1. INTRODUCTION
Nitrogen (N) is one of the essential macronutrients that help in the plant growth and development. It is also an important component of proteins, nucleic acids, protoplasm, chlorophyll, and other nitrogenous compounds [1]. Nitrogen is abundantly available in earth’s atmosphere but cannot be utilized by the plants so it must be converted into ammonia (NH3) or related nitrogenous compounds. This conversion of atmospheric N (N2) is referred as N2-fixation. The N2 fixation process is considered as the most important process influencing primary productivity, after photosynthesis. This process is carried out by physicochemical and biological process (through microbes) that fixes 10% and 90% natural nitrogen, respectively [2,3]. Biological fixation of nitrogen occurs in certain restricted plant species due to microbe’s specific niche. Over 102 years, the process of biological N2 fixation (BNF) is studied in legumes and is restricted to prokaryotes, as N2 fixation is an anaerobic process [4].
To make nitrogen available to non-leguminous plants, it is supplied as external source and is prepared by the Haber-Bosch process. This process of manufacturing ammonia has made the most valuable invention and saved millions people worldwide [5]. About 450 million tonnes of nitrogen fertilizer per year, in the form of anhydrous ammonia, ammonium nitrate, and urea are produced by Haber process. Haber–Bosch is perhaps the most significant invention of the 20th century; yet, it has numerous harmful effects, including reduced biodiversity, formation of aquatic algal blooms, which also contributes to air pollution as well as climate change ultimately contributing to global warming [6]. Therefore, the utilization of alternative resources as microbes in forms of bioinoculant, which are cost effective and eco-friendly, is essential. In recent time, much interest is focused on endophytic microbes as potential biofertilizers. The significance of biofertilizers is to enhance the plant growth, through combination of BNF, along with solubilization of various macronutrients [phosphorus (P) and potassium (K)] and micronutrients such as zinc (Zn) from soil, synthesis of hormones, and inhibiting the growth of various phytopathogens [7].
Microbes residing inside the tissue of plant host for at least part of its life cycle without causing apparent disease are referred to as endophytes [8]. Endophytic microbes are better in comparison to rhizospheric microbes as they provide N directly to the host plant [7]. Endophytic microbes are guarded from abiotic stress condition such as change in pH, temperature, water availability, and nutrient plus biotic stress such as competition [9,10]. Endophytic microbes colonize various parts of plant (roots, stem, leaves, flowers, fruits, and seeds) and transferred from one generation to next one either by vertical or horizontal transmission [11]. Cavalcante and Dobereiner [12] first laid the foundation of isolation of a N2-fixing bacteria Gluconacetobacter diazotrophicus from internal tissues of sugarcane, latter on agriculturally important crops such as rice, wheat, maize, and kallar grass received much attention of scientists for isolation of N2-fixing endophytic microbes. Endophytic microbes associated with leguminous and non-leguminous plants facilitate the growth, crop yield, and soil health. Endophytic microbes have been reported such as Acetobacter diazotrophicus (sugarcane) [13], Acinetobacter lwoffii (wheat) [14], Azoarcus (rice) [15], Azospirillum brasilense (maize) [16], Acinetobacter calcoaceticus (soybean) [17], and Bradyrhizobium (peanut) [18]. Present review presents an overview and recent advances in the understanding mechanism of N2-fixing endophytic bacteria and their biotechnological applications for plant growth and soil health for agricultural sustainability.
2. BIODIVERSITY AND DISTRIBUTIONS
The microbial biodiversity is higher as compared to vertebrates, insects, and plants. Microbes have adapted to diverse habitat and reported from natural as well as from extreme environment. The diverse microbial groups were reported for their association with several host plants as plant microbiome (epiphytic, endophytic, and rhizospheric) [19]. The predominant and abundant group of microorganisms on earth reported as endophytic bacteria belongs to the phylum Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria [Figure 1a and b]. The phylogenetic tree was made using the MEGA 4 software to know the taxonomical affiliations for entophytic microbes isolated from diverse sources. The phylum Proteobacteria is further grouped into different classes such as α-, β-, and γ- proteobacteria. The member of phylum Proteobacteria with nitrogen-fixing capability has been reported as most dominant from diverse host plants. Least number of N2-fixing endophytic bacteria has been reported from the phylum Bacteroidetes, Actinobacteria, and from Firmicutes [Figure 2]. On review of different host plants, the most dominant genera includes Burkholderia, Rhizobium, Pseudomonas, Bradyrhizobium, Bacillus, Frankia, Enterobacter, Azospirillum, Klebsiella, Herbaspirillum, and Acinetobacter which have been regarded as nitrogen-fixing endophytic bacteria. The relative distributions of N-fixing endophytic genera have been analyzed using studies form different host plants [Figure 3].
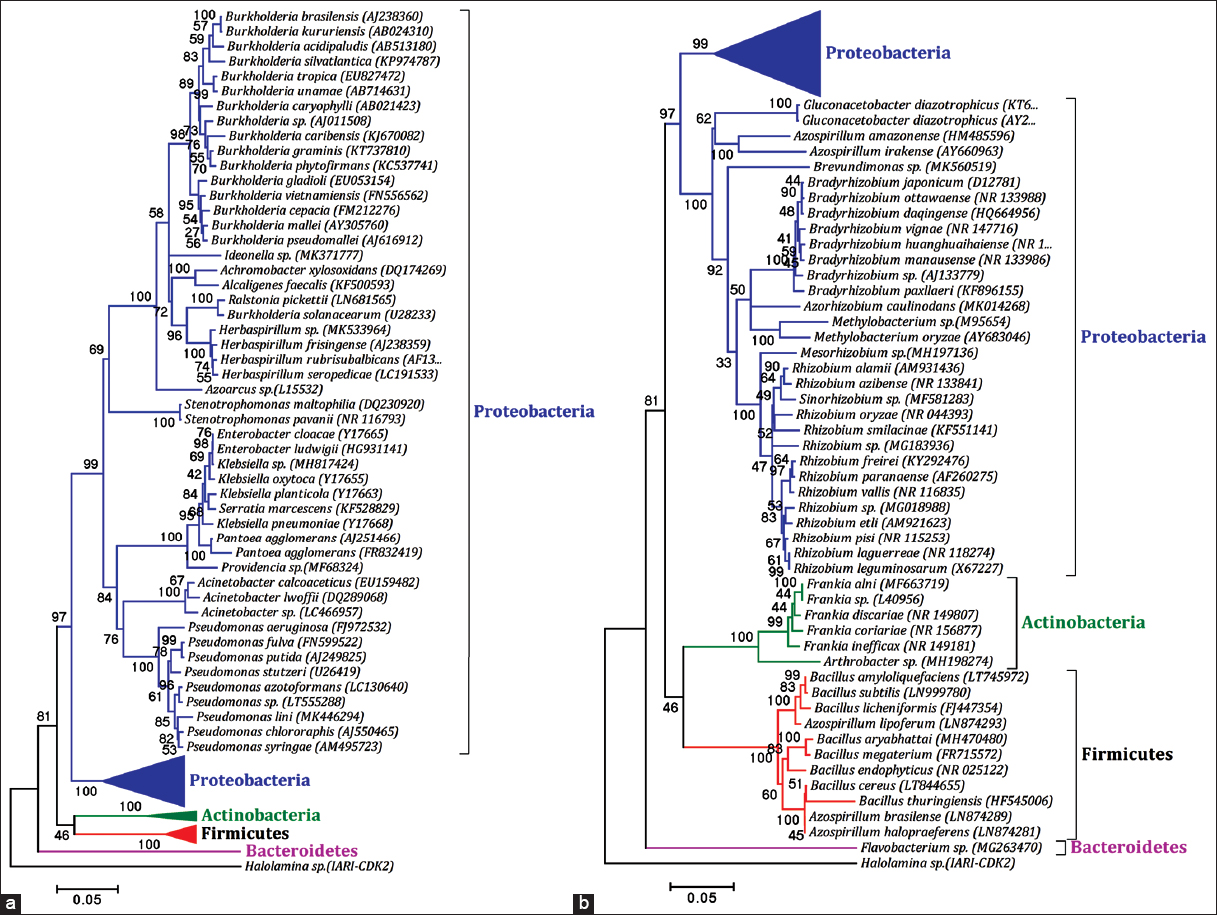 | Figure 1: (a) Phylogenetic tree showing the relationship among different groups of nitrogen-fixing endophytic microbiomes isolated from different host plants, (b) Phylogenetic tree showing the relationship among different groups of nitrogen-fixing endophytic microbiomes isolated from different host plants. [Click here to view] |
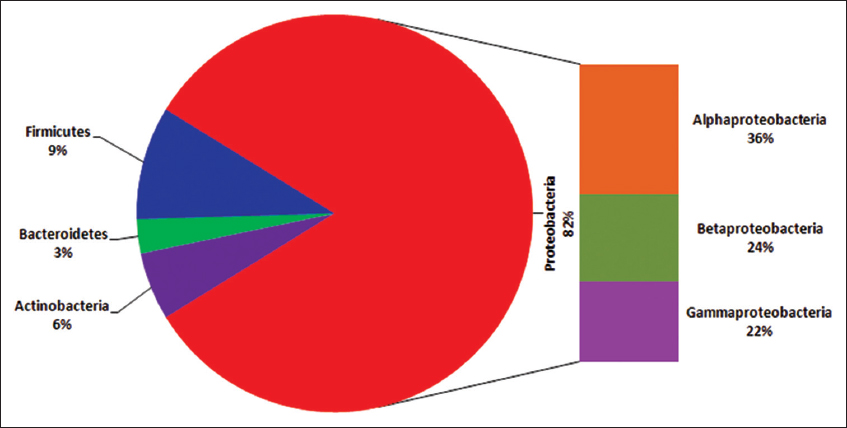 | Figure 2: Abundance and relative distribution of nitrogen-fixing endophytic microbes belonging to diverse phylum. [Click here to view] |
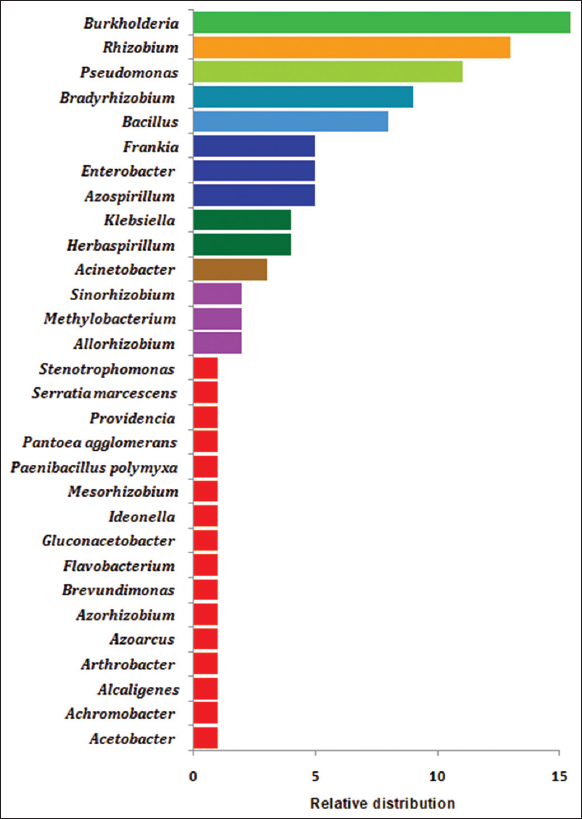 | Figure 3: Relative distribution of predominant genera of nitrogen-fixing endophytic microbes reported from different host plants. [Click here to view] |
Among different genera, Burkholderia belongs to the phylum Proteobacteria and class β-proteobacteria has been reported to colonize maize (Zea mays) [20], soybean (Glycine max) [21], sugarcane (Saccharum officinarum) [22], and rice (Oryza sativa) [23]. Burkholderia sp., in a study, was reported to fix atmospheric N along with solubilization of P and also synthesizes indole-3-acetic acid (IAA) [21]. Further, novel species of genera Burkholderia as Burkholderia unamae MTl-641T and Burkholderia silvatlantica SRMrh-20T were isolated as endophytes from sugarcane [22]. The second most predominant genera Rhizobium also belongs to the phylum Proteobacteria and class α- proteobacteria has been reported to colonize rice [24], soybean [25], faba bean (Vicia faba) [26], clover (Trifolium) [27], beans (Phaseolus vulgaris) [28], and wild rice [29]. The various studies reported the plant growth promoting (PGP) potential of Rhizobium sp. as it plays key role in fixation of atmospheric nitrogen [30]. Rhizobium vallis CCBAU 65647T and Rhizobium hidalguense FH14T were reported as novel N2-fixing endophytic species from P. vulgaris [31]. Another predominant genera Pseudomonas belong to the phylum Proteobacteria and class γ- proteobacteria has been reported to colonize different host such as sugarcane [32], maize [33], rice [34], and soybean [17]. One of the species of Pseudomonas as Pseudomonas putida showed various multifarious PGP attributes N2 fixation, production of phytohormones and siderophores has been reported by Zhao, Xu [17].
