (63.63) | 4 (36.37) | 3 (100) | 0 (00) | 42 (44.69) | 52 (55.31) |
| Macrolides | ATH | 33 (60) | 22 (40) | 11 (44) | 14 (56) | 4 (36.37) | 7 (63.63) | 3 (100) | 0 (00) | 51 (54.26) | 43 (45.74) |
| Folate pathway inhibitors | TS | 30 (54.54) | 25 (45.45) | 14 (56) | 11 (44) | 10 (90.90) | 1 (9.10) | 3 (100) | 0 (00) | 57 (60.63) | 37 (39.37) |
| Oxazolidinones | LZD | 40 (72.72) | 15 (27.28) | 21 (84) | 4 (16) | 9 (81.81) | 2 (18.19) | 3 (100) | 0 (00) | 73 (77.66) | 21 (22.34) |
| Nitrofuran | NI | 3 (5.46) | 52 (94.54) | 10 (40) | 15 (60) | 9 (81.81) | 2 (18.19) | ND | 22 (24.18) | 69 (75.82) | |
GM: Gentamicin, AK: Amikacin, NET: Netilmicin, IMI: Imipenem, MEM: Meropenem, CE: Cephradine, CXM: Cefuroxime sodium, CRO: Ceftriaxone, CAZ: Ceftazidime, CPM: Cefepime, CFM: Cefixime, CIP: Ciprofloxacin, MFX: Moxifloxacin, LEV: Levofloxacin, NA: Nalidixic acid, ATM: Aztreonam, AUG: Amoxicillin+Clavulanic Acid, DXT: Doxycycline, ATH: Azithromycin, TS: Co-trimoxazole (sulfamethoxazole and trimethoprim), LZD: Linezolid, NI: Nitrofurantoin, R: Resistant, S: Sensitive, ND: Not done.
Table 5: Antibiotics susceptibility patterns of Gram-positive bacterial uropathogens isolated from patients with urinary tract infections (%).
| Antibiotic classes | Bacteria | Gram-positive bacteria | |||||
|---|---|---|---|---|---|---|---|
| Enterococcus faecalis (n=31) | Staphylococcus aureus (n=7) | Total (n=38) | |||||
| Antibiotics | R | S | R | S | R | S | |
| Aminoglycosides | GM | 5 (16.13) | 26 (83.87) | 1 (14.29) | 6 (85.71) | 6 (15.79) | 32 (84.21) |
| AK | 3 (9.67) | 28 (90.33) | 1 (14.29) | 6 (85.71) | 4 (10.52) | 34 (89.48) | |
| NET | 3 (9.68) | 28 (90.32) | 1 (14.29) | 6 (85.71) | 4 (10.52) | 34 (89.48) | |
| Carbapenems | IMI | 1 (3.23) | 30 (96.77) | 1 (14.29) | 6 (85.71) | 2 (5.27) | 36 (94.73) |
| MEM | 5 (16.13) | 26 (83.87) | 1 (14.29) | 6 (85.71) | 6 (15.79) | 32 (84.21) | |
| Cephalosporins | CE | 26 (83.87) | 5 (16.13) | 6 (85.71) | 1 (14.29) | 32 (84.21) | 6 (15.79) |
| CXM | 20 (64.51) | 11 (35.49) | 4 (57.14) | 3 (42.86) | 24 (63.16) | 14 (36.84) | |
| CRO | 21 (67.74) | 10 (32.25) | 5 (71.42) | 2 (28.58) | 26 (68.42) | 12 (31.58) | |
| CAZ | 12 (38.70) | 19 (61.30) | 4 (57.14) | 3 (42.86) | 16 (42.10) | 22 (57.90) | |
| CPM | 7 (22.59) | 24 (77.41) | 1 (14.29) | 6 (85.71) | 8 (21.05) | 30 (78.94) | |
| CFM | 28 (90.32) | 3 (9.68) | 6 (85.71) | 1 (14.29) | 34 (89.48) | 4 (10.52) | |
| Fluoroquinolones | CIP | 12 (38.70) | 19 (61.30) | 2 (28.58) | 5 (71.42) | 14 (36.84) | 24 (63.16) |
| MFX | 7 (22.59) | 24 (77.41) | 2 (28.58) | 5 (71.42) | 9 (23.69) | 29 (76.31) | |
| LEV | 8 (25.80) | 23 (74.20) | 1 (14.29) | 6 (85.71) | 9 (23.69) | 29 (76.31) | |
| NA | 25 (80.64) | 6 (19.36) | 3 (42.86) | 4 (57.14) | 28 (73.69) | 10 (26.31) | |
| Monobactams | ATM | 10 (32.26) | 21 (67.74) | 4 (57.14) | 3 (42.86) | 14 (36.84) | 24 (63.16) |
| Penicillins + b-lactamase inhibitors | AUG | 14 (45.17) | 17 (54.83) | 3 (42.86) | 4 (57.14) | 17 (44.73) | 21 (55.27) |
| Tetracyclines | DXT | 17 (54.83) | 14 (45.17) | 2 (28.58) | 5 (71.42) | 19 (50) | 19 (50) |
| Macrolides | ATH | 27 (87.10) | 4 (12.90) | 6 (85.71) | 1 (14.29) | 33 (86.84) | 5 (13.16) |
| Folate pathway inhibitors | TS | 22 (70.97) | 9 (29.03) | 5 (71.42) | 2 (28.58) | 27 (71.06) | 11 (28.94) |
| Oxazolidinones | LZD | 22 (70.97) | 9 (29.03) | 3 (42.86) | 4 (57.14) | 25 (65.79) | 13 (34.21) |
| Nitrofuran | NI | 13 (41.93) | 18 (58.07) | 1 (14.29) | 6 (85.71) | 14 (36.84) | 24 (63.16) |
GM: Gentamicin, AK: Amikacin, NET: Netilmicin, IMI: Imipenem, MEM: Meropenem, CE: Cephradine, CXM: Cefuroxime sodium, CRO: Ceftriaxone, CAZ: Ceftazidime, CPM: Cefepime, CFM: Cefixime, CIP: Ciprofloxacin, MFX: Moxifloxacin, LEV: Levofloxacin, NA: Nalidixic acid, ATM: Aztreonam, AUG : Amoxicillin+Clavulanic Acid, DXT: Doxycycline, ATH: Azithromycin, TS: Co-trimoxazole (sulfamethoxazole and trimethoprim), LZD: Linezolid, NI: Nitrofurantoin, R: Resistant, S: Sensitive.
