1. INTRODUCTION
Earthworms, often hailed as the “farmer’s greatest ally,” are indispensable for enhancing soil fertility and advancing sustainable agriculture. These invertebrates, celebrated as nature’s preeminent soil engineers, significantly enhance soil structure, promote nutrient cycling, and facilitate organic matter decomposition [1–4]. As the agricultural sector grapples with the adverse effects of chemical inputs, the shift towards sustainable alternatives, including biofertilizers, is imperative [5]. Earthworms, through their voracious consumption of organic waste, initiate a cascade of transformations that reduce organic carbon, modify the carbon-to-nitrogen ratio, and improve nutrient retention for essential elements like nitrogen, potassium, phosphorus, and calcium.
Despite their well-recognized benefits, the gut microbial biome of these pivotal invertebrates remains an area of incomplete understanding. Indigenous microbes are acknowledged inhabitants of earthworm guts, thriving due to their unique feeding habits involving soil and organic matter ingestion [6]. Although extensive studies have investigated the gut microbes of earthworms, a significant gap remains in understanding the complex symbiotic relationships that promote mutual growth and development among microorganisms within their digestive tracts.
For over 50 years, bacteria residing in plant tissues, known as “endophytes,” have drawn scientific interest for their remarkable capacity to survive without exhibiting obvious disease symptoms [7–10]. Although the term “endophytes” is commonly associated with fungi, endophytic bacteria are widespread across various plant species and tissue types. Notably, in the context of organic matter decay, certain microorganisms, particularly soil bacteria, have been recognized for their ability to break down complex structures such as cellulose, pectin, and lignin into simpler carbon compounds. Despite their potential as valuable alternative sources of energy, bacteria specializing in the degradation of these complex molecules have not been given sufficient attention [11].
This paper embarks on a journey to unravel the symbiotic interplay between the gut microbiome of Eudrilus eugeniae and the endophytic root bacteria of Beta vulgaris plants. A particular focus lies in discerning the bacterial strains accountable for decomposing diverse organic matter within the soil milieu. This eclectic array of microbes exhibits various enzymatic, organic, and inorganic matter decaying capabilities and salt tolerance activities, all of which hold extensive agricultural importance.
This study addresses the knowledge gap by focusing on the isolation of endophytic bacteria from the roots of B. vulgaris and gut bacteria from E. eugeniae. The overarching goal is to enhance our understanding of the interactions between these bacteria and their potential contributions to agriculture, particularly in biomass degradation. A meticulously designed two-step procedure is employed to isolate and identify specific bacterial colonies from the microbiota of both E. eugeniae and B. vulgaris plant roots. By cultivating these bacteria separately, the study seeks to unravel the symbiotic organisms’ ability to break down organic debris. In essence, this paper endeavors to characterize the intricate interactions between these symbiotic organisms and elucidate their crucial role in the breakdown of organic compounds.
2. MATERIALS AND METHODS
2.1. Isolation of B. vulgaris Root Endophytic Bacteria
Beetroot (Beta vulgaris) seeds were purchased from Gandhi Krishi Vigyana Kendra, UAS, Bangalore. These seeds were cultivated in fertile soil for 4 months at the Indian Academy Degree College-Autonomous. Upon reaching maturity, the roots of B. vulgaris were harvested, washed thoroughly under tap water to remove soil debris, and then transferred to sterile Petri dishes, which were placed subsequently in a Laminar Air Flow (LAF) chamber to maintain a contamination-free environment.
For the isolation of endophytic bacteria, a widely accepted procedure involves surface sterilization of plant tissues using various disinfectants, such as sodium hypochlorite [12,13], ethanol [12,14], mercuric chloride [8,15], or combinations thereof [16,17]. In this study, the B. vulgaris roots were washed with distilled water, followed by chemical sterilization in 1% sodium hypochlorite for 2–3 minutes. This was followed by a brief immersion in 0.1% mercuric chloride for 30 seconds, and then washed three times with distilled water, which is a common practice to ensure all residual disinfectant is removed. These steps were essential to eliminate epiphytic and soil-associated bacteria, thereby minimizing the risk of contamination [18].
