1. INTRODUCTION
Endemic plants’ mutualistic plant-bacteria interactions ensure survival within their restricted geographical distribution. Plants growing as endemics in the Western Ghats region experience abiotic stress in uneven water availability, steep terrain, and high rates of nutrient leaching from the soil [1]. Higher densities of organisms increase competition for sunlight, space, and other significant environmental benefits. To overcome these challenges, organisms such as plants and microorganisms evolved with mutualistic interactions such as mycorrhizae, nodulating root bacteria, or endophytic and rhizosphere bacteria [2]. The dynamics of microbial populations near the plant have produced a distinct microbial ecosystem that contributes to the stability of biogeochemical cycles involving nitrogen, phosphorous, and other elements [3]. The Western Ghats contains more than 1270 endemic angiosperms [4]. Conservation of these plants must be a top priority. Because endemism reasons are unclear, in situ conservation is preferred. A better understanding of plant-microbiome interactions will benefit conservation efforts and produce bacteria suitable for commercial use in this region’s agricultural system.
Phosphorous biofertilization is a significant problem in the Western Ghats for two reasons: The high rates of phosphorus leaching and loss caused by monsoons. On the other hand, the bioavailable phosphorus gets limited by microclimate dynamics [5,6]. The association of plant-bacteria helps to acquire phosphorus by interactions between them. Moreover, plant growth-promoting bacteria that produce growth hormones such as indole acetic acid (IAA) and biocontrol agents such as siderophores favor plant survival [7,8].
Humboldtia brunonis Wall. is an endemic Leguminosae member growing an understory tree. It is a non-nodulating leguminous plant preferably grown as a riparian element in 200–800 ft altitude of the Western Ghats of Karnataka and Kerala [9]. This study aimed at bacterial detection by isolating and characterizing phosphorus solubilizing bacteria based on culture-dependent methods and analysis of its phosphate solubility. Furthermore, bacterial diversity analysis through 16s metagenomic sequencing and prediction of their functional attributes as plant growth-promoting bacterial endophytes.
2. MATERIALS AND METHODS
2.1. Collection of Roots and Soil Samples
The feeder roots of H. brunonis Wall. collected from three sites (13°04’20.7”N 75°19’18.0”E, 13°10’22.8”N 75°20’44.3”E, and 12°43’55.0”N 75°39’39.9”E) belonging to Western Ghats regions of Karnataka state.
2.2. Surface Sterilization of Roots
Collected feeder roots washed with running tap water to remove soil particles. Then, the roots dipped in 2% sodium hypochlorite solution for 2 min. Transferred treated root segments to sterile distilled water, then cut into 1 cm long pieces with a sterile blade and stored in distilled water to get inoculated into the selective media [10].
2.3. Growing the Bacteria in Selective Media
Pikovskaya’s selective medium for screening the phosphate solubilizing bacteria employed in the present study [11,12]. Plates were inoculated with sterile root segments and incubated at 28°C for 7 days. The cut root ends are checked regularly for bacterial growth. The appearance of a colony surrounded by a halo-zone in the media indicated the organism’s growth and phosphate solubilizing ability [13,14]. The organism was subcultured into slants. Furthermore, a pure culture of bacteria got stored in nutrient agar slants for further analysis.
2.4. Determination of Phosphate Solubilizing Ability
The halo-zone formation surrounding the colony in Pikovskaya’s medium determines the phosphate solubilizing ability of the pure bacterial cultures qualitatively. For quantitative analysis, freshly inoculated pure culture in the Pikovskaya’s medium was observed once in 2 days. Then, the colony diameter and halo-zone diameter were measured to incorporate into the formula used to determine the solubilization index [15-17].
2.5. Identification of Bacteria
Biochemical protocols for bacterial identification for Gram’s staining, motility, starch hydrolysis, lipid hydrolysis, casein hydrolysis, gelatin hydrolysis, carbohydrate fermentation, H2S, nitrate reduction, catalase, urease, oxidase, indole production, methyl red, Voges–Proskauer, citrate utilization, and phenylalanine deaminase tests performed following standard protocols [18]. For amplification of the 16S rRNA gene, a reaction mixture of 25 μL, containing 15 ng of template DNA, 0.12 μL of 5U μL−1 Taq DNA polymerase, 2.5 μL of 10× Taq polymerase buffer, 1 μL of 5 mM dNTPs, 2 μL of 25 mM MgCl2, and 1 μL of each 10 μM forward and reverse primers 27F/1492R (27F 5’-AGAGTTTGATCCTGGCTCAG-3’; 1492R 5’-GGTTACCTTGTTACGACTT-3’), were prepared. The PCR amplification of 16S rDNA included the following steps: Initial denaturation at 94°C for 5 min, followed by 30 cycles at 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min, and final extension for 10 min at 72°C [19,20].