On the contrary, the preponderance of the Gram-positive isolates tested was sensitive to imipenem (94.73%), amikacin (89.48%), netilmicin (89.48%), meropenem (84.21%), gentamicin (84.21%), cefepime (78.94%), moxifloxacin (76.31%), levofloxacin (63.16%), ciprofloxacin (63.16%), aztreonam (63.16%), nitrofurantoin (63.16%), ceftazidime (57.90%), and amoxicillin + clavulanic acid (55.27%). In contrast, Gram-positive bacteria were all very resistant to cefixime (89.48%), azithromycin (86.84%), cephradine (84.21%), nalidixic acid (73.69%), co-trimoxazole (71.06%), ceftriaxone (68.42%), linezolid (65.79%), and cefuroxime sodium (63.16%). More than 55% of E. faecalis isolates were resistant to cefixime, azithromycin, cephradine, nalidixic acid, co-trimoxazole, linezolid, ceftriaxone, and cefuroxime sodium. Low resistance rates were found for imipenem (3.23%), amikacin (9.67%), netilmicin (9.68%), meropenem (16.13%), and gentamicin (16.13%). Over 55% of S. aureus was resistant to cephradine, cefixime, azithromycin, ceftriaxone, co-trimoxazole, cefuroxime sodium, ceftazidime, and aztreonam. However, 14.29% of S. aureus were only resistant to gentamicin, amikacin, netilmicin, imipenem, meropenem, cefepime, levofloxacin, and nitrofurantoin.
3.4. MDR Trends and the MAR Index of Uropathogenic Bacteria
From antibiotic susceptibility profiles, 117/132 (88.6%) of the bacterial isolates were MDR, 27/132 (20.4%) were extreme drug resistant (XDR), and 1/132 (0.75%) were PDR [Table 6]. Among MDR strains, only 4 (3%) isolates were resistant to two classes of antibiotics; the rest, 113 (85.6%) were resistant to three or more classes of antibiotics. In addition, a range of MAR indices, from 0.13 to 2.13, were detected in the antibiotic resistance profiles of each bacterial uropathogenic isolate. Among tested Gram-negative bacteria isolates, 80/94 (85.1%) were MDR and 17/94 (18%) were XDR. Five bacterial (5.3%) isolates were antibiotic-sensitive across all antibiotic classes [Table 6]. However, a comparison of multidrug-resistance patterns within species revealed that 47/55 (85.4%) of E. coli isolates, 20/25 (80%) of K. pneumoniae isolates, 10/11 (90.9%) of P. aeruginosa isolates, and 3/3 (100%) of P. mirabilis isolates were all MDR. In addition, 1/11 (9.1%) of P. aeruginosa isolates were extensively drug-resistant, as were 9/55 (16.4%) of E. coli isolates, 5/25 (20%) of K. pneumoniae isolates, and 2/3 (66.6%) of P. mirabilis isolates [Table 6]. According to the results of the study’s antibiotic susceptibility test, the isolates of E. coli had the highest MAR index (2.13), followed by those of K. pneumoniae (0.90), P. aeruginosa (0.45), and P. mirabilis isolates (0.13). 37/38 (97.3%) of the isolates of Gram-positive bacteria were MDR, 10/38 (26.3%) were XDR, and 1/38 (2.6%) were PDR-resistant isolates. Gram-positive bacterial isolates were not susceptible to any of the investigated antibiotic classes. However, of the examined isolates, E. faecalis was MDR in 31/31 (100%) cases, with 9/31 (29%) being XDR isolates. In this analysis, 6/7 (85.7%) and 1/7 (14.2%) of the S. aureus isolates were MDR and XDR, respectively. Only 1/7 (14.2%) of the PDR strain was found in S. aureus isolation. As shown in Table 6, E. faecalis had a MAR index of 1.40, whereas S. aureus had a MAR value of 0.27.
Table 6: Multi-drug resistance patterns of uropathogens isolated from patients with urinary tract infections.