After sterilization, the roots were carefully selected and sliced into thin sections, 1–2 cm segments using a sterile blade to maximize the release of endophytic bacteria into the nutrient broth. The excision of root segments was followed by immediate immersion into 100 ml of nutrient broth to allow sufficient diffusion of bacteria from the root tissues into the liquid medium and incubated at 28°C for 24 hours [19]. This procedure facilitated the growth of endophytic bacteria from the root tissues into the nutrient broth.
2.2. Isolation of E. eugeniae Gut Bacteria
Eudrilus eugeniae earthworms were sourced from Karnataka Compost Development Corporation and maintained under optimal laboratory conditions at the Biochemistry Laboratory, Indian Academy Degree College-Autonomous to ensure acclimatization.
Earthworms were washed and placed in a sterile petri dish to isolate gut bacteria. Once transferred to a LAF chamber, the external parts of the earthworms were sterilized by immersion in 70% ethanol for 1 minute, after cleaning with sterile water to remove any residual debris and contaminants [20]. The gut contents were aseptically collected by compressing the earthworms from the anterior to the posterior end. The worms were then dissected in the midgut region using a sterile blade. The gut contents were sampled with a sterile cotton swab, inoculated in 100 ml Nutrient broth, and then incubated at 28°C for 24 hours.
Following the incubation step, the broths were serially diluted to a final concentration of 10−5, which was then streaked onto Nutrient Agar plates. Due to the variability in bacterial densities across different samples, the 10−5 dilution was selected to ensure an optimal range for colony isolation. These plates were incubated at 28°C for 24 hours. Pure bacterial colonies were obtained through three successive subcultures based on color and morphology.
2.3. Examining Symbiotic Relationship by Perpendicular Cross-Streaking Method
A modified perpendicular cross-streaking technique explored the symbiotic interactions among the bacterial species [21].
The naming of the bacterial colony was done in the following manner:
- P denotes colonies obtained in Pectin media, while C denotes colonies obtained in Cellulose media.
- R denotes endophytic root bacteria and E denotes earthworm gut bacteria.
- The number following P/C denotes the subculture plate that was taken.
- The number succeeding the subculture number denotes the number of the colony that was isolated separately on a plate and was taken further for studying symbiosis.
Four bacterial isolates from the gut of E. eugeniae (C-E12, C-E15, P-E12, P-E25) and four isolates from the roots of B. vulgaris (C-R11, C-R16, P-R21, P-R26) were analyzed. These strains were selected based on their distinct morphology and pigmentation. The images of some sample isolates are shown in Figure 1a and b. Sixteen Nutrient Agar plates were used to assess the interactions among these eight bacterial strains. The cross-streaking method involved streaking bacterial cultures perpendicularly to evaluate the presence of mutualistic relationships between these isolates. The bacterial growth at the intersecting nodes of the cross-streaks indicated a mutualistic symbiotic relationship. All experiments were conducted in duplicate. The study was confirmed in both cellulose and pectin-containing media.
 | Figure 1. (a): Isolated B. vulgaris endophytic bacterial colonies. (b): Isolated E. eugeniae gut bacterial colonies. [Click here to view] |
2.3.1. Symbiotic relationship in cellulose media
To study the symbiotic relationship between B. vulgaris endophytic and E. eugeniae gut bacteria, a modified perpendicular cross-streaking technique was performed on cellulose agar plates with a composition of 0.05% dihydrogen potassium phosphate (KH2PO4), 0.04% magnesium sulfate (MgSO4), 0.2% carboxymethyl cellulose (CMC), 0.2% gelatin, and 2% agar. Growth on the intersecting nodes inferred mutualism. Plates exhibiting positive symbiotic associations (Fig. 2) were selected for further confirmation, identification, and subsequent culturing for stock maintenance.
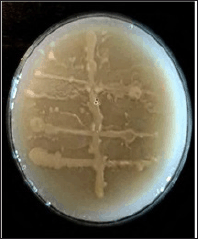 | Figure 2. Symbiotic relationships observed between B. vulgaris endophytic and E. eugeniae gut bacteria in cellulose media (CR11XCE12). [Click here to view] |
2.3.2. Symbiotic relationship in pectin media
Similarly, to study the symbiotic relationship between B. vulgaris endophytic and E. eugeniae gut bacteria, a modified perpendicular cross-streaking technique was performed on pectin agar plates containing a composition of 0.5% peptone, 0.5% yeast extract, 1.5% pectin, and 2% agar. Growth on the intersecting nodes inferred mutualism. Plates exhibiting positive symbiotic associations (Fig. 3) were selected for further confirmation, identification, and subsequent culturing for stock maintenance.