2.6. Identification of Isolates by 16S rRNA Sequence Analysis
Agarose gel eluted PCR products of the 16S rRNA gene are purified and sequenced according to Applied Biosystems’ 16S direct workflow protocol. The SeqStudio Genetic Analyzer capillary electrophoresis system performed both high-precision sizing and analysis of multicolor fluorescent DNA fragments and automated Sanger DNA sequencing [21]. Performed sequence similarity searches using the Basic Local Alignment Search Tool by comparing the sequences obtained to other microbial sequences in the National Center for Biotechnology Information (NCBI) database [22]. MEGA-X constructed the phylogenetic tree for our query sequence by obtaining closely related 16S rRNA gene sequences of other strains from the NCBI database through the Clustal W program, which performed multiple sequence alignments by the maximum-likelihood method based on the Jukes-Cantor model [23].
2.7. Metagenomics of V3-V4 Region
Metagenomic studies analyze the prokaryotic 16S ribosomal RNA gene (16S rRNA), approximately 1500 bp long and containing nine variable regions interspersed between conserved regions. This study used V3-V4 variable regions of 16S rRNA in phylogenetic classifications of the genus present in the plant-associated microbial population obtained through the 16S Metagenomic Sequencing Library Preparation protocol workflow [24]. High-quality DNA was extracted by following the manufacturer’s recommendation of QIAamp® DNA Mini Kit. From the extracted DNA, 40 ng−1 μl of DNA got utilized for the 16S Metagenomic Sequencing Library Preparation of V3-V4 region of 16S RNA gene as per the designated workflow. V13F/V13R (V13F – AGAGTTTGATGMTGGCTCAG and V13R –TTACCGCGGCMGCSGGCAC) are the primers used in this library preparation protocol. AMPure beads and components utilized from the Qubit dsDNA high sensitivity assay kit purified the amplicons. Illumina MiSeq with a 2x300PE v3 sequencing kit used for sequencing.
2.8. Bioinformatics and Statistical Analysis
The GAIA 2.0 analysis workflow, which provides an integrated metagenomic analysis suite, makes till genus-level taxa identification. The overall workflow involves GAIA consisting of different Python, Java, Bash, and R scripts that perform the following five steps: The first step is the quality check and trimming, in that GAIA calls BBDuk to remove both adapter sequences and bad quality portions from the reads. In the second step, BWA is used to map the high-quality reads from Illumina against custom-made databases created from NCBI sequences. Next, an in-house built Lowest Common Ancestor algorithm reads performs specific taxonomy level classification. Minimum identity thresholds are applied to classify reads into strains, species, genus, family, order, class, phylum, and domain levels; finally, phyloseq calculates alpha and beta diversities [25]. Interpreting the taxonomic and phenotypic profiles of microorganisms obtained from the metagenomic workflow conducted by inbuilt statistical functions of METAGENassist. METAGENassist (http://www.metagenassist.ca.) performs an automated taxonomic-to-phenotypic mapping. The taxonomic input data automatically generate phenotypic information covering nearly 20 functional categories such as GC content, genome size, oxygen requirements, energy sources, and preferred temperature range. Univariate data analyses, including clustering and classification, have been done using this phenotypically enriched data. This paper also uses METAGENassist generated graphs [26]. Further, the online database KEGG module to predict plant growth-promoting activities explored the possible plant-microbe interactions [27].
3. RESULTS
Surface disinfected root samples of H. brunonis Wall. were grown on Pikovskaya’s medium to obtain the phosphate solubilizing endophytic bacteria culture. Subculturing isolates producing the halo-zone in this medium gave its pure culture. These steps resulted in one pure culturable, phosphate solubilizing bacterium is identified and assigned to the taxonomy using both molecular method (16sRNA gene sequencing) and biochemical characterization. Result summary of phenotypic characterization of the bacterium is shown in Table 1 and the 16S rRNA sequencing method assigned the bacteria as Brevibacillus brevis (Supplementary data). This phosphate solubilizer produced a halo-zone around the colony to qualitatively determine its phosphate solubilization property [Figure 1]. Quantitative estimation of the phosphate solubility of B. brevis species measured on days 1, 3, 5, and 7, and the average phosphate solubility index obtained was 2.67 [Table 2].