| Uropathogens | Degree of resistance | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R0 | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 | ≥R12 | MDR | XDR | PDR | MAR Index | |
| Gram-negative | 5 (5.3) | 5 (5.3) | 4 (4.2) | 6 (6.4) | 2 (2.1) | 3 (3.2) | 8 (8.5) | 5 (5.3) | 12 (12.8) | 15 (15.9) | 8 (8.5) | 5 (5.3) | 16 (17) | 80 (85.1) | 17 (18) | 0 | 0.90 |
| Escherichia coli (n=55) | 2 (3.6) | 5 (9.1) | 1 (1.8) | 3 (5.4) | 1 (1.8) | 2 (3.6) | 6 (10.9) | 3 (5.4) | 8 (14.5) | 8 (14.5) | 4 (7.3) | 4 (7.3) | 8 (14.5) | 47 (85.4) | 9 (16.4) | - | 2.13 |
| Klebsiella pneumoniae (n=25) | 2 (8) | - | 3 (12) | 2 (8) | - | - | 2 (8) | 2 (8) | 1 (4) | 5 (20) | 2 (8) | 1 (4) | 5 (20) | 20 (80) | 5 (20) | - | 0.90 |
| Pseudomonas aeruginosa (n=11) | 1 (9.1) | - | - | 1 (9.1) | 1 (9.1) | 1 (9.1) | - | - | 3 (27.3) | 2 (18.2) | 1 (9.1) | - | 1 (9.1) | 10 (90.9) | 1 (9.1) | - | 0.45 |
| P. mirabilis (n=3) | - | - | - | - | - | - | - | - | - | - | 1 (33.3) | - | 2 (66.6) | 3 (100) | 2 (66.6) | - | 0.13 |
| Gram-positive | 0 | 1 (2.6) | 0 | 0 | 1 (2.6) | 0 | 2 (5.3) | 2 (5.3) | 10 (26.3) | 3 (7.9) | 8 (21) | 3 (7.9) | 8 (21) | 37 (97.3) | 10 (26.3) | 1 (2.6) | 0.83 |
| Enterococcus faecalis (n=31) | - | - | - | - | 1 (3.2) | - | 2 (6.4) | 2 (6.4) | 5 (16.1) | 3 (9.7) | 8 (25.8) | 3 (9.7) | 7 (22.6) | 31 100) | 9 (29) | - | 1.40 |
| Staphylococcus aureus (n=7) | - | 1 (14.2) | - | - | - | - | - | - | 5 (71.4) | - | - | - | 1 (14.2) | 6 (85.7) | 1 (14.2) | 1 (14.2) | 0.27 |
| Total (n=132) | 5 (3.7) | 6 (4.5) | 4 (3) | 6 (4.5) | 3 (2.3) | 3 (2.3) | 10 (7.6) | 7 (5.3) | 22 (16.7) | 18 (13.6) | 16 (12.1) | 8 (6) | 24 (18.1) | 117 (88.6) | 27 (20.4) | 1 (0.75) | |
Unless otherwise specified, all data are presented as numbers (%). R0: susceptible to all antibiotics, R1-11: resistance to 1-11 respectively, antibiotics, ≥R12: resistance to 12 or more antibiotics, MDR: multidrug resistant, XDR: extreme drug resistant, PDR: pan drug-resistant, MAR: multiple antibiotic resistance.
3.5. Associated Risk Factors for MDR Bacteria (MDRB) among Study Participants
By contrasting patients with and without UTIs caused by MDRB, risk factors related to these infections were examined. The Chi-square test analysis revealed a significant relationship between MDRB UTI infections and age (P = 0.006), place of residence (P = 0.000), marital status (P = 0.029), and prior antibiotic use (P = 0.000). As shown in Table 7, the risk variables for MDR bacteria (MDRB)-associated UTIs were residential location (rural vs. urban, odds ratio = 0.09, 95% confidence interval = 0.02–0.36, P = 0.000) and marital status (P = 0.042).
Table 7: Multidrug-resistant bacteria-associated risk factors among urinary tract infection suspects in the northern part of Bangladesh.
| Risk factors | MDRB | χ2 | P-value | Bivariate regression analysis | ||
|---|---|---|---|---|---|---|
| Yes (n=117) | No (n=15) | OR (95% CI) | P-value | |||
| Age (years) | ||||||
| ≤15 | 11 | 5 | 14.36 | 0.006 | 0.24 (0.00–9.45) | 0.226 |
| 16–30 | 26 | 2 | 5.90 (0.99–35.21) | |||
| 31–45 | 23 | 2 | 5.22 (0.87–31.31) | |||
| 46–60 | 22 | 6 | 1.66 (0.41–6.69) | |||
| ≥61 | 35 | 0 | 2.61 (0.00–6.60) | |||
| Sex | ||||||
| Male | 42 | 5 | 0.038 | 0.845 | 1.12 (0.36–3.49) | 0.845 |
| Female | 75 | 10 | ||||
| Residence | ||||||
| Rural | 85 | 3 | 16.58 | 0.000 | 0.09 (0.02–0.36) | 0.000 |
| Urban | 32 | 12 | ||||
| Disease acquired from | ||||||
| Outpatient | 94 | 14 | 1.50 | 0.219 | 0.29 (0.03–2.33) | 0.246 |
| Inpatient | 23 | 1 | ||||
| Marital status | ||||||
| Single | 31 | 9 | 7.06 | 0.029 | 0.23 (0.07–0.77) | 0.042 |
| Married | 72 | 5 | 0.97 (0.10–8.96) | |||
| Widowed | 14 | 1 | 4.06 (0.46–35.25) | |||
| Prior antibiotic use | ||||||
| Yes | 99 | 4 | 26.04 | 0.000 | 6.19 (0.00–2.21) | 0.946 |
| No | 18 | 11 | ||||
| Educational status | ||||||
| Illiterate | 30 | 1 | 6.25 | 0.100 | 0.150 | |
| Primary school | 28 | 2 | 0.46 (0.04–5.43) | |||
| Secondary school | 29 | 4 | 0.51 (0.02–2.29) | |||
| University | 30 | 8 | 0.51 (0.01–1.06) | |||
MDRB: Multidrug-resistant bacteria, CI: Confidence interval, OR: Odds ratio
4. DISCUSSION
UTIs have significantly burdened the healthcare system because of their high frequency in community and hospital settings [1,26]. The development of uropathogens, their pathogenesis, and their patterns of antibiotic susceptibility have evolved during the intervening period and will proceed in this manner in the future [27]. By continuously monitoring the sensitivity to antibiotics of urinary bacteria in specific locations, it is usually possible to identify the pathogens and choose an appropriate antibiotic for treating bacterial UTIs [28].