2.1. Rhizobia
During the 17th century, on roots of legume, people observed “bumps” which laid the foundation for discovery of microorganisms inside nodules responsible for fixing atmospheric N. The organism inside nodule has been identified as vibrio-like or bacteria-like named as rhizobia [35]. Rhizobium, Bradyrhizobium, and Azorhizobium, are collectively referred to as rhizobia. Symbiotic N2-fixing rhizobia belong to the family Rhizobiaceae. In 1889, the first species of genus rhizobia was identified as Rhizobium leguminosarum. Rhizobia are diverse group of soil bacteria established Rhizobium-legume symbiosis. The symbiotic association of rhizobia with leguminous plants is a complex process which results in the development of nodule like structure. The nodules are the house for N2-fixing endophytic microorganisms. It has been already documented in leguminous plants that Rhizobia species (Azorhizobium, Allorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium) act as natural endophytes and perform BNF to great extent [36]. The species of rhizobia is not only limited as an endophyte to leguminous plants, the member of rhizobia has been reported to colonize the roots region of non-legumes wheat, maize, and barley. Roots of rice were found to be attached by rhizobia. The bean plant inoculated with transformed Rhizobium resulted in an increased N content of seeds, nitrogenase activity, and leaf content [28]. Azorhizobium caulinodans has been reported in roots of rice [36]. Azorhizobium is symbiotic bacteria form nodule on the stem, fixes N2, and produces large amount of IAA [37]. Bradyrhizobium reported to enhance the P, K, N, and organic content in soil and fixes N2 efficiently in Mucuna seeds [38]. R. pisi (Trifolium) [27] and R. smilacinae (Smilacina japonica) [39] have been isolated from different hosts.
2.2. Frankia
The member of Frankia belongs to family Frankiaceae in the order Actinomycetales, which has been reported as symbiotic association with higher plants. The genus Frankia consisting of high G + C% are Gram-positive grows at a temperature of 28–30°C and are reported as slow growing N2-fixing endophytes [40]. Frankia inhabits vital ecological niche in the living plant tissue and also express nitrogenase enzyme, thereby fix the atmospheric N2 and transferred fixed N2 to their host [41]. In the recent time, various studies revealed nodule of actinorhizal plants harbor a broad range of bacteria [42]. In non-leguminous plants, Frankia was known for development of N2 -fixing nodules [43]. Frankia mostly established symbiotic association with higher plants such as Alnus, Elaeagnus, Coriaria, Casuarina, Datisca glomerata, Elaegnaceae, and Myrica species allows the availability of element such as N and P [44]. Different species of Frankia such as F. elaegni, F. alni, F. inefficax, F. discariae, F. coriariae, and F. discariae has been reported from more than 24 host plants [45]. In a study, Frankia has been reported as N2-fixing bacteria isolated using culture-independent methods from O. sativa [46], G. max [47]. In a study, inoculation of Casuarina equisetifolia, Acacia nilotica, and Eucalyptus tereticornis plant with Frankia, arbuscular mycorrhizal fungi, Rhizobium, and Azospirillum has been done as a single inoculation or as consortium which finally revealed increased output with consortium than single inoculation. The plants absorb more P from soil, which developed more nodules with high nitrogenase activity as compared to control [48].
2.3. Azospirillum
Azospirillum is associative N2 -fixing rhizobacteria also known to synthesize various antifungal, antibacterial, and siderophore compounds that are a Gram negative, aerobic, N2-fixing, and non-nodule forming bacteria belonging to family Spirilaceae [49]. Azospirillum is a potential N2-fixing microsymbiont as well as associative diazotrophs of grasses which also penetrate the root of gramineous plant species [50]. The endophytic nature of Azospirillum in rice plant has been demonstrated by Rodrigues, Rodrigues [51]. Under the genus Azospirillum, many species were discovered such as A. lipoferum, A. amazonense, A. brasilense, and A. halopraeferens [52]. The inoculation of plant with Azospirillum increases the water absorption and nutrient uptake which happen due to more development of root system. The inoculation of plants with Azospirillum results in the better root system development with highest efficiency in water and nutrients absorption from soil [53]. The Azospirillum genera demonstrated its potential application in agricultural systems, up to 30% in productivity [54]. The endophyte A. lipoferum reported to produce phytohormones such as abscisic acid and gibberellins which result in the alleviation of water scarcity [55]. O. sativa inoculated with bacterial endophyte, Azospirillum sp. B510, resulted in improved resistance against phytopathogens Magnaporthe oryzae and Xanthomonas oryzae [56]. The application of rice plants with Azospirillum resulted with an improved aerial biomass along with an increased content of N by 16 and 50 kg ha−1 [57]. The Azospirillum promoted the plant growth by secretion of various growth promoters (IAA, gibberellins, and cytokinin), uptake of various nutrients and enhances the development of root of plant [58].
The inoculation of non-leguminous plant with growth-promoting N2-fixing endophytic bacteria is achieving its value globally. The Z. mays treated with A. brasilense strain Ab-V5 designated that the treatment was proficient in stimulating growth of plant and also increases crop productivity [59]. In an experiment, the rice plant co-inoculated with Rhizobia and A. brasilense efficiently promoted seed germination, increased wetland rice production which may, further, decrease production costs and negative impacts on environment caused by usage of N fertilizers [60]. The inoculation of legumes with Rhizobia and azospirilla could over-improve the activity of plants, in comparison with a single inoculation. Over the year, significant attainments were gained from the research that is the employment of azospirilla as a commercial inoculants for cereals or legumes as well as non-legume crops [61]. Azospirillum sp. B510 originally isolated from rice demonstrated its skill to activate the innate immune system in tomato plant against Pseudomonas syringae and Botrytis cinerea the causal agent of bacterial leaf spot and gray mold, respectively [62]. Azospirillum sp. B510 induces systemic disease resistance in the host and further study signified that involvement of ethylene (ET) signaling is necessary for endophyte-mediated ISR in rice [63]. For increasing the production of different agricultural product, Azospirillum could be used as a potential biocontrol agents and biofertilizers [64].