Table 1: Morphological and biochemical test results.
| S. No. | Morphological parameter | Result |
|---|---|---|
| 1. | Cell morphology | Rods |
| 2. | Colony morphology | Smooth, round |
| 3. | Oxygen requirements for growth | + |
| 4. | Motility | + |
| 5. | Gram staining | + |
| 6. | Starch hydrolysis test | - |
| 7. | Lipid hydrolysis test | + |
| 8. | Casein hydrolysis test | + |
| 9. | Gelatin hydrolysis test | + |
| 10. | Carbohydrate fermentation test | + |
| 11. | H2S test | - |
| 12. | Nitrate reduction test | + |
| 13. | Catalase test | - |
| 14. | Urease test | - |
| 15. | Oxidase test | - |
| 16. | Indole production test | + |
| 17. | Methyl red test | - |
| 18. | Voges–Proskauer test | - |
| 19. | Citrate utilization test | - |
| 20. | Phenylalanine deaminase test | - |
| 21. | Presence of endospores in a culture | + |
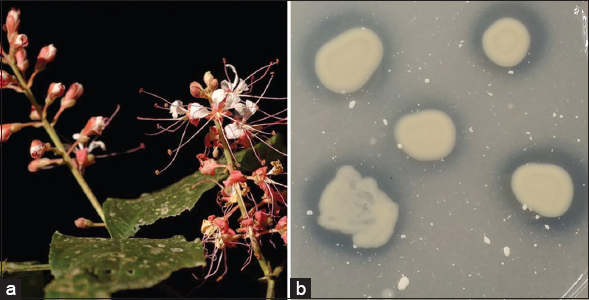 | Figure 1: (a) Humboldtia brunonis Wall. (b) Phosphate solubilization; exhibited by cultured Brevibacillus brevis in Pikovskaya’s medium. [Click here to view] |
Table 2: Phosphate solubilizing ability of bacteria measured qualitatively.
| Trial | Day 1 | Day 3 | Day 5 | Day 7 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDa | HDb | SIc | CD | HD | SI | CD | HD | SI | CD | HD | SI | |
| (B) | (A) | (A/B) | (B) | (A) | (A/B) | (B) | (A) | (A/B) | (B) | (A) | (A/B) | |
| 1 | 0.4 | 0.7 | 1.75 | 0.5 | 0.9 | 1.80 | 0.6 | 1.2 | 2.00 | 0.7 | 1.2 | 1.71 |
| 2 | 0.2 | 0.5 | 2.50 | 0.35 | 0.8 | 2.29 | 0.3 | 0.7 | 2.33 | 0.3 | 0.9 | 3.00 |
| 3 | 0.35 | 0.55 | 1.57 | 0.4 | 0.8 | 2.00 | 0.4 | 1.0 | 2.5 | 0.4 | 1.1 | 2.75 |
a – Colony diameter, b – halo-zone diameter, c – solubilizing index
The metagenomics studies have been done directly from root samples targeting the V3-V4 region of 16S rRNA and identified up to 43 endophytes, as shown in Figure 2 (supplementary data). Domain to genus taxonomic profile in Figure 3 shows the bacteria as dominant domain (99.8%) than Archea. Phylum Proteobacteria shares 98.9%, where Pseudomonas is the dominant genus. The members screened belong to Rhizobiales (3.3%), Burkholderiales (1.8%), and other orders. METAGENassist is uploaded in the form of data input with the Bacterial OTUs generated from metagenomic analysis obtained from the samples are summarised in [Table 3]. It is also provided with metadata of these samples in [Table 4] with discrete, qualitative labeled data values to generate well-annotated tables and colorful, labeled graphs. METAGENassist generated [Figure 4] statistically shows the role of the bacteria as dinitrogen and nitrogen fixers, carbon fixers, ammonia oxidizers, and nitrite reducers. The figure also emphasizes their energy sources, oxygen requirement, sporulation, and their nature of interaction with the plant.
 | Figure 2: Taxonomy plot analysis at genus level as obtained in V3–V4 metagenomics. [Click here to view] |
 | Figure 3: Taxonomic profile from 16S metagenomic sequencing method. [View full-size image] [Click here to view] |
Table 3: Bacterial OTUs from metagenomic analysis used in METAGENassist studies.