Bangladesh lacks data on how antimicrobial resistance (AMR) is being tracked, so the research results provide insight into how AMR is changing in that country. This study aimed to determine how resistant uropathogens are to antibiotics and to look at the situation in the northern Bangladeshi cities of Rangpur and Bogra. Of the 256 urine samples used in this investigation, 132 (51.56%) showed noticeable bacterial growth, while 124 (48.43%) showed no growth. The frequency rate was found to be higher in this studied population compared to the previous reports in Bangladesh, in a range between 42.66% and 30.9% [6,29-31]. However, it is also lower than that reported in other studies in Bangladesh, such as 62% [32], 60% [33], and 71% [34]. The research approach, demographics, sample size, and spatial variation may influence these discrepancies.
In agreement with the previous reports that found women are more likely to experience UTIs than men [32-34,35], our findings also show a higher prevalence of UTIs in females (64.40%) than in males (35.60%). There may be more female cases of UTIs than male cases due to factors such as the shorter urethra, the proximity of the urethral meatus to the anus, sexual activity, incontinence, and unhygienic or unsanitary toilet habits. Our study found that young females of potential reproductive age between 16 and 45 years had a higher incidence of UTIs, which is consistent with the previous research in Meerut (90.7% in 26–36 years), Jaipur (41.3% in 21–50 years), and Ethiopia (37.5% in 20–29 years) [36-38]. This is because their anatomical makeup makes them more susceptible to this condition. However, our study found that older males (61-years-old) had a higher frequency of UTIs (16.67%) than older females (9.84%). This finding is consistent with research results from Jaipur, Rajasthan (47.3%), Meerut, Uttar Pradesh (71.2%), and Sonipat, and Haryana (58.3%) in India [36,37,39]. The increased prevalence of neurogenic bladder and benign prostatic enlargement in older men may be the primary cause of increased UTI incidence [40].
This study also found a significant association between the prevalence of UTIs and pregnancy. Our study shows that pregnant patients are more likely to get UTIs than non-pregnant patients. In addition, individuals with prior therapy with antibiotics are at a higher risk for UTIs. Changes in the immune system and urine composition during pregnancy and improper antibiotic administration may contribute to these differences [41,42].
This study shows that Gram-negative bacteria (71.21%) are more prevalent than gram-positive bacteria (28.79%). Similar results have also been reported in India [35], Ethiopia [13,43], and other regions [44]. In agreement with several published reports globally [45-48], we found that the gram-negative bacterium E. coli (41.66%) was the most prevalent. In our investigation, K. pneumoniae (18.93%) and P. aeruginosa (8.33%) were the next two most often seen bacteria, followed by P. mirabilis (2.27%). The predominance of Gram-negative bacteria from the Enterobacteriaceae family that cause UTIs has increased for several reasons, including the colonization of the urogenital mucosa through integrins, pili, and fimbriae and the P-1 blood group phenotypic receptor [49]. E. faecalis, which accounted for up to 23.48% of isolated cases in this investigation, was the second most common species of UTI bacterium. The discovery of E. faecalis isolation as an uropathogen was consistent with investigations conducted in Dhaka, Bangladesh [50].
Antibiotic resistance is the most significant public health concern in UTIs. Our antibiogram study revealed higher resistance to commonly prescribed antimicrobial drugs. Overall, more than 54% of Gram-negative bacterial isolates were found to be resistant to linezolid, cephradine, nalidixic acid, cefuroxime sodium, co-trimoxazole, cefixime, and azithromycin, which is in line with the previous studies [51]. However, a low level of resistance has been documented for gram-negative agents against meropenem, imipenem, amikacin, netilmicin, nitrofurantoin, gentamicin, moxifloxacin, levofloxacin, ceftazidime, aztreonam, ciprofloxacin, ceftriaxone, amoxicillin plus clavulanic acid, doxycycline, and cefepime [Table 4]. In contrast, E. coli, K. pneumoniae, and P. aeruginosa showed the highest resistance (>70%) to cephradine, nalidixic acid, and linezolid. Similar findings were reported in Bangladesh [51] and Pakistan [52], where the resistance rate of cephradine and nalidixic acid was about 90%. Due to the high levels of antibiotic resistance found in Gram-negative bacterial isolates against commonly prescribed antimicrobial drugs, clinicians must use reservations and evidence-based prescribing practices to treat UTIs and stop spreading antibiotic resistance.
None of the isolates showed sensitivity to these antibiotics in the case of P. mirabilis. In this study, all the reported Gram-negative organisms were resistant to co-trimoxazole was 60.63%. This increase in resistance may result from the indiscriminate use of antibiotics as well as their easy access without a prescription in Bangladesh [53]. In December 2022, a WHO report exposed the growing issue of antibiotic resistance [54]. According to these results, more than 20% of E. coli isolates, which are frequently responsible for UTIs, are resistant to first-line medications such as ampicillin and co-trimoxazole and second-line medications such as fluoroquinolones [55]. In our study, the lowest observed resistance for E. coli was 1.81%, 5.46%, 7.28%, and 9.10% against meropenem, nitrofurantoin, netilmicin, and amikacin, respectively. A similar pattern has been reported for other Gram-negative uropathogens. However, imipenem was 100% effective against E. coli, P. aeruginosa, and P. mirabilis, except for K. pneumoniae (96%). Among the tested Gram-positive bacteria, more than 70% resistance was observed to cefixime, azithromycin, cephradine, nalidixic acid, co-trimoxazole, and linezolid in E. faecalis and S. aureus. This result is being investigated in Pakistan [52] and Bangladesh [51]. Due to the potential misuse of these antibiotics in our area to treat various diseases without first determining their culturing sensitiveness, an enormous rise in antibiotic resistance has been seen [53]. This worrying circumstance accounts for the majority of MDR infections among UTIs.