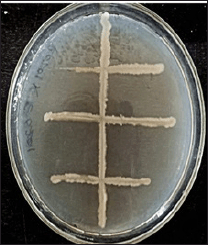 | Figure 3. Symbiotic relationships observed B. vulgaris endophytic and E. eugeniae gut bacteria in pectin media (PR21XPE12). [Click here to view] |
2.3.3. Confirmation of cellulolytic and pectin degradation assays
Numerous studies have highlighted the complex symbiotic interactions within ecosystems, notably the cooperation between earthworm gut bacteria and endophytic root bacteria, with a focus on enzymatic activities such as cellulase, pectinase, and amylase [22–24].
The assays were conducted on cellulose media containing Congo Red as an indicator dye [25]. The media composition included 0.05% dihydrogen potassium phosphate (KH2PO4), 0.04% magnesium sulfate, 0.2% CMC, 0.2% gelatin, and 2% agar and 0.02% Congo red [26]. The isolated colonies exhibiting symbiotic relationships were inoculated in the agar and incubated at 37ºC for 24 hours. Congo Red binds to cellulose, and its degradation results in the formation of clear zones around cellulolytic bacterial colonies. These clear zones indicate cellulase activity, allowing for the identification of cellulose-degrading bacteria (Fig. 4). Colonies exhibiting such clear zones were selected for further analysis to confirm their cellulolytic activity through additional enzymatic assays [27]. Similarly, pectin degrading potential was determined using pectin media [27], The pectin media consisted of 0.5% peptone, 0.5% yeast extract, 1.5% pectin, and 2% agar [28].
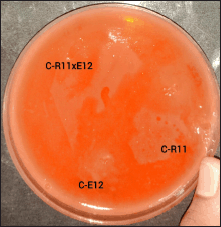 | Figure 4. Congo red test for cellulolytic activity. [Click here to view] |
2.4. Dinitro Salicylic Acid (DNS) Method
Broth solutions containing 0.5% cellulose and pectin as substrates were incubated with the respective bacterial isolates for 10 days. After incubating for 10 days, 0.5 ml aliquots were sampled for analysis and the degradation products (reducing sugars) were confirmed by the DNS method. The DNS method was employed to quantify the enzymatic activities of pectinase and cellulase [28]. Standard calibration curves were generated using aliquots of glucose solutions ranging from 0.2 to 1 ml, prepared from a 100 mg/ml standard glucose solution. To each aliquot, 1 ml of DNS solution was added, followed by incubation in a boiling water bath for 15 minutes. The samples were then diluted with 4 ml of distilled water. Absorbance was measured at 540 nm using a colorimeter. A blank was prepared with 1 ml of distilled water, 1 ml of DNS solution, and 4 ml of distilled water. All experiments were conducted in duplicate. The formation of an orange–red complex indicated a positive result for enzymatic activity.
2.5. 16S rRNA Sequencing
To elucidate the microbial diversity of the isolated bacteria, 16S rRNA sequencing was conducted [29,30]. The 16S rRNA gene, a universally conserved marker in bacterial genomes, serves as a critical tool for phylogenetic and taxonomic classification. Genomic DNA from the bacterial isolates was extracted for sequencing. This method not only provided an in-depth analysis of the bacterial community structure but also facilitated the identification of rare or novel species. The insights gained from 16S rRNA sequencing enhanced our knowledge of microbial ecology within the studied environments and the complex interactions among bacterial communities.
3. RESULTS
3.1. Isolation of B. vulgaris Root Endophytic and E. eugeniae Gut Bacterial Colonies
Endophytic bacterial colonies were isolated from the root tissues of B. vulgaris and the gut of E. eugeniae using standard microbiological techniques. The isolation process relied on identifying distinct morphological features of the colonies, such as pigmentation, size, shape, and texture. These features helped in the initial identification of unique bacterial isolates. Subsequently, the colonies were sub-cultured on nutrient agar plates to obtain pure cultures for further biochemical and molecular analyses. The distinct morphology of the colonies indicates successful isolation, paving the way for studying their enzymatic activities and symbiotic interactions. Visual representations of the colonies and their morphological traits can be found in Figure 1a and b.