| Bacterial OTU | HB1 | HB2 | HB3 | HB4 | HB5 | HB6 | HB7 | HB8 | HB9 |
|---|---|---|---|---|---|---|---|---|---|
| Acidobacterium | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| Acinetobacter | 1545 | 1545 | 1545 | 100 | 24 | 24 | 24 | 24 | 24 |
| Bradyrhizobium | 35 | 35 | 35 | 24 | 24 | 24 | 24 | 24 | 24 |
| Brevundimonas | 11 | 11 | 11 | 24 | 24 | 24 | 24 | 24 | 24 |
| Burkholderia | 69 | 69 | 69 | 24 | 24 | 24 | 24 | 24 | 24 |
| Candidatus Solibacter | 33 | 33 | 33 | 24 | 24 | 257 | 24 | 24 | 24 |
| Duganella | 5 | 5 | 5 | 24 | 24 | 24 | 24 | 24 | 24 |
| Ensifer | 5 | 5 | 5 | 24 | 24 | 24 | 24 | 24 | 24 |
| Enterobacter | 20 | 20 | 20 | 24 | 24 | 24 | 24 | 24 | 24 |
| Escherichia | 11 | 15 | 15 | 24 | 308 | 24 | 24 | 24 | 24 |
| Fusobacterium | 19 | 19 | 19 | 24 | 24 | 24 | 24 | 24 | 24 |
| Geobacter | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Halobacterium | 10 | 10 | 10 | 24 | 24 | 24 | 24 | 24 | 24 |
| Herbaspirillum | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Hyphomicrobium | 8 | 8 | 8 | 24 | 24 | 24 | 24 | 24 | 24 |
| Janthinobacterium | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Klebsiella | 33 | 33 | 33 | 24 | 24 | 24 | 24 | 24 | 24 |
| Labrys | 6 | 6 | 6 | 24 | 24 | 24 | 24 | 24 | 24 |
| Leptothrix | 7 | 7 | 7 | 24 | 24 | 24 | 24 | 24 | 24 |
| Leptotrichia | 9 | 9 | 9 | 24 | 24 | 24 | 24 | 24 | 24 |
| Massilia | 7 | 9 | 9 | 24 | 24 | 24 | 24 | 24 | 24 |
| Mesorhizobium | 6 | 6 | 6 | 24 | 24 | 24 | 24 | 24 | 24 |
| Methylobacterium | 6 | 6 | 6 | 24 | 24 | 24 | 24 | 24 | 24 |
| Nitrospira | 4 | 4 | 4 | 24 | 24 | 24 | 24 | 24 | 24 |
| Ochrobactrum | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Pantoea | 43 | 43 | 43 | 24 | 24 | 10 | 24 | 24 | 24 |
| Paraburkholderia | 28 | 28 | 28 | 24 | 24 | 24 | 24 | 24 | 24 |
| Paracoccus | 61 | 61 | 61 | 24 | 24 | 24 | 24 | 1 | 24 |
| Pectobacterium | 5 | 5 | 5 | 24 | 24 | 24 | 24 | 24 | 24 |
| Pseudomonas | 3468 | 5879 | 4582 | 24 | 24 | 1 | 24 | 24 | 24 |
| Psychrobacter | 6 | 6 | 6 | 24 | 24 | 24 | 24 | 24 | 24 |
| Rhizobium | 73 | 73 | 73 | 24 | 24 | 24 | 24 | 24 | 1 |
| Rhodomicrobium | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Roseomonas | 7 | 7 | 7 | 24 | 24 | 24 | 24 | 24 | 24 |
| Salmonella | 5 | 5 | 5 | 24 | 24 | 24 | 2 | 24 | 24 |
| Serratia | 10 | 10 | 10 | 24 | 24 | 24 | 24 | 24 | 24 |
| Shinella | 40 | 40 | 40 | 24 | 24 | 24 | 24 | 24 | 24 |
| Sphingomonas | 16 | 16 | 16 | 24 | 24 | 24 | 24 | 24 | 24 |
| Stenotrophomonas | 20 | 20 | 20 | 24 | 24 | 24 | 24 | 24 | 24 |
| Streptobacillus | 16 | 16 | 16 | 24 | 24 | 24 | 24 | 28 | 24 |
| Terriglobus | 3 | 3 | 3 | 24 | 24 | 24 | 24 | 24 | 24 |
| Variovorax | 8 | 8 | 8 | 24 | 24 | 24 | 24 | 24 | 24 |
Table 4: Metadata file used for METAGENassist studies.