However, as the number of treatments utilizing these medications rises, the likelihood of the disease acquiring resistant strains also rises. The exorbitant expenses associated with medical treatment and the inherent risk of ineffective outcomes may result in severe adverse effects or even fatalities for individuals. In our study, the frequency of MDR, XDR, and PDR strains in UTIs emphasizes the urgent need for all-encompassing strategies to maintain antibiotics’ efficacy and protect the public’s health. The medical community must work together to combat antibiotic resistance, implement efficient measures to prevent infections, and support research into new management strategies to manage UTIs. If this issue is not passably addressed, patients may experience severe consequences, such as fewer treatments, increased health-care costs, and possibly fatal outcomes. MDR and MAR-caused UTIs are a severe threat to public health worldwide. Alarmingly, the overall prevalence of MDR was 88.6% among bacterial isolates found in UTI patients, which is comparable to the findings of other research in Gondar (85.7%) and Mozambique (88.2%) [56,57]. It was, however, lower than reports from Pakistan (90.7%) and many regions of Ethiopia, including Jimma (100%), Bahirdar (95.6%) [52,58,59]. The increasing tendency of MDR strains over time, variations in the study period, and variances in the study population could all contribute to the fluctuation in the prevalence of MDR isolates. This study showed that 85.4% of E. coli, 80% of K. pneumoniae, and 90.9% of P. aeruginosa isolates were MDR, which is almost identical to the previous study’s findings [52,60]. On the contrary, 100% of E. faecalis and 85.7% of S. aureus were MDR, which is higher than the earlier studies [52,13]. In line with the previous research, the high number of MDR strains in E. coli, K. pneumoniae, and P. aeruginosa isolates shows how hard it is to treat UTIs because bacteria are becoming resistant to antibiotics. Furthermore, the worrying rise in MDR rates in E. faecalis and S. aureus compared to earlier studies shows how urgently we need effective antimicrobial stewardship and cutting-edge strategies to deal with the growing threat of MDR infections caused by these bacteria. For instance, a range of MAR indices, from 0.13 to 2.13, were detected in the antibiotic resistance profiles of each bacterial uropathogenic isolate. According to the MAR index, antibiotics were frequently employed to treat patients in the region where urinary tract pathogens were identified, indicating an alarming factor for MDR and MAR bacteria. These bacteria, resistant to multiple drugs, could spread through the environment and transfer their resistance genes to other bacteria.
The rising irrational use of antibiotics, the spread of resistance genes from person to person or from animal to person, and the consumption of animal products treated with antibiotics may all contribute to the observed high resistance. This study’s rise in antibiotic resistance may be due to self-medication, non-compliance with prescriptions, and sales of substandard drugs [53]. In this study, age, place of residence, marital status, and prior antibiotic use were linked to MDR UTI. Similarly, in bivariate analysis, the risk factors associated with MDR bacteria (MDRB) in UTI patients were marital status and place of residence. This can be because of the geographical variation and the often poor hygiene standards in rural areas [61]. Due to regional differences in antibiotic usage and medical procedures, the place of residence can be a risk factor for MDRB in UTI patients. Due to poor access to medical facilities in rural locations, patients may have delayed diagnoses and overused antibiotics. In such areas, poor hygiene standards may further encourage the growth of resistant bacteria and foster an environment favorable for MDRB. The interaction between the environment and resistance is complicated; environmental factors may influence the selection and spread of antibiotic-resistant genes among bacteria, resulting in the formation and persistence of MDRB in the community.
5. CONCLUSIONS
The overall prevalence of UTIs was 51.56% in this study, where female and older patients were mostly affected. The most common bacterial uropathogens identified were E. coli, E. faecalis, K. pneumoniae, P. aeruginosa, S. aureus, and P. mirabilis. However, alarming levels of resistance were observed against several antimicrobials, posing a significant public health concern, limiting treatment options and necessitating the adoption of evidence-based prescribing practices to combat antibiotic resistance effectively. A concerning 88.6% of the bacterial isolates were found to be MDR. The study also revealed significant associations between MDR UTIs and patients’ age, place of residence, marital status, and prior antibiotic use. These findings emphasize the widespread issue of antibiotic resistance in Bangladesh. To address this problem, reducing the incidence of UTIs and continually monitoring the susceptibility of common uropathogens to commonly used antibacterial drugs is crucial. Major health organizations are urged to regularly assess and monitor emerging patterns and trends of AMR to prioritize and implement effective antimicrobial stewardship policies and recommendations at health facilities. Finally, comprehensive surveys and research are necessary to fully understand the national situation regarding antibiotic resistance and develop effective management strategies.
6. ACKNOWLEDGMENTS
We want to thank all of the study participants. The author would like to thank Professor M. Sawkat Anwer (Tufts University, USA) for his constructive criticism of the manuscript.
7. AUTHOR CONTRIBUTIONS
MFI and MHH designed the experiment. MFI, DKD, BI, and MHH experimented. MFI, DKD, BI, MBR, SS, and MHH collected, analyzed, and interpreted data. MFI, DKD, BI, and MHH wrote the draft. The text has undergone considerable modifications by MFI, DKD, BI, MBR, SS, and MHH. All authors read and approved the final draft of the article.
8. FUNDING
The study was supported by research funding from the University Grants Commission of Bangladesh (UGC) and Grants No. 37.01.0000.073.03.023.21.1048.
9. CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
The research methods and guidelines employed in this study were approved by the Institutional Animal, Medical Ethics, Biosafety, and Biosecurity Committee of the Institute of Biological Science at the University of Rajshahi (Memo No. 56/321/IAMEBBC/IBSc).