2.4. Herbaspirillum
Herbaspirillum is an endophytic N2-fixing organism known to colonize the intercellular spaces of roots of maize, rice, sorghum, and sugarcane. Herbaspirillum seropedicae has been first reported as N2 -fixing bacterium associated with the roots of O. sativa, Sorghum bicolor, and Z. mays [65]. The endophytic nature of Herbaspirillum was confirmed by fluorescence and electron microscopy as the DNA sequence of bacteria was tagged with green fluorescent protein (GFP) [66]. Eighty different strains of H. seropedicae fix 31–45% of N in rice from atmosphere. Under aseptic condition, N2 fixation by Herbaspirillum was 33–58 mg tube−1 [67]. In wheat plant, H. seropedicae acts as an endophytic diazotrophs and also utilizes diverse range carbon sources [68]. H. seropedicae is well known diazotrophic endophytic bacteria that interact with various species of plant belonging to family Graminiae. In addition to diazotrophy, H. seropedicae possesses complete machinery for siderophores, auxin, and ACC deaminase synthesis which makes it suitable for enhancement of plant growth [69]. H. seropedicae is an endophytic diazotrophs reported for colonization with various plants and also increase their productivity [70]. H. seropedicae colonizes several non-leguminous plants, uses various mechanisms for BNF, solubilization of minerals, synthesis of phytohormones, and by acting as a biocontrol agent. The treatment of Z. mays L., with combination of humic substances and H. seropedicae resulted with variation of sugar and N metabolism, and better net photosynthesis along with better production of maize about 65% under field conditions [71].
Miscanthus sinensis inoculated with Herbaspirillum frisingense GSF30T significantly promoted root and shoot growth affected the architecture of root by modulating phytohormone signaling pathways which ultimately affected the architecture of root [72]. The wetland rice inoculated with biological fertilizer (mycorrhizal fungi and H. seropedicae) extensively improved the productivity and also enhanced fortification and root growth. Integrated nutrient management with biological and chemical fertilizers advances the yield of O. sativa L., also health and fertility of soil [73]. In a study, H. seropedicae has been demonstrated for its colonization with the roots of Triticum aestivum. The transcriptomic analysis of H. seropedicae attached to roots of wheat did not show iron metabolism genes up-regulated; further, suggesting the effect of bacteria on plant iron metabolism is more complex [74].
2.5. Gluconacetobacter
Firstly, the diazotrophs responsible for N2 fixation in root and stem of sugarcane was named as Saccharobacter nitrocaptans [12] and then transformed to A. diazotrophicus [75]. However, later on, A. diazotrophicus was reclassified into a new genus Gluconacetobacter [76]. The bacterium is a small, Gram-negative, aerobic, and rod shaped, which showed pellicle formation in N-free semi-solid medium. G. diazotrophicus reported as diazotrophic endophyte provides significant amount of N2 to plants and also shows some other physiological properties of diazotrophs such as tolerance to high sucrose and salt and low pH conditions [77]. G. diazotrophicus is acid-tolerant endophyte need high concentration of sugar in the medium to grow [78] and that requirement was fulfilled in the sugarcane [79]. The endophytic nature of G. diazotrophicus as it was labeled with immunogold defined its occurrence on exterior and interior of S. officinarum plant tissues was confirmed by James, Reis [80]. The intercellular spaces of sugarcane stems were filled with the N2-fixing endophytic bacterium G. diazotrophicus [81]. The close relationship between addition of N fertilizer to sugarcane and isolation frequencies of G. diazotrophicus was suggested by Fuentes-Ramirez, Jimenez-Salgado [82]. When sugarcane plant was fertilized with low amount of 120 kg of N/ha, densities of isolation elevated up to 70%. [67,83].
Further, Munoz-Rojas, Fuentes-Ramírez [84] analyzed that the antagonistic activity between 55 G. diazotrophicus strain bacteriocin-like molecules (approximately 3400 Da) with low molecular weight is responsible for such antagonism. Bacteriocin suppresses the growth of Xanthomonas albilineans [85]. The antagonistic property of some strains of G. diazotrophicus is responsible for colonization of sugarcane plant. Within the plant, there may be antagonistic interaction between endophytic bacteria [84]. G. diazotrophicus produces a Bacteriocin (lysozyme) that controlled the growth of the Xanthomonas albilineans and showed antifungal activity against Helminthosporium carbonum and Fusarium sp. [86].
Inoculation of micropropagated sugarcane plant with G. diazotrophicus and Burkholderia vietnamiensis resulted in improved yield of sugarcane [87]. In an investigation Sevilla, Burris [13] investigated the ability of G. diazotrophicus to promote the growth of rice, wheat, and maize. They noticed that the G. diazotrophicus colonization is restricted to only root tissues in rice, wheat, and maize. The major supplier of fixed N2 to sugarcane is A. diazotrophicus. A. diazotrophicus fixes N2 when sucrose concentration high and it helps in enhancement of hormone production such as auxin [88]. A. diazotrophicus also found associated with sweet potato and Cameroon grass [89]. Inoculation with A. diazotrophicus resulted in a significant improvement in height of N-limited sugarcane plants in comparison with un-inoculated plants [13]. The unique characteristic of this bacterium is its capability to fixed up 48% of its N and make it available to the yeast. This reveals that the association A. diazotrophicus may directly benefit its host [90].
The constitutive exoenzyme levansucrase (LsdA) was identified in G. diazotrophicus strains. The enzyme hydrolyzed sucrose to produce oligofructans and levan. Further, G. diazotrophicus grows on sucrose, signifying that the G. diazotrophicus as an endophytic species utilizes plant sucrose through levansucrase and for the catabolism of glucose in G. diazotrophicus extracellular glucose oxidation is considered as the foremost route [91]. About 150 Kg N ha-1 yr-1 fixed by G. diazotrophicus in sugarcane has been reported by Muthukumarasamy, Cleenwerck [92]. The colonization of G. diazotrophicus strain PAL5 in the interior of root tissue of rice was confirmed by light microscopy as the PAL 5 marked with the GFP plasmid pHRGFPTC. The plasmids pHRGFPTC are valid tools for supervising the colonization of sugarcane and hydroponic rice with strain PAL5 Rouws, Meneses [93]. The colonization of rice roots endophytically by strain PAL5 was specified by Alquéres, Meneses [94]. In response to phytopathogens attack, plants activate there defense system by secretion of reactive oxygen species (ROS). At an early stage of colonization, of rice plants with the strain PAL5, the bacterium upregulated the ROS-detoxifying genes transcript levels. This ROS scavenging enzymes of PAL5 also participate in the colonization of rice plants. G. diazotrophicus endophytically colonizes the roots of Arabidopsis thaliana. Inoculation by this strain extensively promoted plant biomass leaves number, total leaf area, and improved water use efficiency in A. thaliana [95].
2.6. Burkholderia
The genus Burkholderia belongs to the β-proteobacteria class of phylum Proteobacteria. The genus Burkholderia divided into two main clusters on the basis of phylogenetically: the first one comprises chiefly of human, animal, and plant pathogens e.g. Burkholderia cepacia complex (Bcc) and the second one comprises plant-beneficial-environmental (PBE) [96]. In the early 1980s, the strains of B. cepacia were recovered from cystic fibrosis patients [97]. B. cepacia species includes seven closely related genomic species, now termed as “cepacia complex” [98]. Species of environment origin was, then, further joined to this genus including B. graminis, B. caribensis, and Burkholderia kururiensis. In recent times, the PBE cluster has been the center of study, due to their versatile ability of N2 fixation, nodulation in legume, and degradation of recalcitrant compounds [99]. The three species of Burkholderia as B. caryophylli, B. gladioli, and B. solanacearum are identified as phytopathogens, whereas; B. pseudomallei, B. mallei, and B. pickettii are primary pathogens of animals and humans [75].
B. acidophilus, B. kururiensis, B. phytofirmans, and B. unamae have been isolated from surface sterilized plants [100-103]. During 1991, B. phytofirmans PsJN isolated from roots of onion infected with fungus Glomus vesiculiferum by Dr. Jerzy Nowak. Initially, the strain was classified as Pseudomonas sp. [104]. Later on, PsJN strain was reclassified as B. phytofirmans, recent research reported plant inoculated with strain PsJN that produces enhanced root system, large amount of chlorophyll, and phenolics compound in the plant [102]. B. phytofirmans found to be antagonistic to potato pathogen Rhizoctonia solani. Colonization of Vitis vinifera L., Solanum tuberosum, and Solanum lycopersicum L. by PsJN reported to enhanced the root system, leaf content, and water use efficiency [105-107]. B. phytofirmans PsJN reported to stimulate resistance in grapevine against the pathogen Botrytis cinerea causing gray mold diseases [108]. B. phytofirmans PsJN has been recovered from roots of Saccharum sp., and agricultural soil in Netherlands [109]. The gfp-marked strain PsJN visualized as an endophyte inside young berries of V. vinifera L. [110]. B. phytofirmans PsJN has the largest genome comprising two chromosomes and one plasmid. The strain PsJN comprises quorum-sensing system that manages the switch of bacterium from free-living to the symbiotic system [111]. In another interesting study, B. phytofirmans strain PsJN transformed with nif operon containing genomic DNA. The gene was horizontally transferred from one of the known N2-fixing bacterium, B. phymatum STM 815 [112].
A massive number of endophytic bacteria have been isolated from the roots of rice. Burkholderia sp. strains revealed that bacteria colonizes to the root surface and then invade into the intercellular spaces [113]. The isolation of Burkholderia from the roots of rice was demonstrated by Singh, Mishra [114]. Further, the B. cepacia from the stem of Eucalyptus was proficient in inhibiting Magnaporthe grisea, Fusarium moniliforme, and Rhizoctonia solani [115]. B. vietnamiensis (MGK3) had the maximum nitrogenase activity determined by its ability to reduce acetylene [23]. As reported by Govindarajan, Balandreau [87] sugarcane plant inoculated with Burkholderia MG43 resulted in increase in the yield of sugarcane saving the cost of ~140 kg ha−1 N fertilizer, later in a study by Govindarajan, Balandreau [23] B. vietnamiensis (MGK3) inoculated to rice plants resulted with enhanced grain yield of rice by 5.6-12.6%. Grain yield of rice plant in a field trial increased up to 8 t ha−1 when inoculated with B. vietnamiensis [116].