| OUT | Fungi | Plant | Season | Depth | Temp. | Moisture |
|---|---|---|---|---|---|---|
| HB1 | Yes | NO | Summer | Surface | High | Less |
| HB2 | Yes | NO | Winter | Subsoil | Moderate | Moderate |
| HB3 | Yes | No | Monsoon | O horizon | Moderate | Very high |
| HB4 | Yes | NO | Summer | Subsoil | High | Less |
| HB5 | Yes | NO | Summer | Surface | High | Less |
| HB6 | Yes | NO | winter | Subsoil | Moderate | Moderate |
| HB7 | Yes | No | Monsoon | O horizon | Moderate | Very high |
| HB8 | Yes | NO | Summer | Subsoil | High | Less |
| HB9 | Yes | NO | Summer | Surface | High | Less |
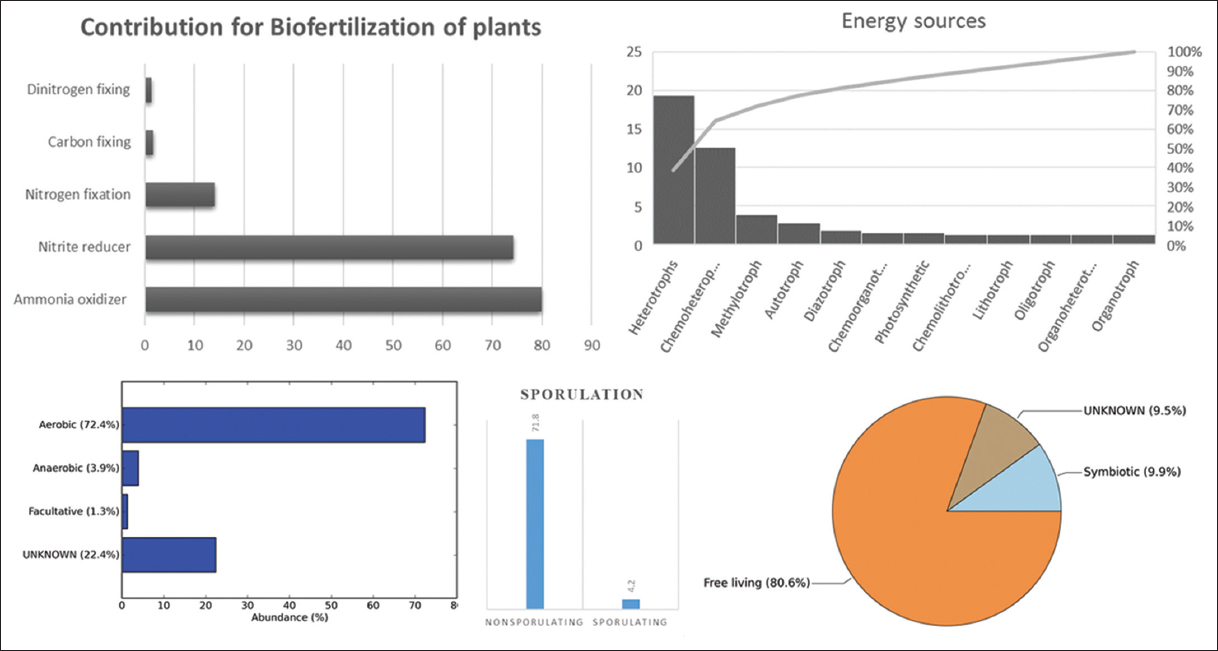 | Figure 4: Phenotypic functional profile of endophytic bacteria present in Humboldtia brunonis Wall. [Click here to view] |
4. DISCUSSION
The identified B. brevis, which belongs to the phylum Firmicutes, act as a phosphorus solubilizing endophyte in the endosphere region of H. brunonis Wall. This endophytic nature may be a fact due to its adaptability to extreme environments. This understory plant experience floods during monsoon and drought during hot summer in its confined distributed area. The association of B. brevis appeared in culture-based studies, and other endophytes reported from metagenomic analysis of 16S RNA play an essential role in plant adaptation. The genus-level plot analysis obtained from the metagenomic studies, which then combined with the KEGG module data (M00579, M00175, M00529, and M00116) resultant phosphate solubilizers are Acidobacterium, Bradyrhizobium, Brevundimonas, Burkholderia, Candidatus Solibacter, Duganella, Ensifer, Enterobacter, Escherichia, Fusobacterium, Geobacter, Herbaspirillum, Hyphomicrobium, Janthinobacterium, Klebsiella, Labrys, Leptothrix, Leptotrichia, Massilia, Mesorhizobium, Methylobacterium, Ochrobactrum, Pantoea, Paraburkholderia, Paracoccus, Pectobacterium, Pseudomonas, Psychrobacter, Rhizobium, Roseomonas, Salmonella, Serratia, Shinella, Sphingomonas, Stenotrophomonas, Streptobacillus, and Variovorax, but are non-responsive to culture-based techniques. Metagenomic report-based prediction of plant growth-promoting traits of these obtained genera in this plant endosphere zone suggests their role as biocontrol agents, biofertilizers, and phytostimulants. Biofertilization by the obtained taxa is quite evident as it confirms the presence of bacteria belonging to Acinetobacter, Burkholderia, Caulobacter, Devosia, Rhizobium, Herbaspirillum, and Bradyrhizobium as these genera can contribute to nitrogen, phosphorus, and potassium bioavailability. Bradyrhizobium, Burkholderia, Ensifer, Mesorhizobium, Methylobacterium, Paraburkholderia, and Rhizobium are capable of nodule formation here appear as endophytes of this non-nodulating legume. Enterobacter, Geobacter, Herbaspirillum, Hyphomicrobium, Klebsiella, Leptothrix, Magnetospirillum, Methylocystis, Pantoea, Pectobacterium, Pseudomonas, Rhodomicrobium, Serratia, Sphingomonas, and Vibrio are free-living diazotrophs taking shelter of this plant. Many of these bacteria are also known for their biocontrol abilities.