11. DATA AVAILABILITY
The raw data are available from the corresponding author and can be provided upon reasonable request.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections:Epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015;13:269-84. [https://doi.org/10.1038/nrmicro3432]
2. Lee JB, Neild GH. Urinary tract infection. J Med 2007;35:423-8. [https://doi.org/10.1016/j.mpmed.2007.05.009]
3. Foxman B. Urinary tract infection syndromes:Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014;28:1-13. [https://doi.org/10.1016/j.idc.2013.09.003]
4. Wagenlehner FM, Lichtenstern C, Rolfes C, Mayer K, Uhle F, Weidner W, et al. Diagnosis and management for urosepsis. Int J Urol 2013;20:963-70. [https://doi.org/10.1111/iju.12200]
5. Davenport M, Mach KE, Shortliffe LM, Banaei N, Wang TH, Liao JC. New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol 2017;14:296-310. [https://doi.org/10.1038/nrurol.2017.20]
6. Haque R, Akter ML, Salam MA. Prevalence and susceptibility of uropathogens:A recent report from a teaching hospital in Bangladesh. BMC Res Notes 2015;8:416. [https://doi.org/10.1186/s13104-015-1408-1]
7. Munita JM, Cesar AC. Mechanisms of antibiotic resistance. Microbiol Spectr 2016;4:10. [https://doi.org/10.1128/microbiolspec.VMBF-0016-2015]
8. Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 2018;31:e00088-17. [https://doi.org/10.1128/CMR.00088-17]
9. Algammal AM, Hetta HF, Elkelish A, Alkhalifah DH, Hozzein WN, Batiha GE, et al. Methicillin-resistant Staphylococcus aureus (MRSA):One health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist 2020;13:3255-65. [https://doi.org/10.2147/IDR.S272733]
10. Abolghait SK, Fathi AG, Youssef FM, Algammal AM. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int J Food Microbiol 2020;328:108669. [https://doi.org/10.1016/j.ijfoodmicro.2020.108669]
11. Algammal AM, Hetta HF, Batiha GE, Hozzein WN, El Kazzaz WM, Hashem HR, et al. Virulence-determinants and antibiotic-resistance genes of MDR-E. Coli isolated from secondary infections following FMD-outbreak in cattle. Sci Rep 2020;10:19779. [https://doi.org/10.1038/s41598-020-75914-9]
12. Algammal AM, Mohamed MF, Tawfiek BA, Hozzein WN, El Kazzaz WM, Mabrok M. Molecular typing, antibiogram and PCR-RFLP based detection of Aeromonas hydrophila complex isolated from Oreochromis niloticus. Pathogens 2020;9:238. [https://doi.org/10.3390/pathogens9030238]
13. Addis T, Mekonnen Y, Ayenew Z, Fentaw S, Biazin H. Bacterial uropathogens and burden of antimicrobial resistance pattern in urine specimens referred to Ethiopian public health institute. PLoS One 2021;16:e0259602. [https://doi.org/10.1371/journal.pone.0259602]
14. World Health Organization. Anti-infective Drug Resistance Surveillance and Containment Team. WHO Global Strategy for Containment of Antimicrobial Resistance. Geneva:World Health Organization;2001. Available from:https://apps.who.int/iris/handle/10665/66860
15. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance:A global multifaceted phenomenon. Pathog Glob Health 2015;109:309-18. [https://doi.org/10.1179/2047773215Y.0000000030]
16. CDC. Antibiotic Resistance Threats in the United States. United States:Centers for Disease Control and Prevention;2019. 1-113. Available from:https://www.cdc.gov/drugresistance/biggest_threats.html
17. Holloway KA, Rosella L, Henry D. The impact of WHO essential medicines policies on inappropriate use of antibiotics. PLoS One 2016;11:e0152020. [https://doi.org/10.1371/journal.pone.0152020]
18. Kish L. Survey Sampling. Vol. 59. New York:John Wiley and Sons, Inc. 1965. 1025. [https://doi.org/10.1017/S0003055400132113]
19. Yasmeen BH, Islam S, Islam S, Uddin MM, Jahan R. Prevalence of urinary tract infection, its causative agents and antibiotic sensitivity pattern:A study in Northern international medical college hospital, Dhaka. North Int Med Coll 2015;7:105-9. [https://doi.org/10.3329/nimcj.v7i1.25704]
20. Aspevall O, Osterman B, Dittmer R, Stén L, Lindbäck E, Forsum U. Performance of four chromogenic urine culture media after one or two days of incubation compared with reference media. J Clin Microbiol 2002;40:1500-3. [https://doi.org/10.1128/JCM.40.4.1500-1503.2002]
21. Bergey DH, Buchanan RE, Gibbons NE. Bergey's Manual of Determinative Bacteriology. 8th ed. Washington, DC, USA:American Society for Microbiology;1974. 966-1097.
22. Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966;45:493-6. [https://doi.org/10.1093/ajcp/45.4_ts.493]
23. CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100-S28. Wayne, PA:Clinical and Laboratory Standards Institute;2018.
24. Sweeney MT, Lubbers BV, Schwarz S, Watts JL. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother 2018;73:1460-3. [https://doi.org/10.1093/jac/dky043]
25. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 1983;46:165-70. [https://doi.org/10.1128/aem.46.1.165-170.1983]
26. Medina M, Castillo-Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urol 2019;11:17562∑9832172. [https://doi.org/10.1177/1756287219832172]
27. Ahmed SS, Shariq A, Alsalloom AA, Babikir IH, Alhomoud BN. Uropathogens and their antimicrobial resistance patterns:Relationship with urinary tract infections. Int J Health Sci (Qassim) 2019;13:48-55.