3.2. Examining Symbiotic Relationship by Perpendicular Cross-Streaking Method
3.2.1. Types of symbiotic relationships observed in cellulose media
Two types of interactions were observed between B. vulgaris root endophytic bacteria and E. eugeniae gut bacteria mutualism and competition, using the perpendicular cross-streaking method on cellulose media. The observation of bacterial growth at the intersecting nodes of the cross-streaks, as shown in Figure 2, indicated a mutualistic symbiotic relationship. This mutualism was particularly evident between the B. vulgaris endophyte (CR11) and the E. eugeniae gut bacterium (CE12) in the cellulose media (CR11XCE12) and was further sent for identification by 16S-RNA gene sequencing.
3.2.2. Types of symbiotic relationships observed in pectin media
Similarly, two types of interactions were also observed in pectin media using the same cross-streaking method. As observed in Figure 3, the growth of bacteria at the intersecting nodes of the streaks indicated mutualism, signifying a mutual symbiotic relationship between the B. vulgaris endophyte (PR21) and the E. eugeniae gut bacterium (PE12) in the pectin media (PR21XPE12) was further sent for identification by 16S-RNA gene sequencing.
3.3. Pectinase Activity Confirmation
Pectinase activity was confirmed using the DNS (3,5-Dinitrosalicylic Acid) method following a 10-day incubation period [28]. The pure bacterial cultures P-R21 and P-E12 each demonstrated significant pectinase activity, with reducing sugar concentrations measured at 76 mg/ml. Notably, when these cultures were co-cultivated, the combined reducing sugar concentration increased to 112 mg/ml, as presented in Table 1. This marked increase highlights the synergistic effect of the mutual symbiotic relationship between the two bacterial strains, leading to a substantial enhancement in pectinase activity.
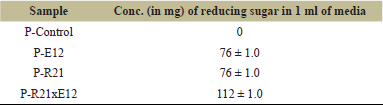 | Table 1. Pectinase activity. [Click here to view] |
The cultures P-R21 and P-E12, which demonstrated a mutual symbiotic relationship by cross streaking technique, were further identified through 16S rRNA gene sequencing, confirming their taxonomic classification.
3.4. Cellulase Activity Confirmation
Cellulase activity was assessed using the Congo Red CMC agar assay, a widely accepted method for detecting cellulolytic activity in microbial isolates [26]. The presence of clear zones around the bacterial colonies, as illustrated in Figure 4, indicated cellulase production. These colonies were subsequently subjected to quantitative analysis using the DNS (3,5-Dinitrosalicylic Acid) method to measure the concentration of reducing sugars released during cellulose degradation.
In the quantitative assay, pure cultures C-R11 and C-E12 individually produced 6 and 4 mg/ml of reducing sugars, respectively. Upon co-cultivation of these cultures, a synergistic effect was observed, resulting in an enhanced yield of 10 mg/ml of reducing sugars, as displayed in Table 2. This indicates a mutualistic symbiotic relationship between the two bacterial strains, which effectively enhances their cellulase activity.
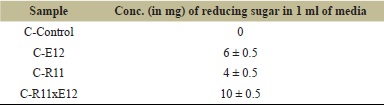 | Table 2. Cellulolytic activity. [Click here to view] |
Further confirmation of cellulolytic potential was obtained through the positive Congo Red inhibition observed in the CMC agar assay for both individual and co-cultivated cultures C-R11 and C-E12. Following a 10-day incubation period, the DNS method was applied to CMC broths of the respective cultures to quantitatively confirm cellulose degradation. All experiments were performed in duplicate to ensure the reliability of the results [26].
The synergistic enhancement of cellulase activity observed in the symbiotic relationship between C-R11 and C-E12 was further investigated and validated through 16S rRNA sequencing, leading to the precise identification of the bacterial species involved.