2.7. Bacillus and Bacillus-Derived Genera
Bacillus and Bacillus-derived genera (BBDG) are ubiquitous in nature and have been reported worldwide as plant microbiome, soil microbiome, and extremophiles. Bacillus is a genus of Gram-positive, endospore-forming, and rod shaped bacterium. Under unfavorable environmental conditions, it is identified that Bacillus species can form dormant spores (endospore). The endospores remain viable for longer period of time and are resistant to heat, sunlight, and chemicals. Bacillus species might be obligate aerobes or facultative anaerobes and is well known for its agricultural application (as an effective biocontrol agent) such as Bacillus thuringiensis prove as a biocontrol agent globally and industrial applications. Co-inoculation of pea and lentil plant with B. thuringiensis KR1 with R. leguminosarum PR1 resulted in enhanced growth of plant, increases the number of nodules [117]. Bacillus amyloliquefaciens as an endophyte in the internal tissues of T. aestivum was described firstly by Verma, Yadav [14].
One of the significant applications of BBDG in agricultural field is used as a biocontrol agent. Cabbage seedlings inoculated with B. thuringiensis resulted as a biocontrol agent against Xanthomonas campestris pv. campestris causing black root of cabbage [118]. One of the most destructive diseases in the production of coconut is basal stem rot caused by Ganoderma lucidum. The Bacillus subtilis EPC8 secluded from roots tissues of coconut (Cocos nucifera) showed strong inhibition of pathogen [119]. The ability of Bacillus subtilis EDR4 also reported to suppress the hyphal growth of Sclerotinia sclerotiorum [120]. In another study, B. licheniformis CHM1 has been reported to protect Z. mays and V. faba against Bipolaris maydis and Rhizoctonia solani. Under in vitro conditions, the culture filtrate of B. licheniformis CHM1 also inhibited the growth of B. cinereapers, Colletotrichum gossypii, Dothiorella gregaria, Fusarium oxysporum, Gibberella zeae, and Rhizoctonia solani [121]. Bacillus megaterium was reported as first quinclorac degrading endophytic bacterium and can be used as good bioremediation bacterium. The degradation of quinclorac naturally in soil is very slow process [122]. B. megaterium BMN1 isolated from root nodules of Medicago sativa exhibited promotion of plant growth by synthesis of IAA and by solubilization of inorganic phosphate [123,124].
2.8. Pseudomonas
Pseudomonas is ubiquitous in nature and is a Gram-negative, rod-shaped, aerobic, non-spore forming, and polar-flagellated bacterium. Several studies described the association of Pseudomonas endophytically with variety of plants host for example Arachis, Artemisia, Populus, Brassica, Triticum, Pennisetum, and Trifolium [125-131]. Pseudomonas reported to enhance the growth of plant through a variety of mechanism such as phosphorous solubilization, production of phytohormones, BNF, and stimulation of induced systemic resistance [132]. Pseudomonas has been reported for exhibiting biocontrol activity as by synthesizing different compounds for example HCN, pyoluteorin, pyrrolnitrin, and phenazines chiefly phenazine-1-carboxamide and phenazine-1-carboxylic acid [133].
A diazotrophic endophyte, P. chlororaphis, isolated from the Sophora alopecuroides root nodules which have been confirmed through microscopic analysis expression of the gfp gene. The endophyte P. chlororaphis reported as potential growth promoting agent. The combined inoculation of P. chlororaphis with Mesorhizobium sp. showed significant effect on growth of plant such as production of siderophore, IAA, and solubilization of phosphorous [134]. Looking at the plant growth enhancing abilities of genus Pseudomonas, Tariq, Shafiqe [135] reported that endophytic P. aeruginosa showed significant inhibition of the root rotting fungi Macrophomina phaseolina, Rhizoctonia solani, and Fusarium solani, both under in vivo and in vitro conditions in wheat. In a study conducted on P. stutzeri strain, A15 under greenhouse experiment reported to induce five-fold enhancement in the biomass of O. sativa seedlings compared to uninoculated seedlings [136].
Other endophytes associated to genus Pseudomonas were isolated from root and seeds of maize plants. The strain P. putida FMZR9, P. lini MRR2, P. aeruginosa NFTR, P. fulva MRC41, P. aeruginosa FTR, and P. montelli FMZR2 showed tolerance against drought, salinity, and temperature [137]. P. azotoformans one of the drought resistant strains reported to nurture phytoremediation effectiveness of Trifolium arvense grown on contaminated soil containing multi-metal Cu, Zn, and Ni under stress of drought [138]. Rice seeds reported to harbor bacterial endophytes identified as P. putida (VWB3) by 16S rDNA sequencing. The bacterial endophyte prominently influenced the growth of seedlings, length of root and shoot, formation of secondary root hair, and also acted as biocontrol agent [139].
2.9. Other N2-Fixing Endophytic Bacteria
The other endophytic N2-fixing bacteria belong to genera Acinetobacter, Achromobacter, Azoarcus, and Stenotrophomonas. Acinetobacter belongs to a genus of Gram-negative bacteria that are an important soil microorganism and A. lwoffii was reported as an endophyte from wheat [14]. A. calcoaceticus DD161 isolated from G. max L. reported for its strongest inhibitory activity against Phytophthora sojae 01 which, further, causes the morphological abnormal changes of fungal mycelia [17]. Achromobacter xylosoxidans isolated from wheat variety Malviya-234, which demonstrated considerable nitrogenase activity, production of IAA, and solubilization of P [140].
In a study by Ladha and Reddy [141], association of Azoarcus with Kallar grass (Leptochloa fusca) roots reported for enhancing the yield up to 20–40 t ha-1yr- and Azoarcus also expresses nif genes and nitrogenase protein was demonstrated by [142]. Malik and co-workers reported by utilization of 15N isotope dilution and 15N natural abundance, kallar grass fix up to 26% of content of N [143]. Stenotrophomonas maltophilia described as endophytic association exerting advantageous result on the growth of plant (N2 fixation and phytohormone induction) and anti-fungal activity [144]. Further, the species of the genus Stenotrophomonas that fixes N2 in Sugarcane is Stenotrophomonas pavanii that is Gram-negative and non-motile and do not form spores [145].
3. N2-FIXING NOVEL ENDOPHYTIC BACTERIA
BNF is one of the possible biological alternatives to N-fertilizers (urea) and could lead to more productive and agricultural sustainability without harming the environment. Several endophytic bacteria are now known to increase the availability of N for their host plants. Different novel N2-fixing endophytic bacteria have been isolated from several crops, which contribute fixed N2 to the associated plants. Acetobacter belonging to phylum proteobacteria reported as novel species A. diazotrophicus LMG 7603 and Acetobacter nitrogenifigens RG1T which have been isolated from S. officinarum and Kombucha tea [146]. Novel species of N-fixing endophytic bacteria belonging to different genera as A. diazotrophicus LMG 7603 [147], B. unamae MTl-641T [148], B. silvatlantica SRMrh-20T [22], S. pavanii ICB 89T [145], Enterobacter sacchari SP1T [149], and Klebsiella variicola strain DX120E [150] has been reported from sugarcane.
Novel bacterial species such as Gluconacetobacter sp. PA12 [151], Ochrobactrum oryzae MTCC 4195T [152], Phytobacter diazotrophicus LS 8T[153], Acinetobacter oryzae B23T [154], Enterobacter oryzendophyticus, and E. oryziphilus [155] has been reported as N2-fixing endophytic bacteria from rice. Rhizobia are N2-fixing soil bacteria that fully or partially satisfy the N demand of the host plant belonging to leguminous family plant. The diversity of rhizobia from Sesbania rostrata [156], alfalfa [157], P. vulgaris L. and Leucaena sp. [158], and Pisum sativum [159] has been studied and characterized for N-fixing attributes. Further, novel species of genera Rhizobium as R. vallis CCBAU 65647T [160] and R. hidalgonense FH14T [31] has been reported for forming effective nodules in P. vulgaris [Table 1].
Table 1: Nitrogen-fixing novel species of microbes reported from diverse crop species.