The overall results show a great diversity of bacterial species, and their statistical analysis shows that nearly 10% of these species [Figure 4] live in a symbiotic regime within plants. Moreover, other organisms can be opportunistic. The presence of about 72% aerobic bacteria suggests that they live inside the roots using the plant’s oxygen supply and are involved in recycling nitrogen and carbon, which are related to different pathways involved in phosphate solubilization, such as the pentose phosphate pathway [28]. Analysis of genomic diversity revealed that endophytes not only exhibit P solubility but also participate in other PGP traits; namely, biological nitrogen fixation (14%) and carbon fixation (3%) through the production of growth promoters such as IAA (+ for indole test); biological control functions such as subcellular production and antagonistic interactions were also evident.
5. CONCLUSION
H. brunonis Wall., a non-nodulating endemic legume, is belonging to the subfamily Detarioideae, seems to establish plant-microbe interactions in its endosphere with many groups of bacteria in various manners to utilize their services. This microbial ecology contributes significantly to the survival of this endemic plant. Detritus-driven Western Ghats forest floor dynamics create unique microbial ecology. Higher endemic taxa keep unique microbial associations to fulfill their survival requirements, as shown by this plant. The current research has shown that complex interactions between plants and microorganisms are necessary for coexistence. It is imperative to understand the local microbiota’s remarkable ability in exhibiting bio fertilization, biocontrol, and phytostimulation traits as endophytes of these endemic plants. The dynamics of endophytic biomes also remain an important area for future studies. Further studies examining the performance of these bacteria and their mutants on plant growth will help explore the mechanism and potential of these plant growth-promoting traits.
6. ACKNOWLEDGMENTS
The authors are thankful to Sri Dharmasthala Manjunatheshwara College, Ujire, for providing the laboratory facility to conduct the experiments.
7. Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. Funding
There is no funding to report.
9. Conflicts of interest
The authors report no financial or any other conflicts of interest in this work.
10. Ethical approvals
This study does not involve experiments on animals or human subjects.
11. Data availability
All data generated and analyzed are included within this research article.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Radhakrishna T, Mohamed AR, Venkateshwarlu M, Soumya GS, Prachiti PK. Mechanism of rift flank uplift and escarpment formation evidenced by Western Ghats, India. Sci Rep 2019;9:1-7. [CrossRef]
2. Rashid MI, Mujawar LH, Shahzad T, Almeelbi T, Ismail IM, Oves M. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol Res 2016;183:26-41. [CrossRef]
3. Moënne-Loccoz Y, Mavingui P, Combes C, Normand P, Steinberg C. Microorganisms and biotic interactions. Environ Microbiol 2014;395-444. [CrossRef]
4. Nayar T, Beegam A, Sibi M. Flowering Plants of the Western Ghats, India. Vol. 1. Dicots;2014.
5. El Habil-Addas F, Aarab S, Rfaki A, Laglaoui A, Bakkali M, Arakrak A. European Journal of Biotechnology and Screening of phosphate solubilizing bacterial isolates for improving the growth of wheat. J Biotechnol Biosci 2017;2:7-11.
6. Kalayu G. Phosphate solubilizing microorganisms:Promising approach as biofertilizers. Int J Agron 2019;2019:4917256. [CrossRef]
7. Bhutani N, Maheshwari R, Suneja P. Isolation and characterization of plant growth-promoting endophytic bacteria isolated from Vigna radiata. Indian J Anim Res 2018;52:596-603.