28. Grigoryan L, Trautner BW, Gupta K. Diagnosis and management of urinary tract infections in the outpatient setting:A review. JAMA 2014;312:1677-84. [https://doi.org/10.1001/jama.2014.12842]
29. Abedin MZ, Akter KF, Quruni MO, Aktar MB, Islam MI, Rahaman MA, et al. Prevalence and Antimicrobial Susceptibility Pattern of Pathogens Isolated from Different Age Groups with Urinary Tract Infection in Bangladesh. Durham, UK:Research Square;2022. [https://doi.org/10.21276/SSR-IIJLS.2022.8.4.2]
30. Rustom S, Zaman RF, Barua P, Khanum H. Urinary tract infection among the outpatients of a diagnostic center in Dhaka, Bangladesh. Bangladesh J Zool 2021;48:347-56. [https://doi.org/10.3329/bjz.v48i2.52374]
31. Parveen R, Yusuf MA, Sharmin I, Islam MS, Rahim I. Antibiotic sensitivity of bacteria causing urinary tract infection. Bangladesh J Infect Dis 2017;2:13-8. [https://doi.org/10.3329/bjid.v2i1.31324]
32. Mollick S, Dasgupta T, Hasnain MJ, Ahmed M. Isolation and characterization of pathogens responsible for urinary tract infection in Bangladesh and determination of their antibiotic susceptibility pattern. J Appl Pharm Sci 2016;6:72-6. [https://doi.org/10.7324/JAPS.2016.60410]
33. Hossain G, Hossain E, Ahammed F, Kabir MR, Karmaker G, Chowdhury IA, et al. Bacteriological profile and sensitivity pattern of urinary tract infection patients in North East part of Bangladesh. Int J Adv Med 2020;7:1614-8. [https://doi.org/10.18203/2349-3933.ijam20204510]
34. Islam MA, Islam MR, Khan R, Amin MB, Rahman M, Hossain MI, et al. Prevalence, etiology and antibiotic resistance patterns of community-acquired urinary tract infections in Dhaka, Bangladesh. PLoS One 2022;17:e0274423. [https://doi.org/10.1371/journal.pone.0274423]
35. Bhargava K, Nath G, Bhargava A, Kumari R, Aseri GK, Jain N. Bacterial profile and antibiotic susceptibility pattern of uropathogens causing urinary tract infection in the Eastern part of Northern India. Front Microbiol 2022;13:965053. [https://doi.org/10.3389/fmicb.2022.965053]
36. Sood S, Gupta R. Antibiotic resistance pattern of community acquired uropathogens at a tertiary care hospital in Jaipur, Rajasthan. Indian J Community Med 2012;37:39-44. [https://doi.org/10.4103/0970-0218.94023]
37. Prakash D, Saxena RS. Distribution and antimicrobial susceptibility pattern of bacterial pathogens causing urinary tract infection in urban community of Meerut city, India. ISRN Microbiol 2013;2013:749629. [https://doi.org/10.1155/2013/749629]
38. Seifu WD, Gebissa AD. Prevalence and antibiotic susceptibility of uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect Dis 2018;18:30. [https://doi.org/10.1186/s12879-017-2911-x]
39. Malik S, Rana JS, Nehra K. Prevalence and antibiotic susceptibility pattern of uropathogenic Escherichia coli strains in Sonipat region of Haryana in India. Biomed Biotechnol Res J 2021;5:80-7. [https://doi.org/10.4103/bbrj.bbrj_212_20]
40. Lee CL, Kuo HC. Pathophysiology of benign prostate enlargement and lower urinary tract symptoms:Current concepts. Ci Ji Yi Xue Za Zhi 2017;29:79-83. [https://doi.org/10.4103/tcmj.tcmj_20_17]
41. Gessese YA, Damessa DL, Amare MM, Bahta YH, Shifera AD, Tasew FS, et al. Urinary pathogenic bacterial profile, antibiogram of isolates and associated risk factors among pregnant women in Ambo town, central Ethiopia:A cross-sectional study. Antimicrob Resist Infect Control 2017;6:132. [https://doi.org/10.1186/s13756-017-0289-6]
42. Khawcharoenporn T, Vasoo S, Singh K. Urinary tract infections due to multidrug-resistant Enterobacteriaceae:Prevalence and risk factors in a Chicago emergency department. Emerg Med Int 2013;2013:258517. [https://doi.org/10.1155/2013/258517]
43. Gebremariam G, Legese H, Woldu Y, Araya T, Hagos K, GebreyesusWasihun A. Bacteriological profile, risk factors and antimicrobial susceptibility patterns of symptomatic urinary tract infection among students of Mekelle university, Northern Ethiopia. BMC Infect Dis 2019;19:950. [https://doi.org/10.1186/s12879-019-4610-2]
44. Abdullah AM, Abdullah RM, Salman SL. Use of chromogenic Agar in detection of urinary tract pathogens and antimicrobial susceptibility. J Fac Med Baghdad 2009;51:39-42.