| Novel microbes | Host/association | References |
|---|---|---|
| A. nitrogenifigens RG1T | Kombucha tea | Dutta and Gachhui [146] |
| A. melinis TMCY 0552T | M. minutiflora | Peng, Wang [164] |
| B. mimosarum PAS44T | M. pigra | Chen, James [165] |
| B. phytofirmans PsJNT | A. cepa | Sessitsch, Coenye [102] |
| B. silvatlantica SRMrh-20T | S. officinarum | Perin, Martínez-Aguilar [22] |
| E. oryzae Ola 51T | O. latifolia | Peng, Zhang [166] |
| E. sacchari SP1T | S. officinarum | Zhu, Zhou [149] |
| F. endophyticum 522T | Z. mays | Gao, Lv [167] |
| F. coriariae BMG5.1T | C. japonica | Nouioui, Ghodhbane-Gtari [45] |
| G. kombuchae RG3T | Kombucha tea | Dutta and Gachhui [168] |
| Gluconacetobacter sp. PA12 | O. sativa | Loganathan and Nair [151] |
| H. lusitanum P6-12T | P. vulgaris | Valverde, Velazquez [169] |
| K. phytohabitans KLBMP 1111T | J. curcas L. | Xing, Bian [170] |
| K. arsenatis CM1E1T | P. laevigata | Román-Ponce, Wang [171] |
| M. phragmitis MP23T | P. karka | Behera, Ramana [172] |
| M. nidulans LMG 21967T | C. glaucoides | Rozahon, Ismayil [173] |
| O. oryzae MTCC 4195T | O. sativa | Tripathi, Verma [152] |
| P. herberti R33T | H. sendtneri | Guo, Zhou [174] |
| P. wenxiniae 373T | Z. mays | Gao, Lv [175] |
| P. diazotrophicus LS 8T | O. sativa | Zhang, Peng [153] |
| P. diazotrophica R5-392T | J. curcas | Madhaiyan, Jin [176] |
| R. hidalgonense FH14T | P. vulgaris | Yan, Yan [31] |
| R. oryzae Alt 505T | O. alta | Peng, Yuan [24] |
| R. pongamiae VKLR-01T | P. pinnata | Kesari, Ramesh [177] |
| R. populi K-38T | P. euphratica | Rozahon, Ismayil [173] |
| R. smilacinae PTYR-5T | S. japonica | Zhang, Shi [39] |
| R. vallis CCBAU 65647T | P. vulgaris | Wang, Wang [160] |
| S. kummerowiae CCBAU 25048T | K. stipulacea | Lin, Wang [178] |
| S. pavanii ICB 89T | S. officinarum | Ramos, Van Trappen [145] |
B. silvatlantica: Burkholderia silvatlantica, A. nitrogenifigens: Acetobacter nitrogenifigens, A. melinis: Azospirillum melinis, B. mimosarum: Burkholderia mimosarum, B. phytofirmans: Burkholderia phytofirmans, E. oryzae: Enterobacter oryzae, E. sacchari: Enterobacter sacchari, F. endophyticum: Flavobacterium endophyticum, F. coriariae: Frankia coriariae, G. kombuchae: Gluconacetobacter kombuchae, H. lusitanum: Herbaspirillum lusitanum, K. phytohabitans: Kibdelosporangium phytohabitans, K. arsenatis: Kocuria arsenatis, M. phragmitis: Mangrovibacter phragmitis, M. nidulans: Methylobacterium nidulans, O. oryzae: Ochrobactrum oryzae, P. herberti: Paenibacillus herberti, P. wenxiniae: Paenibacillus wenxiniae, P. diazotrophicus: Phytobacter diazotrophicus, P. diazotrophica: Pleomorphomonas diazotrophica, R. hidalgonense: Rhizobium hidalgonense, R. oryzae: Rhizobium oryzae, R. pongamiae: Rhizobium pongamiae, R. populi: Rhizobium populi, R. smilacinae: Rhizobium smilacinae, R. vallis: Rhizobium vallis, S. kummerowiae: Shinella kummerowiae, S. pavanii: Stenotrophomonas pavanii, M. minutiflora: Melinis minutiflora, M. pigra: Mimosa pigra, A. cepa: Allium cepa, S. officinarum: Saccharum officinarum, O. latifolia: Oryza latifolia, Z. mays: Zea mays, C. japonica: Coriaria japonica, O. sativa: Oryza sativa, P. vulgaris: Phaseolus vulgaris, J. curcas: Jatropha curcas, P. laevigata: Prosopis laevigata, P. karka: Phragmites karka, C. glaucoides: Crotalaria glaucoides, H. sendtneri: Herbertus sendtneri, O. alta: Oryza alta, P. pinnata: Pongamia pinnata, P. euphratica: Populus euphratica, S. japonica: Smilacina japonica, K. stipulacea: Kummerowia stipulacea.
4. GENOMICS AND MECHANISMS OF N2 FIXATION
Symbiotic microbes from leguminous plants provide substantial benefit in the growth through N metabolism. “Symbiome” is the protein family involved in N2 fixation, nodulation, production of exopolysaccharides, and transport of oxygen. Sevilla, Meletzus [161] identified various genes involved in the N2-fixing process such as nifHDK, nifA, nifB, nifE, nifV, and ntrBC. Rhizobium carries plasmid genes for nodulation (nod, nol, and noe) in close proximity with nif and fix genes [162]. Rhizobium secretes the lipochitin oligosachcharides as the nodulation factor responsible for maintaining symbiotic association of rhizobia with legume [163].
Many endophytes reported to fix atmospheric N2 as demonstrated by the transcriptional activator nifA, involved in activation of N2-fixing (nif) operon, and regulator modE, involved sensing and uptake of molybdate at nanomolar concentrations [179]. About 26% of investigated community of endophytic microbes were detected with harboring the nifA activator, which concluded that endophytes have the higher capacity to promote growth of plant by N2 fixation process. Non-symbiotic microbes mostly include endophytes and free-living microbes such as Azotobacter and Azospirillum [180]. Different approaches have been used to study the interaction of endophytes with host plant in fixing atmospheric N2 and providing fixed N2 to plant such as by acetylene reduction assay, 15N isotope dilution experiments, and 15N2 reduction assays. BNF takes place under normal temperature and pressure and is catalyzed by the enzyme nitrogenase. A new N2-fixing endophyte, that is, Pantoea isolated from sugarcane, able to generate H2, develop in aerobic and anaerobic conditions, show, adjustability to higher pH, higher temperature, higher osmotic pressure, reduced acetylene, and also revealed the development of pellicle in semisolid LGI-P medium [181]. Verma, Yadav [14] reported diverse N2-fixing bacteria belonging to genera Acinetobacter, Bacillus, Pseudomonas, and Stenotrophomonas. The gene association with N2 fixation such as nif HDK, nifV, nifE, and nifA has already been sequenced and detected in A. diazotrophicus [161]. The amplification of nifH gene fragment by polymerase chain reaction (PCR) the N2-fixing endophytes was identified [140, 182].
Biological fixation of atmospheric N2into ammonia was achieved by an enzyme complex called as nitrogenase present in the diazotrophic bacteria. Nitrogenase is a complex enzyme structure consisting of two subunits dinitrogenase and dinitrogenase reductase. Dinitrogenase/Component I (Larger component protein of the Mo-dependent nitrogenase is called as MoFe protein) containing molybdenum, iron and inorganic sulfur, and dinitrogenase reductase/Component II (The smaller component protein, called the Fe protein) containing iron and inorganic sulfur. The Fe protein transfers electrons from a reducing agent, such as ferredoxin to the MoFe protein [183]. The nif gene encodes the nitrogenase complex. The nitrogenase enzyme is synthesized by prokaryotes. It is found in both main domains of prokaryotes, in the archaea and in the bacteria. Pan and Vessey [184] described in term of changes of atmospheric pO2 G. diazotrophicus that has a switch-off/switch-on mechanism for the protection of nitrogenase enzyme system.
About 20 genes were found to be responsible in the complex N2 fixation mechanism for both genetic regulation of the process and nitrogenase protein synthesis [185,186]. To control the expression of genes including the nif system in the diazotrophs, a general N regulation (ntr) system is important. Key proteins which are involved in N control system are as follows: A uridylyltransferase (UTase) encoded by glnD, a small tetrameric effector protein (PII) encoded by glnB, NtrB encoded by ntrB, and a transcriptional activator NtrC encoded by ntrC [187]. Ureta and Nordlund [188] presented that the evidence for conformational protection of nitrogenase against O2 in G. diazotrophicus, in which protein is involved, is putative FeSII Shethna. To confirm the microbe in sample immunolabeling, in situ hybridization, and reporter genes have been used to identify the expression of nitrogenase proteins and/or nitrogenase encoding (nif) genes. The endophytic expression of nifH by Alcaligenes faecalis A15 and Azospirillum irakense within epidermal cells of roots has been shown by Vermeiren, Willems [189] and similar results have been obtained with Azoarcus [190].
Reis and Döbereiner [191], in his study, observed that nitrogenase enzyme was protected against inhibition by oxygen, ammonium, and some amino acids by 10% sucrose. Further, Madhaiyan, Poonguzhali [192] described the reduction in nitrogenase activity of pure cultures of G. diazotrophicus by the addition of pesticides to the growth medium. N-fixing system is sensitive to molecular oxygen. This sensitivity results from the instability of nitrogenase when exposed to oxygen. Diazotrophs use protective mechanism to protect nitrogenase [193]. Leghemoglobin is the protective protein associated nitrogenase. Leghemoglobin bind with the oxygen and keep nitrogenase enzyme active from oxygen. During the process of N2 fixation, sufficient amount of oxygen is required by rhizobia for respiration and prevention of oxygen supply to the nitrogenase complex take place at same time as leghemoglobin has high affinity for binding to oxygen [194].
During symbiotic association of rhizobia with host plant, flavonoids and phytohormones act as signaling molecule is produced by legume plant and activates the nodulation factor (nod) in rhizobia. N-fixing bacteria initiate the division of cortical cell of root and lead to nodule organogenesis [37]. Through the process of transformation the gene of Hb transferred from Vitreoscilla sp. to Rhizobium etli resulted in higher respiration rate of rhizobia [195]. Without the defense of leghemoglobin, the Rhizobium bacteriods will only fix N2 when external oxygen concentration is decreased to about 0.01 atm [196]. The structural genes for nitrogenase nif HDK in G. diazotrophicus are organized in a cluster (GDI0425–GDI0454), along with other N2 fixation-related genes, for instance fix ABCX, mod ABC and nif AB. In the regulation and metabolism of oxygen fix, gene family is involved. Other related genes, ntrX, ntrY, and ntrC (GDI2263, GDI2264, and GDI2265) are confined somewhere else in the chromosome in a 5.2 kb cluster. Thaweenut, Hachisuka [197] described the expression of nifH genes in the stems and roots of sugarcane plants by reverse transcription (RT)–PCR. The inoculation of Oryza officinalis with endophytic Herbaspirillum sp. B501 and in planta N2 fixation was studied using acetylene reduction assay and 15N2 gas incorporation approaches [66]. The colonization of rice plant with endophytic Burkholderia resulted in 372 mg N per plant an increase in N content in grains. The N2 was derived by biological N2fixation [113]. Roncato-Maccari, Ramos [198] reported in the intercellular region of vascular tissue and root cortex of T. aestivum, S. bicolor, O. sativa, and Z. mays roots observed with the expression of nifH gene encoding for iron protein of nitrogenase.