8. Mitter EK, Tosi M, Obregón D, Dunfield KE, Germida JJ. Rethinking crop nutrition in times of modern microbiology:Innovative biofertilizer technologies. Front Sustain Food Syst 2021;5:606815. [CrossRef]
9. Sanjappa M. A revision of the genus Humboldtia Vahl (Leguminosae-Caesalpinioideae). Blumea 1986;31:329-39.
10. Yamaji K, Watanabe Y, Masuya H, Shigeto A, Yui H, Haruma T. Root fungal endophytes enhance heavy-metal stress tolerance of Clethra barbinervis growing naturally at mining sites via growth enhancement, promotion of nutrient uptake, and decrease of heavy-metal concentration. PLoS One 2016;11:169089. [CrossRef]
11. Yulianti E, Rakhmawati A. Screening and Characterization of Phosphate Solubilizing Bacteria from an Isolate of Thermophilic Bacteria. In:AIP Conference Proceedings. College Park, Maryland:American Institute of Physics Inc.;2017. [CrossRef]
12. Das S, Sultana KW, Chandra I. Isolation and characterization of a plant growth-promoting bacterium Acinetobacter sp. SuKIC24 From in vitro-Grown Basilicum polystachyon (L.) Moench. Curr Microbiol 2021;78:2961-9. [CrossRef]
13. Rao NS. Soil Microbiology. Enfield:Science Publishers;1999.
14. Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, et al. Plant growth-promoting rhizobacteria:Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci 2018;871:1473. [CrossRef]
15. Edi Premono M, Moawad AM, Vlek PL. Effect of phosphate-solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones J Crop Sci 1996;11:13-23.
16. Paul D, Sinha SN. Isolation and characterization of phosphate solubilizing bacterium Pseudomonas aeruginosa KUPSB12 with antibacterial potential from river Ganga. India Ann Agrar Sci 2017;15:130-6. [CrossRef]
17. Kantarian C, Topoonyanont N, Pumisutapon P, Niamsup P, Klayraung S. Screening of bioactive compounds from bacterial endophytes isolated from micropropagated plants. Suranaree J Sci Technol 2016;23:59-67.
18. Cappuccino JG, Welsh C. Microbiology. A Laboratory Manual. London, United Kingdom:Pearson Education Limited;2017.
19. Pérez E, Sulbarán M, Ball MM, Yarzábal LA. Isolation and characterization of mineral phosphate-solubilizing bacteria naturally colonizing a limonitic crust in the South-Eastern Venezuelan region. Soil Biol Biochem 2007;39:2905-14. [CrossRef]
20. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory:Pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761-4. [CrossRef]
21. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now:Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 2008;14:908-34. [CrossRef]
22. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403-10. [CrossRef]
23. Tamura K, Stecher G, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [CrossRef]
24. Pichler M, Coskun ÖK, Ortega-ArbulúAS, Conci N, Wörheide G, Vargas S, et al. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiologyopen 2018;7:e00611. [CrossRef]
25. PaytuvíA, Battista E, Scippacercola F, Cigliano RA, Sanseverino W. GAIA:An integrated metagenomics suite. bioRxiv 2019;?804690. [CrossRef]
26. Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, et al. METAGENassist:A comprehensive web server for comparative metagenomics. Nucleic Acids Res 2012;40:W88-95. [CrossRef]
27. Tanabe M, Kanehisa M. Using the KEGG database resource. Curr Protoc Bioinformatics 2012;Chapter 1:Unit1.12. [CrossRef]
28. Kandel SL, Firrincieli A, Joubert PM, Okubara PA, Leston ND, McGeorge KM, et al. An in vitro study of bio-control and plant growth promotion potential of Salicaceae endophytes. Front Microbiol 2017;8:386. [CrossRef]
SUPPLEMENTARY DATA
16S rRNA sequences obtained from Humboldtia brunonis Wall.