45. Ali AH, Reda DY, Ormago MD. Prevalence and antimicrobial susceptibility pattern of urinary tract infection among pregnant women attending Hargeisa group hospital, Hargeisa, Somaliland. Sci Rep 2022;12:1419. [https://doi.org/10.1038/s41598-022-05452-z]
46. Huang L, Huang C, Yan Y, Sun L, Li H. Urinary tract infection etiological profiles and antibiotic resistance patterns varied among different age categories:A retrospective study from a tertiary general hospital during a 12-year period. Front Microbiol 2022;12:813145. [https://doi.org/10.3389/fmicb.2021.813145]
47. Jagadeesan S, Tripathi BK, Patel P, Muthathal S. Urinary tract infection and diabetes mellitus-etio-clinical profile and antibiogram:A North Indian perspective. J Family Med Prim Care 2022;11:1902-6. [https://doi.org/10.4103/jfmpc.jfmpc_2017_21]
48. Komagamine J, Yabuki T, Noritomi D, Okabe T. Prevalence of and factors associated with atypical presentation in bacteremic urinary tract infection. Sci Rep 2022;12:5197. [https://doi.org/10.1038/s41598-022-09222-9]
49. Terlizzi ME, Gribaudo G, Maffei ME. UroPathogenic Escherichia coli (UPEC) infections:Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol 2017;8:1566. [https://doi.org/10.3389/fmicb.2017.01566]
50. Akhter J, Ahmed S, Anwar S. Antimicrobial susceptibility patterns of Enterococcus species isolated from urinary tract infections. Bangladesh J Med Microbiol 2014;8:16-20. [https://doi.org/10.3329/bjmm.v8i1.31069]
51. Majumder MM, Mahadi AR, Ahmed T, Ahmed M, Uddin MN, Alam MZ. Antibiotic resistance pattern of microorganisms causing urinary tract infection:A 10-year comparative analysis in a tertiary care hospital of Bangladesh. Antimicrob Resist Infect Control 2022;11:156. [https://doi.org/10.1186/s13756-022-01197-6]
52. Muhammad A, Khan SN, Ali N, Rehman MU, Ali I. Prevalence and antibiotic susceptibility pattern of uropathogens in outpatients at a tertiary care hospital. New Microbes New Infect 2020;36:100716. [https://doi.org/10.1016/j.nmni.2020.100716]
53. Hoque R, Ahmed SM, Naher N, Islam MA, Rousham EK, Islam BZ, et al. Tackling antimicrobial resistance in Bangladesh:A scoping review of policy and practice in human, animal and environment sectors. PLoS One 2020;15:e0227947. [https://doi.org/10.1371/journal.pone.0227947]
54. WHO. Reports Signals Increasing Resistance to Antibiotics in Bacterial Infections in Humans and Need for Better Data;2022. Available from:https://www.who.int/news/item/09-12-2022-report-signals-increasing-resistance-to-antibiotics-in-bacterial-infections-in-humans-and-need-for-better-data
55. Warren JW, Abrutyn E, Hebel JR, Johnson JR, Schaeffer AJ, Stamm WE. Guidelines for antimicrobial treatment of uncomplicated acute bacterial cystitis and acute pyelonephritis in women. Infectious Diseases Society of America (IDSA). Clin Infect Dis 1999;29:745-58. [https://doi.org/10.1086/520427]
56. Tessema B, Kassu A, Mulu A, Yismaw G. Prdominant isolates of urinary tract pathogens and their antimicrobial susceptibility patterns in Gondar University teaching hospital, Northwest Ethiopia. Ethiop Med J 2007;45:61-7.
57. Van der Meeren BT, Chhaganlal KD, Pfeiffer A, Gomez E, Ferro JJ, Hilbink M, et al. Extremely high prevalence of multi-resistance among uropathogens from hospitalised children in Beira, Mozambique. S Afr Med J 2013;103:382-6. [https://doi.org/10.7196/SAMJ.5941]
58. Agersew AM, Meseret A, Mucheye G. Uropathogenic bacterial isolates and their antimicrobial susceptibility patterns among HIV/AIDS patients attending Gondar university specialized hospital Gondar, Northwest Ethiopia. J Microb Res Rev 2013;1:42-51.
59. Biadglegne F, Abera B. Antimicrobial resistance of bacterial isolates from urinary tract infections at Felge Hiwot referral hospital, Ethiopia. Ethiop J Health Dev 2009;23:236-8. [https://doi.org/10.4314/ejhd.v23i3.53248]
60. Haque MH, Islam MZ, Miah ML, Das SK, Sarker S. Molecular characterization of multidrug-resistant Escherichia coli isolated from human urine infections with their antibiogram profile. J Adv Biotechnol Exp Ther 2022;6:172-83. [https://doi.org/10.5455/jabet.2023.d116]
61. Shaifali I, Gupta U, Mahmood SE, Ahmed J. Antibiotic susceptibility patterns of urinary pathogens in female outpatients. N Am J Med Sci 2012;4:163-9. [https://doi.org/10.4103/1947-2714.94940]
SUPPLEMENTARY TABLE
Table S1: Sociodemographic features of research participants in Bangladesh’s northern region (n=256).
| Factors | Frequency | Percentage |
|---|---|---|
| Age (years) | ||
| ≤15 | 27 | 10.54 |
| 16–30 | 57 | 22.27 |
| 31–45 | 56 | 21.88 |
| 46–60 | 57 | 22.27 |
| ≥61 | 59 | 23.04 |
| Sex | ||
| Male | 92 | 35.93 |
| Female | 164 | 64.07 |
| Residencem | ||
| Rural | 172 | 67.19 |
| Urban | 84 | 32.81 |
| Disease acquired from | ||
| Outpatient | 216 | 84.38 |
| Inpatient | 40 | 15.62 |
| Marital status | ||
| Single | 73 | 28.51 |
| Married | 157 | 61.33 |
| Widowed | 26 | 10.16 |
| Prior antibiotic use | ||
| Yes | 131 | 51.18 |
| No | 125 | 48.82 |
| Educational status | ||
| Illiterate | 50 | 19.53 |
| Primary school | 59 | 23.05 |
| Secondary school | 75 | 29.30 |
| University | 72 | 28.12 |
| Pregnancy | ||
| Yes | 15 | 5.86 |
| No | 241 | 94.14 |