In a study, 343 endophytes were isolated from stem, root, and leaves of soybean cultivar. The two methodologies were used for screening N2-fixing endophytes. About 60% of endophytes grow on NFb medium and 21% of endophytes revealed containing nifH gene by PCR analysis. The endophytic isolates identified as A. calcoaceticus, Burkholderia sp., Pseudomonas sp., and Ralstonia sp. [21]. Further, Jha and Kumar [140] selected only those wheat plants showing acetylene reduction assay positive for endophytic bacteria isolation. One T. aestivum variety Malviya-234 revealed the occurrence of endophytic diazotrophic bacteria the highest nitrogenase activity was found in A. xylosoxidans (WM234C-3) which was 1.70 mmol C2H4 mg-1 protein h-1. Enterobacter ludwigii and Enterobacter cloacae reported to fix N2 by acetylene reduction assay and also have the potential of solubilization of phosphorous [199]. Hongrittipun, Youpensuk [200] reported B. cepacia (CS5) and Citrobacter sp. (CR9) isolated from rice as an endophyte and their inoculation in rice seedlings increases the concentration of N2 in roots of rice. Endophytic species Azoarcus sp. (Kallar grass), Burkholderia sp. (Rice), G. diazotrophicus (Sugarcane), and Herbaspirillum sp. (Rice) were reported to benefit their host plant by BNF. The commercially available N2-fixing biofertilizers are Rhizobium, Bradyrhizobium, Azospirillum, and Azotobacter [201].
5. PGP ATTRIBUTES OF ENDOPHYTIC N2-FIXING MICROBES
Endophytic microbes, along with N2-fixing attributes, also help in the uptake of various nutrients from soil and transfer to the plants and also synthesize various growth promoting compounds. The endophytic microbes directly or indirectly affect the growth of plants. The endophytic microbes in a direct way effectively enhance the growth of plants by solubilizing nutrients such as P, K, and Zn from soil as well as stimulate the better development of plant root which absorbs nutrients from soil, synthesize phytohormones which enhance the plant growth at different stages [Table 2]. Indirectly endophytic microbes enhances the plant growth by functioning as biocontrol agent, that is, suppresses various phytopathogens, release of siderophores, production of hydrogen cyanide, antibiotics and enzymes such as chitinases, and β-1,3-glucanase [202-204].
Table 2: Endophytic microbe from cereal crops with multifarious plant growth promoting properties.
| Endophytic microbes | Host | NF | P | K | Zn | IAA | Sid | References |
|---|---|---|---|---|---|---|---|---|
| A. diazotrophicus | Sugarcane | Sevilla, Burris [13] | ||||||
| A. xylosoxidans | Wheat | Jha and Kumar [140] | ||||||
| A. calcoaceticus | Soybean | Kuklinsky-Sobral, Araújo [21] | ||||||
| A. lwoffii | Wheat | Verma, Yadav [14] | ||||||
| A. faecalis | Rice | You and Zhou [217] | ||||||
| A. amazonense | Sugarcane | Oliveira, Urquiaga [218] | ||||||
| A. brasilense | Maize | Sw?drzy?ska and Sawicka [16] | ||||||
| A. brasilense | Sugarcane | de Bellone and Bellone [52] | ||||||
| A. brasilense | Wheat | Saubidet, Fatta [219] | ||||||
| B. amyloliquefaciens | Rice | Verma, Yadav [14] | ||||||
| B. amyloliquefaciens | Soybean | Zhao, Xu [17] | ||||||
| B. amyloliquefaciens | Wheat | Verma, Yadav [14] | ||||||
| B. aryabhattai | Rice | Ji, Gururani [220] | ||||||
| B. cereus | Soybean | Zhao, Xu [17] | ||||||
| B. megaterium | Soybean | Subramanian, Kim [30] | ||||||
| B. subtilis | Soybean | Bai, Zhou [221] | ||||||
| B. thuringiensis | Soybean | Bai, Zhou [221] | ||||||
| B. japonicum | Soybean | Subramanian, Kim [30] | ||||||
| Brevundimonas | Maize | Montañez, Abreu [33] | ||||||
| B. silvatlantica | Sugarcane | Perin, Martínez-Aguilar [22] | ||||||
| B. vietnamiensis | Rice | Govindarajan, Balandreau [23] | ||||||
| E. agglomerans | Rice | James, Gyaneshwar [222] | ||||||
| E. cloacae | Rice | Ladha, Barraquio [223] | ||||||
| E. cloacae | Soybean | Zhao, Xu [17] | ||||||
| E. cloacae | Sugarcane | Mirza, Ahmad [224] | ||||||
| Gluconacetobacter | Maize | Eskin [225] | ||||||
| Gluconacetobacter | Sugarcane | Suman, Shrivastava [226] | ||||||
| Herbaspirillum | Maize | de Bellone and Bellone [52] | ||||||
| H. seropedicae | Maize | Oliveira, Urquiaga [218] | ||||||
| H. seropedicae | Rice | Baldani, Baldani [113] | ||||||
| H. seropedicae | Sugarcane | Ladha, Barraquio [227] | ||||||
| K. oxytoca | Soybean | Mirza, Ahmad [224] | ||||||
| K. oxytoca | Sugarcane | Roesch, Camargo [228] | ||||||
| K. planticola | Rice | Ladha, Barraquio [223] | ||||||
| K. pneumonia | Maize | Govindarajan, Kwon [32] | ||||||
| K. pneumonia | Sugarcane | Gyaneshwar, James [229] | ||||||
| M. oryzae | Rice | Subramanian, Kim [30] | ||||||
| M. oryzae | Soybean | Kumar, Tomar [208] | ||||||
| P. polymyxa | Maize | Subramanian, Kim [30] | ||||||
| P. agglomerans | Rice | Puri, Padda [230] | ||||||
| P. agglomerans | Soybean | Verma, Ladha [209] | ||||||
| P. fluorescens | Rice | Montaíez, Abreu [33] | ||||||
| P. putida | Soybean | Mbai, Magiri [34] | ||||||
| R. leguminosarum | Rice | Govindarajan, Kwon [32] | ||||||
| S. marcescens | Rice | Dakora, Matiru [25] |
NF: Nitrogen fixation, P: Phosphorus solubilization, K: Potassium solubilization, Zn: Zinc solubilization, IAA: Indole-3-acetic acid, Sid: Siderophores, A. diazotrophicus: Acetobacter diazotrophicus, A. xylosoxidans: Achromobacter xylosoxidans, A. calcoaceticus: Acinetobacter calcoaceticus, A. lwoffii: Acinetobacter lwoffii, A. faecalis: Alcaligenes faecalis, A. amazonense: Azospirillum amazonense, A. brasilense: Azospirillum brasilense, B. amyloliquefaciens: Bacillus amyloliquefaciens, B. aryabhattai: Bacillus aryabhattai, B. cereus: Bacillus cereus, B. megaterium: Bacillus megaterium, B. subtilis: Bacillus subtilis, B. thuringiensis: Bacillus thuringiensis, B. japonicum: Bradyrhizobium japonicum, B. silvatlantica: Burkholderia silvatlantica, B. vietnamiensis: Burkholderia vietnamiensis, E. agglomerans: Enterobacter agglomerans, E. cloacae: Enterobacter cloacae, H. seropedicae: Herbaspirillum seropedicae, K. oxytoca: Klebsiella oxytoca, K. planticola: Klebsiella planticola, K. pneumonia: Klebsiella pneumonia, M. oryzae: Methylobacterium oryzae, P. polymyxa: Paenibacillus polymyxa, P. agglomerans: Pantoea agglomerans, P. fluorescens: Pseudomonas fluorescens, P. putida: Pseudomonas putida, R. leguminosarum: Rhizobium leguminosarum, S. marcescens: Serratia marcescens.
5.1. Solubilization of Macronutrients and Micronutrients
The most essential macronutrients for growth of plant are P and K. In soil, the phosphorous exists in two different forms organic and inorganic. The most important attributes performed by PGP endophytic microbes are the conversion of insoluble phosphates into soluble form [205]. In the soil, the concentration of P and K is very low, mineral compounds of phosphorus usually contain aluminium, iron, manganese, and calcium. The K in the soil exists in the form of feldspar (90–98%) and mica [206]. The phenomenon of solubilization of phosphate from soil by endophytic microbes mostly dependent on the pH and type of soil [207]. Endophytic N2-fixing bacteria belonging to genera Achromobacter [140], Acinetobacter [21], Bacillus [14], Burkholderia [21], Methylobacterium [208], and Pantoea [209] have been reported for phosphate-solubilization. The phosphorous solubilizing microbe dissolves the phosphorous through synthesis of different types of acids such as (acetate, lactate, oxalate, tartatate, succinate, citrate, gluconate, keto-gluconate, and glycolate) [210]. In case of Gram-negative bacteria, the enzyme glucose dehydrogenase contributes significantly to mineral phosphate solubilization [211]. The soil contains P in the form of organic substrates, which can be hydrolyzed to inorganic P by the enzyme alkaline phosphatases [212], phytase [213], phosphonoacetate hydrolase [214], D-α-glycerophosphatase [215], and C-P lyase [216] and subsequently available for plant nutrition. Endophytic bacteria A. calcoaceticus and Burkholderia sp. isolated from soybean cultivars showed significant growth promotion of plant by solubilization of mineral phosphate [21].
In a study Jha and Kumar [140] reported, A. xylosoxidans one of the diazotrophic endophytic bacteria isolated from surface-sterilized roots and culms of wheat variety Malviya-234 showed appreciable level of P solubilization ability. Verma, Yadav [14], in their findings, isolated 247 bacteria from wheat from five different sites of northern hills zone of India. The isolated strains were screened for P-, K-, and Zn-solubilization. The P, K, and Zn- solubilizing isolate was identified as B. amyloliquefaciens. Singh, Rajawat [231], in a study, evaluated the role of endophytes in the Zn fortification in wheat genotypes. Two endophytic bacteria Bacillus subtilis DS-178 and Arthrobacter sp. DS-179 inoculated in wheat resulted in two folds higher yield as compared to un-inoculated control. These Zn solubilizing endophytic bacteria also enhance the translocation and improvement of Zn to grains in wheat. In the modern strategies for biofortification, the endophytic microbes resulted as a better option.