3.5. Identification of Bacterial Isolates via 16S rRNA Sequencing
The bacterial isolates were identified by using the technique of 16S rRNA gene sequencing, a reliable molecular technique widely employed for bacterial classification and phylogenetic analysis. This sequencing was conducted by Credora Life Sciences, Bangalore, and facilitated the precise identification of four distinct bacterial species (Table 3).
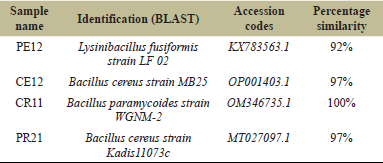 | Table 3. Identification of bacteria based on 16S rRNA sequencing. [Click here to view] |
Among the isolates, those exhibiting significant pectinase activity (P-E12) were identified as Lysinibacillus fusiformis, isolated from the gut of the earthworm E. eugeniae, and Bacillus cereus strain Kadis11073c, which was extracted from the root of the plant B. vulgaris (commonly known as beetroot) (P-R21).
The isolates demonstrating cellulase activity (C-E12) were identified as B. cereus strain MB25, also sourced from the gut of E. eugeniae, and Bacillus paramycoides, obtained from the root of B. vulgaris (C-R11).
To gain further insights into the evolutionary relationships and genetic relatedness of these bacterial species, a phylogenetic tree was constructed using the Clustal Omega tool, a robust bioinformatics tool for multiple sequence alignment and phylogenetic inference. The analysis, corresponding to job ID ClustalO-I20230625-172248-0690-70135694-p1m, provided a comprehensive visualization of the phylogenetic relationships among the four bacterial species, thereby enhancing our understanding of their evolutionary history and potential functional similarities as shown in Figure 5 (Table 3).
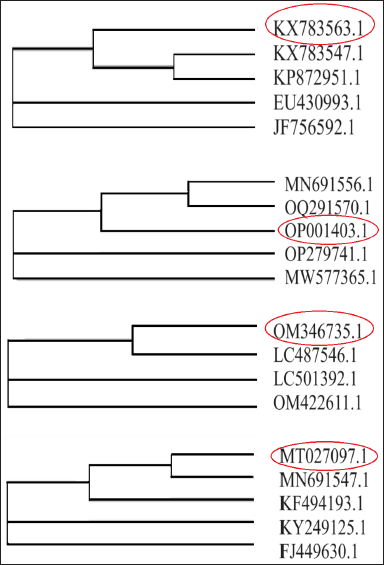 | Figure 5. (a): Phylogenetic tree analysis for pectin degrading E. eugeniae gut bacteria (PE12). (b): Phylogenetic tree analysis for cellulose degrading E. eugeniae gut bacteria (CE12). (c): Phylogenetic tree analysis for cellulose degrading B. vulgaris root endophytic bacteria (CR11). (d): Phylogenetic tree analysis for pectin degrading B. vulgaris root endophytic bacteria (PR21). [Click here to view] |
This molecular identification and phylogenetic analysis underscore the ecological diversity and functional capabilities of the gut bacteria of E. eugeniae and the root environment of B. vulgaris, highlighting their potential roles in pectin and cellulose degradation within their respective niches.
4. DISCUSSION
The present study provides insights into the symbiotic relationships between the root endophytes of B. vulgaris and the gut bacteria of E. eugeniae, with a focus on their roles in organic matter degradation. Through the application of cross-streaking techniques on cellulose and pectin media, we identified mutualistic interactions that enhanced both cellulase and pectinase activity. These findings align with existing studies that highlight the role of earthworms and their associated microbiota in promoting organic matter breakdown and nutrient cycling [1,4,6,22].
The synergistic effect observed in the co-cultivation experiments suggests that the interaction between these bacterial isolates results in enhanced enzymatic activities. Specifically, co-cultivation led to a significant increase in the production of reducing sugars in both pectinase and cellulase assays, corroborating previous findings on microbial cooperation in the degradation of complex polysaccharides [5,27]. This observation strengthens the hypothesis that the interaction between endophytes and gut bacteria is critical for optimizing the breakdown of cellulose and pectin, which are vital components of plant cell walls.