Forward sequence:
>HB_27F_G04.ab1
GGATGCACATCTATCTGCAGTCGAGCGAGGCTCTTCGGACG CTAGCGGCGGAGGGTGAGTAACACGTAGGCAACCTGCCTC TCAGACTGGGATAACATAGGGAAACTTATGCTAATACCGGA TAGGTTTTTGGATCGCATGATCCGAAAAGAAAAGGCGGCT TCGGCTGTCACTGGGAGATGGGCCTGCGGCGCATTAGCTAG TTGGTGGGGTAACGGCCTACCAAGGCGACGATGCGTAGCCG ACCTGAGAGGGTGACCGGCCACACTGGGACTGAGACACGG CCCAGACTCCTACGGGAGGCAGCAGTAGGGAATTTTCCACA ATGGACGAAAGTCTGATGGAGCAACGCCGCGTGAACGATG AAGGTCTTCGGATTGTAAAGTTCTGTTGTTAGGGACGAATA AGTACCGTTCGAATGGGGCGGCACCTTGACGGCCCCTGAC GAGAAAGACCCGGTTTATTTTA
Reverse sequence:
>HB_1492R_D05.ab1
CGCTAACCATCTTCGGCGGCTGGCTCCTTGCGGTTACTCACC GACTTCGGGTGTTGCAAACTCCCGTGGTGTGACGGGCGGTG TGTACAAGGCCCGGGAACGTATTCACCGCGGCATGCTGATC CGCGATTACTAGCGATTCCGACTTCATGTAGGCGAGTTGCAG CCTACAATCCGAACTGAGATTGGTTTTAAGAGATTGGCGTC CCCTCGCGAGGTAGCATCCCGTTGTACCAACCATTGTAGCA CGTGTGTAGCCCAGGTCATAAGGGGCATGATGATTTGACGT CATCCCCGCCTTCCTCCGTCTTGTCGACGGCAGTCTCTCTAG AGTGCCCAACTGAATGCTGGCAACTAAAGATAAGGGTTGCG CTCGTTGCGGGACTTAACCCAACATCTCACGACACGAGCTG ACGACAACCATGCACCACCTGTCACCGCTGCCCCGAAGGG AAGCTCTGTCTCCAGAGCGGTCAGCGGGATGTCAAGACCT GGTAAGGTTCTTCGCGTTGCTTCGAATTAAACCACATGCTC CACCGCTTGTGCGGGCCCCCGTCAATTCCTTTGAGTTTCACT CTTGCGAGCGTACTCCCCAGGCGGAGTGCTTATTGCGTTAG CTGCGGCACTGAGGGTATTTGAAACCCCCAACACCTAGCAC TCATCGTTTTACAGCGTGGACTACCAGGGTATCTAATCCTGT TTTGCCTCCCCCACGCTTTCGCGCCTCAGCGGGAGATACAG ACCAGAAAAAACCCCTTCGCCACTGGGGGTTCCCTCCACA ATGTCTTACACCATGTTCTC
Contig:
>HB_Query.ab1
GGATGCACATCTATCTGCAGTCGAGCGAGGCTCTTCGGACG CTAGCGGCGGAGGGTGAGTAACACGTAGGCAACCTGCCTC TCAGACTGGGATAACATAGGGAAACTTACGCTAACCATCTT CGGCGGCTGGCTCCTTGCGGTTACTCACCGACTTCGGGTG TTGCAAACTCCCGTGGTGTGACGGGCGGTGTGTACAAGGC CCGGGAACGTATTCACCGCGGCATGCTGATCCGCGATTACT AGCGATTCCGACTTCATGTAGGCGAGTTGCAGCCTACAATC CGAACTGAGATTGGTTTTAAGAGATTGGCGTCCCCTCGCGA GGTAGCATCCCGTTGTACCAACCATTGTAGCACGTGTGTAGC CCAGGTCATAAGGGGCATGATGATTTGACGTCATCCCCGCC TTCCTCCGTCTTGTCGACGGCAGTCTCTCTAGAGTGCCCAA CTGAATGCTGGCAACTAAAGATAAGGGTTGCGCTCGTTGCG GGACTTAACCCAACATCTCACGACACGAGCTGACGACAAC CATGCACCACCTGTCACCGCTGCCCCGAAGGGAAGCTCTG TCTCCAGAGCGGTCAGCGGGATGTCAAGACCTGGTAAGGT TCTTCGCGTTGCTTCGAATTAAACCACATGCTCCACCGC TTGTGCGGGCCCCCGTCAATTCCTTTGAGTTTCACTCTTG CGAGCGTACTCCCCAGGCGGAGTGCTTATTGCGTTAGCTGC GGCACTGAGGGTATTTGAAACCCCCAACACCTAGCACTCA TCGTTTTACAGCGTGGACTACCAGGGTATCTAATCCTGTTTT GCCTCCCCCACGCTTTCGCGCCTCAGCGGGAGATACAGACC AGAAAAAACCCCTTCGCCACTGGGGGTTCCCTCCACAATGT CTTACACCATGTTCTC
Hit1: Brevibacillus brevis strain ZLynn1000-15 16S ribosomal RNA gene, partial sequence
Query coverage: 97%
E value: 0.0
Percentage identity: 97.68%
Accession no.: KY316484.1
Phylogenic tree:

METAGENassist data used to interpret relevant phenotype and functional characteristics.
Biotic interaction

Energy metabolism

Metabolism

Oxygen requirement

Habitat
Sporulation