5.2. Phytohormones Production
The hormones are the chemical messengers or an organic substance being produced in any one part and are transferred to another part that there influences a specific physiological process which included auxins, gibberellins, cytokinins, abscisic acid, and ethylene [232]. The various reports have demonstrated the ability of endophytic bacteria such as A. xylosoxidans [140], A. calcoaceticus, B. amyloliquefaciens [17], Burkholderia sp. [21], E. cloacae [17], Klebsiella oxytoca [224], Methylobacterium oryzae [30], Pantoea agglomerans [233], and Pseudomonas fluorescence [34] isolated from different host plant to synthesize different phytohormones. Ambawade and Pathade [234] reported Bacillus siamensis as a newly isolated endophytic bacteria from the banana have the ability to produce a significant amount of gibberellic acid with and without tryptophan-supplemented in media. One of the beneficial endophytic bacteria reported for its potential in N2 fixation along with production of phytohormones especially IAA with important crops such as sugarcane [80], rice [93], A. thaliana [95], and sorghum [235], and various other important crops are G. diazotrophicus. In a study by Rodrigues, Soares [236] identified the genes and pathways involved in biosynthesis of IAA by G. diazotrophicus and finally concluded that G. diazotrophicus PAL5T synthesizes IAA through the IPyA pathway in cultures supplemented with tryptophan and provides data for the contribution of an L-amino acid oxidase gene cluster in the IAA biosynthesis.
In a study by Defez, Andreozzi [182] reported endophytic bacteria E. cloacae RCA25 and K. variicola RCA26 which were isolated from roots and leaves of rice plants. K. variicola RCA26 had the highest nitrogenase activity. The two isolates, E. cloacae RCA25 and K. variicola RCA26, and two model bacteria (H. seropedicae z67 and Sinorhizobium fredii NGR234) were transformed to enhance the IAA biosynthesis. In addition to IAA, the endophytic bacteria Bacillus aryabhattai (MF693121.1), B. megaterium, and Bacillus cereus were isolated from nodules of Vigna radiata. Further, the study confirmed the presence of trp-dependent pathway for IAA production in the isolates and the in vitro application of bacterial isolates on plant roots resulted in increase in root length as well as number of lateral roots [237]. One of the most widely studied and best-characterized phytohormone is IAA. Bradyrhizobium sp. confirmed as an endophyte in rice plant and demonstrated its potential in enhancing plant growth by N2 fixation, production of ACC deaminase, and IAA.
5.3. Fe-Chelating Compounds Production
Siderophores (hydroxamate, catacholate and carboxylates) are high-affinity, iron-chelating organic compounds with low molecular masses, synthesized by microbes, and plants under low iron conditions. In the recent years, siderophores producing microbes have gained much attention due to their potential applications in environmental research. Microbes synthesizing the siderophores provide the bioavailability of Fe to plants for enhancing their growth [238]. The production of siderophores is generally specific at the genus level, for example, Pseudomonas sp. produces only pyoverdins, ornibactin by Burkholderia sp., and mycobactin by Mycobacterium sp. [239]. The production of siderophores by microbes confers the benefits to inhabit plant and eliminate other microbes from the similar ecological niche. Siderophores-producing bacteria under in vitro inhibition assays showed that Burkholderia was good antagonistic as they suppress fungal pathogens [240]. Various researches have shown the practicality of using eco-friendly approaches as microbes for improving the health and productivity of crop. From the roots of tomato plant, about 12 endophytic bacteria have been isolated and screened for PGP attributes and most of the root endophytes Pseudomonas, Rhizobium, Rhodococcus, and Agrobacterium have been detected for siderophores production. As, production of siderophores is considered as one of the biocontrol trait as lower availability of iron limits the growth of phytopathogens [241].
One of the endophytic bacterium, Pseudomonas brassicacearum strain Zy-2-1 was reported for producing siderophores and its inoculation with Sinorhizobium meliloti on Medicago lupulina resulted in enhancement of fresh weight of roots, total dry weight, nodule number and fresh weight and N content. The results demonstrated the usage of P. brassicacearum for remediation of heavy metal-contaminated soils [242]. A total of 276 endophytic bacteria were isolated from the root nodules of G. max L. Out of 276 endophytic bacteria, A. calcoaceticus resulted in the production of 54.33 ± 0.093 mg mL-1 siderophores and also causes the morphological abnormal changes of P. sojae 01 by acting as strongest inhibitor [17]. The contamination of arsenic in rice plants causes many negative impacts. In a study by Dolphen and Thiravetyan [243] reported, Bacillus pumilus had a capability to reduce accumulation of arsenic in rice grains to levels below those achieved by Pseudomonas sp. and B. thuringiensis.
5.4. ACC Deaminase
In agriculture practices, the plant hormone ethylene performs a significant function. Ethylene is one of the organic molecule and at a very low concentration (0.05 mL/L), ethylene functions as a proficient plant growth regulator [244]. The decreased concentration of ethylene produces significant increase in promoting the growth of roots [245]. In V. vinifera and S. tuberosum, the endophyte Burkholderia sp. synthesizes high levels of ACC deaminase [107]. Burkholderia strain PsJNT shows high ACC deaminase activity [246]. The novel species B. phytofirmans PsJNT showed highest activity of 1-aminocyclopropane-1-carboxylate deaminase ACC and lower the level of ethylene [102]. Plant seeds and roots treated with ACC deaminase containing bacteria demonstrated with 2–4 folds decrease in ACC and ethylene synthesis [247]. Pseudomonas sp. producing ACC deaminase resulted in enhanced height and weight of root and total biomass of maize plant [248]. The use of ACC deaminase producing bacteria reduces the level of ethylene along with mitigates the salt stress reported in maize plant [249]. H. frisingense strain GSF30T was isolated from M. sinensis C4 as an endophytic diazotrophs [250]. A. xylosoxidans AUM54 isolated from Catharanthus roseus was identified as highly salt tolerant endophytic bacteria [251]. The inoculation of two rice cultivar IR42 and IAC4440 with isolates of A. brasilense sp245 and B. kururiensis M130 resulted in a significant increase in the number of roots after 10 days. The cultivar IR42 was colonized more with bacteria and shows more BNF. The different ethylene receptors (ER) were observed in two cultivars of rice. Data suggested different bacteria could trigger expression of different ER in rice genotypes [252].
One of the useful tools used by microbes is the production of ACC deaminase enzyme to lessen the adverse effect of environmental condition on plants. Infection of legume plant with Rhizobia resulted in increased production of ethylene; increased ethylene level inhibited the nodule formation on root. Some Rhizobial strain produces “rhizobitoxine” a phytotoxin that inhibits the production of enzyme ACC synthase. ACC is precursor for ethylene production [253]. R. leguminosarum RP2 strain isolated from nodules of pea synthesized detectable amount of ACC deaminase [254]. Psychrotolerant endophytic bacteria Arthrobacter, Bacillus, Methylobacterium, Providencia, Pseudomonas, and Flavobacterium isolated from T. aestivum show ACC deaminase activity [14]. During the process of nodulation in the roots of plant, the hormone ethylene and its precursor 1aminocyclopropane-1- carboxylate (ACC) were observed to get accumulated in the root [30]. Heavy metal toxicity in soil decreased the fertility of soil and is detrimental for plants growth. The heavy metals are rapidly absorb by the roots of plants and translocate to shoot and leave leading to reduced growth of plant and finally the death of plant [255].
The ACC deaminase enzyme secreted by microbes lowers the ethylene level. The ethylene accumulation significantly increases during the communication of pathogen with plant. PGP bacteria synthesizes the enzyme ACC deaminase, hence reduces the level of ethylene [256]. The decreased concentration of ethylene in combination with the hormone auxin has beneficial effect on the host plant. R. leguminosarum (RPN5) produces intracellular ACC deaminase and utilizes ACC as a sole source of N [257]. In a study, two ACC deaminase expressing endophytic bacteria Pseudomonas sp. strains, OFT2 and OFT5, were studied as they uphold the growth, photosynthesis, water content in the leaf, and ionic balance of tomato plants. The inoculated OFT5 decreased salt-induced ethylene production by tomato seedlings and also enhanced shoot uptake of P, K, and Mg as well as Mn, Fe, Cu, and Zn [258].