Our results also underscore the ecological significance of these microbial interactions within the root environment of B. vulgaris and the gut ecosystem of E. eugeniae. Endophytic bacteria have been shown to enhance plant growth and resilience by facilitating nutrient acquisition and protecting against pathogens [9,31]. Similarly, earthworm gut bacteria are known to contribute to organic matter mineralization, thereby improving soil health and plant growth [2,20]. The mutualistic relationships observed in this study likely contribute to these ecological benefits, particularly in systems where earthworms and plants co-occur.
The identification of the bacterial species involved, including L. fusiformis and B. cereus strains in the pectinase assay, and B. cereus MB25 and B. paramycoides in the cellulase assay, further supports the functional diversity of these microbial communities. These species have been previously reported to play roles in cellulose and pectin degradation, as well as in promoting plant health [13,20,24,32].
Our study adds to the growing body of evidence that microbial symbiosis in the rhizosphere and gut systems can enhance nutrient cycling, organic matter degradation, and overall soil fertility [6,20,23]. The enhanced enzymatic activities observed in co-cultivation suggest that these interactions could be harnessed for sustainable agricultural practices, including the development of biofertilizers that leverage the natural capabilities of these microbial communities [5,31].
Future research should aim to elucidate the underlying molecular mechanisms governing these symbiotic interactions, as well as their broader implications for ecosystem functioning and agricultural sustainability. The application of advanced omics technologies could provide deeper insights into the metabolic pathways involved and further clarify the roles of these bacteria in the degradation of plant-derived polysaccharides [32].
5. CONCLUSION
In conclusion, the mutualistic relationships between B. vulgaris root endophytes and E. eugeniae gut bacteria significantly enhance cellulase and pectinase activities, emphasizing their potential roles in organic matter decomposition. These findings highlight the importance of microbial symbiosis in ecological processes and sustainable agriculture. The bacterial isolates identified in this study could be further explored for their application in biofertilizer formulations aimed at improving soil health and crop productivity. Future studies should continue to investigate the potential of these microbial interactions in promoting sustainable farming practices.
6. ACKNOWLEDGMENT
The authors extend their heartfelt gratitude to the management of Indian Academy Degree College—Autonomous for their unwavering support and provision of inhouse—seed grant, which has been instrumental in the successful completion of this research work. Their continuous encouragement and financial assistance have been invaluable throughout the duration of this study, enabling us to pursue our research goals effectively. The authors deeply appreciate their commitment to fostering academic research and innovation within their institution.
7. AUTHORSHIP CONTRIBUTION
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
Not Applicable.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Edwards CA, Bohlen PJ. Biology and ecology of earthworms. 4th edition, Springer Science & Business Media, New York, NY, 1996.
2. Lavelle P, Bignell D, Lepage M, Wolters V, Roger P, Ineson PO, et al. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur J Soil Biol 1997;33(04):159–93.
3. Beck T, Joergensen RG, Kandeler E, Makeschin F, Nuss E, Oberholzer HR, et al. An inter-laboratory comparison of ten different ways of measuring soil microbial biomass C. Soil Biol Biochem 1997;29(7):1023–32; CrossRef
4. Edwards CA, Arancon NQ. The role of earthworms in organic matter and nutrient cycles. In: Biology and ecology of earthworms, Springer, New York, NY, pp 233–74, 2022; CrossRef
5. Fasusi OA, Cruz C, Babalola OO. Agricultural sustainability: microbial biofertilizers in rhizosphere management. Agriculture 2021;11(2):163; CrossRef
6. Hoeffner K, Monard C, Santonja M, Cluzeau D. Feeding behaviour of epi-anecic earthworm species and their impacts on soil microbial communities. Soil Biol Biochem 2018;125:1–9.
7. Tervet IW, Hollis JP. Bacteria in the storage organs of healthy plants. Phytopathology 1948;38:960–7.
8. Hollis JP. Bacteria in healthy potato tissue. The University of Nebraska-Lincoln, Lincoln, NE, 1949.
9. Yadav G, Meena M. Biological control of plant diseases by endophytes. In: Shah M, Deka D (eds.). Endophytic association: what, why and how, Academic Press, Cambridge, MA, pp 119–35, 2023.
10. Sapkota R, Santos S, Farias P, Krogh PH, Winding A. Insights into the earthworm gut multi-kingdom microbial communities. Sci Total Environ 2020;727:138301.
11. Kameshwar AK, Qin W. Lignin degrading fungal enzymes. Production of biofuels and chemicals from lignin. pp 81–130, 2016; CrossRef
12. Fisher PJ, Petrini O. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.). N Phytol 1992;120(1):137–43; CrossRef
13. Dong Z, Canny MJ, McCully ME, Roboredo MR, Cabadilla CF, Ortega E, et al. A nitrogen-fixing endophyte of sugarcane stems (a new role for the apoplast). Plant Physiol 1994;105(4):1139–47; CrossRef
14. Hinton DM, Bacon CW. Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia 1995;129:117–25; CrossRef
15. Sriskandarajah S, Kennedy IR, Yu D, Tchan YT. Effects of plant growth regulators on acetylene-reducing associations between Azospirillum brasilense and wheat. Plant Soil 1993;153:165–78; CrossRef
16. Caetano-Anollés G, Favelukes G, Bauer WD. Optimization of surface sterilization for legume seed. Crop Sci 1990;30(3):708–12; CrossRef
17. Pleban S, Ingel F, Chet I. Control of rhizoctonia solani and Sclerotium rolfsii in the greenhouse using endophytic Bacillus spp. Eur J Plant Pathol 1995;101:665–72; CrossRef
18. Khan TJ, Badshah N, Jan N. Effect of various levels of mercuric chloride as surfactant on the explant in tissue culture. Sarhad J Agric 2000;16(1):45–7.
19. Sahu PK, Tilgam J, Mishra S, Hamid S, Gupta A, Jayalakshmi K, et al. Surface sterilization for isolation of endophytes: ensuring what (not) to grow. J Basic Microbiol 2022;62(6):647–68; CrossRef
20. Banerjee A, Biswas JK, Pant D, Sarkar B, Chaudhuri P, Rai M, et al. Enteric bacteria from the earthworm (Metaphire posthuma) promote plant growth and remediate toxic trace elements. J Environ Manage 2019;250:109530; CrossRef
21. Kumar GVP, Pooja G, Nagaraju GV, Malyadri Y. Primary and secondary metabolite profiling, unravelling the antibiotic susceptibility from culture-lysed symbiotic colonies-of diazotroph bacteria (Rhizobium leguminosarum) isolated from root nodules of Dolichos Lab Lab. Int J Curr Microbiol App Sci 2018;7(7):3162–8; CrossRef
22. Béguin P. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal Biochem 1983;131(2):333–6; CrossRef
23. Lu WJ, Wang HT, Nie YF, Wang ZC, Huang DY, Qiu XY, et al. Effect of inoculating flower stalks and vegetable waste with ligno-cellulolytic microorganisms on the composting process. J Environ Sci Health Part B 004;39(5-6):871–87; CrossRef
24. Flack FM, Hartenstein R. Growth of the earthworm Eisenia foetida on microorganisms and cellulose. Soil Biol Biochem 1984;16(5):491–5; CrossRef
25. Edwards CA, Fletcher KE. Interactions between earthworms and microorganisms in organic-matter breakdown. Agric Ecosyst Environ 1988;24(1-3):235–47; CrossRef
26. Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol 1997;43(10):895–914; CrossRef
27. Biz A, Farias FC, Motter FA, de Paula DH, Richard P, Krieger N, et al. Pectinase activity determination: an early deceleration in the release of reducing sugars throws a spanner in the works!. PLoS One 2014;9(10):e109529; CrossRef
28. Sievers F, Higgins DG. The clustal omega multiple alignment package. In: Katoh K (ed.). Multiple sequence alignment: methods and protocols, Humana, New York, NY, pp 3–16, 2021; CrossRef
29. Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 1989;17(19):7843–53; CrossRef
30. Zhang YP, Hong J, Ye X. Cellulase assays. In: Mielenz J (ed.). Biofuels: methods and protocols. Humana Press, Totowa, NJ, pp 213–31, 2009; CrossRef
31. Vandana UK, Rajkumari J, Singha LP, Satish L, Alavilli H, Sudheer PDVN, et al. The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology (Basel) 2021;10(2):101; CrossRef
32. Dang H, Zhang T, Li G, Mu Y, Lv X, Wang Z, et al. Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol 2020;20:1–8; CrossRef