6. CONCLUSION
Over the worldwide, improvements are required regarding the inputs of organics and BNF. Endophytic bacteria have been isolated from a large number of both leguminous and non-leguminous plants and play an integral role in the growth and development of plants and also help in restoration of available nutrient. For the enhancement of delivering fixed N2 to plants, BNF has been proven to be an effective mechanism. The symbiotic association of rhizobia is practically limited to legumes and more interest is focused on N2 fixation in non-legume crops. Over the past few decades, mostly, the research is focused on studying the traits and characteristics of bacterium, and recently, advancement in molecular biology and biochemical analysis opened the new gateway for scientist to study bacterium at its gene level and further provide more details about the process of bacterial endophyte colonization. In the future, development of genetically modified microbes will be a better strategy over transgenic plants for enhancing plant performance and genetic modification of bacterium is better option as modification of higher organism is a complex phenomenon. It is a vital need to develop more effective endophytic strains with longer shelf lives for achieving better sustainable agriculture. More focus should be laid on designing the tools through which identification of genes responsible for endophytic association in non-legumes can be made possible. The creation of artificial symbiosis and endophytic association among N2-fixing microbes and crops of agricultural importance is an important criterion for reducing the usage of chemical fertilizers. Recent advancement in the fields of biotechnology, microbiology, molecular biology, and bioinformatics has unlocked the way to discover novel genes involved in N2 fixation.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. FUNDING
There is no funding to report.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. do Vale Barreto Figueiredo M, do Espírito Santo Mergulhão AC, Sobral JK, de Andrade Lira M, de Araújo AS. Biological nitrogen fixation:Importance, associated diversity, and estimates. In:Arora NK, editor. Plant Microbe Symbiosis:Fundamentals and Advances. New Delhi:Springer;2013. 267-89. [CrossRef]
2. Rondon MA, Lehmann J, Ramírez J, Hurtado M. Biological nitrogen fixation by common beans (
3. Kaur T, Devi R, Kour D, Yadav A, Yadav AN, Dikilitas M,
4. Kahindi JH, Karanja NK. Essentials of nitrogen fixation biotechnology. Biotechnology 2009;8:54.
5. Rockström J, Steffen W, Noone K, Persson Å, Chapin F 3rd, Lambin E,
6. Bhattacharjee RB, Singh A, Mukhopadhyay S. Use of nitrogen-fixing bacteria as biofertiliser for non-legumes:Prospects and challenges. Appl Microbiol Biotechnol 2008;80:199-209. [CrossRef]
7. Cocking EC. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 2003;252:169-75. [CrossRef]
8. Hardoim PR, Van Overbeek LS, Berg G, PirttiläAM, Compant S, Campisano A,
9. Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microb Interact 2006;19:827-37. [CrossRef]
10. Yadav AN, Kour D, Kaur T, Devi R, Yadav A. Endophytic fungal communities and their biotechnological implications for agro-environmental sustainability. Folia Microbiol 2022;67:203-32. [CrossRef]
11. Rana KL, Kour D, Sheikh I, Yadav N, Yadav AN, Kumar V,
12. Cavalcante VA, Dobereiner J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988;108:23-31. [CrossRef]
13. Sevilla M, Burris RH, Gunapala N, Kennedy C. Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with
14. Verma P, Yadav AN, Khannam KS, Panjiar N, Kumar S, Saxena AK,
15. Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. Root colonization and systemic spreading of
16. Sw?drzy?ska D, Sawicka A. Effect of inoculation with
17. Zhao L, Xu Y, Lai X. Antagonistic endophytic bacteria associated with nodules of soybean (
18. Castro S, Permigiani M, Vinocur M, Fabra A. Nodulation in peanut (
19. Yadav AN, Kumar V, Dhaliwal HS, Prasad R, Saxena AK. Microbiome in crops:Diversity, distribution, and potential role in crop improvement. In:Prasad R, Gill SS, Tuteja N, editors. Crop Improvement Through Microbial Biotechnology. San Diego:Elsevier;2018. 305-32. [CrossRef]
20. Estrada P, Mavingui P, Cournoyer B, Fontaine F, Balandreau J, Caballero-Mellado J. A N2-fixing endophytic
21. Kuklinsky?Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani?Kleiner AA, Azevedo JL. Isolation and characterization of soybean?associated bacteria and their potential for plant growth promotion. Environ Microbiol 2004;6:1244-51. [CrossRef]
22. Perin L, Martínez-Aguilar L, Paredes-Valdez G, Baldani J, Estrada-De Los Santos P, Reis V,
23. Govindarajan M, Balandreau J, Kwon SW, Weon HY, Lakshminarasimhan C. Effects of the inoculation of
24. Peng G, Yuan Q, Li H, Zhang W, Tan Z.
25. Dakora F, Matiru V, King M, Phillips D. Plant growth promotion in legumes and cereals by lumichrome, a rhizobial signal metabolite. In:Finan TM, O'Brian MR, Layzell DB, Vessey K, Newton WE, editors. Nitrogen Fixation:Global Perspectives. Wallingford, UK:CABI Publishing;2002. 321-2.
26. Tian CF, Wang ET, Wu LJ, Han TX, Chen WF, Gu CT,
27. Rami?rez-Bahena MH, Garci?a-Fraile P, Peix A, Valverde A, Rivas RL, Igual JM,
28. Glick BR. Resource acquisition. In:Glick BR, editor. Beneficial Plant-bacterial Interactions. Cham:Springer International Publishing;2015. 29-63. [CrossRef]
29. Zhang X, Sun L, Ma X, Sui XH, Jiang R.
30. Subramanian P, Kim K, Krishnamoorthy R, Sundaram S, Sa T. Endophytic bacteria improve nodule function and plant nitrogen in soybean on co-inoculation with
31. Yan J, Yan H, Liu LX, Chen WF, Zhang XX, Verástegui-Valdés MM,
32. Govindarajan M, Kwon SW, Weon HY. Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph
33. Montañez A, Abreu C, Gill PR, Hardarson G, Sicardi M. Biological nitrogen fixation in maize (
34. Mbai F, Magiri E, Matiru V, Nganga J, Nyambati V. Isolation and characterization of bacterial root endophytes with potential to enhance plant growth from Kenyan Basmati rice. Am Int J Contemp Res 2013;3:25-40.
35. Hirsch AM. Brief History of the Discovery of Nitrogen Fixing Organisms;2009. Available from:http://wwwmcdbuclaedu/research/hirsch/imagesb/historydiscoveryn2fixing organisms pdf [Last accessed on 2022 Sep 29].
36. Gopalakrishnan S, Sathya A, Vijayabharathi R, Varshney RK, Gowda CL, Krishnamurthy L. Plant growth promoting rhizobia:Challenges and opportunities. 3 Biotech 2015;5:355-77. [CrossRef]
37. Gourion B, Berrabah F, Ratet P, Stacey G.
38. Youssef M, Eissa M. Biofertilizers and their role in management of plant parasitic nematodes. A review. E3 J Biotechnol Pharm Res 2014;5:1-6.
39. Zhang L, Shi X, Si M, Li C, Zhu L, Zhao L,
40. Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. The Prokaryotes:
41. Soe KM, Bhromsiri A, Karladee D, Yamakawa T. Effects of endophytic actinomycetes and
42. Lechevalier MP. Taxonomy of the genus
43. Coombs JT, Franco CM. Visualization of an endophytic
44. Benson DR, Silvester W. Biology of
45. Nouioui I, Ghodhbane-Gtari F, Rohde M, Klenk HP, Gtari M.
46. Tian X, Cao L, Tan H, Han W, Chen M, Liu Y,
47. Ikeda S, Okubo T, Kaneko T, Inaba S, Maekawa T, Eda S,
48. Shah S, Shah R, Xu H, Aryal U. Biofertilizers:An alternative source of nutrients for sustainable production of tree crops. J Sustain Agric 2007;29:85-95. [CrossRef]
49. Bohlool B, Ladha J, Garrity D, George T. Biological nitrogen fixation for sustainable agriculture:A perspective. Plant Soil 1992;141:1-11. [CrossRef]
50. Ladha J, Barraquio W, Watanabe I. Immunological techniques to identify
51. Rodrigues EP, Rodrigues LS, de Oliveira AL, Baldani VL, dos Santos Teixeira KR, Urquiaga S,
52. de Bellone SC, Bellone C. Presence of endophytic diazotrophs in sugarcane juice. World J Microbiol Biotechnol 2006;22:1065-8. [CrossRef]
53. Reis VM, Baldani JI, Baldani VL, Dobereiner J. Biological dinitrogen fixation in gramineae and palm trees. Crit Rev Plant Sci 2000;19:227-47. [CrossRef]
54. Dalla Santa OR, Hernández RF, Alvarez GL, Ronzelli P Jr., Soccol CR.
55. Cohen AC, Travaglia CN, Bottini R, Piccoli PN. Participation of abscisic acid and gibberellins produced by endophytic
56. Yasuda M, Isawa T, Shinozaki S, Minamisawa K, Nakashita H. Effects of colonization of a bacterial endophyte,
57. De Salamone IE, Di Salvo LP, Ortega JS, Sorte PM, Urquiaga S, Teixeira KR. Field response of rice paddy crop to
58. Trabelsi D, Mhamdi R. Microbial inoculants and their impact on soil microbial communities:A review. Biomed Res Int 2013;2013:863240. [CrossRef]
59. Matsumura EE, Secco VA, Moreira RS, dos Santos OJ, Hungria M, de Oliveira AL. Composition and activity of endophytic bacterial communities in field-grown maize plants inoculated with
60. Hahn L, SáEL, Filho BD, Machado RG, Damasceno RG, Giongo A. Rhizobial inoculation, alone or coinoculated with
61. Cassán F, Díaz-Zorita M. The Contribution of the use of
62. Fujita M, Kusajima M, Okumura Y, Nakajima M, Minamisawa K, Nakashita H. Effects of colonization of a bacterial endophyte,
63. Kusajima M, Shima S, Fujita M, Minamisawa K, Che FS, Yamakawa H,
64. Mohapatra D, Singh N, Rath S. Prospects and application of
65. Saikia SP, Jain V. Biological nitrogen fixation with non-legumes:An achievable target or a dogma?Curr Sci 2007;92:317-22.
66. Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T,
67. Dos Reis FB Jr., Reis VM, Urquiaga S, Döbereiner J. Influence of nitrogen fertilisation on the population of diazotrophic bacteria
68. Catalán AI, Ferreira F, Gill PR, Batista S. Production of polyhydroxyalkanoates by
69. Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB,
70. Chubatsu LS, Monteiro RA, de Souza EM, de Oliveira MA, Yates MG, Wassem R,
71. Canellas LP, Balmori DM, Médici LO, Aguiar NO, Campostrini E, Rosa RC,
72. Straub D, Yang H, Liu Y, Tsap T, Ludewig U. Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte
73. Hoseinzade H, Ardakani M, Shahdi A, Rahmani HA, Noormohammadi G, Miransari M. Rice (
74. Pankievicz V, Camilios-Neto D, Bonato P, Balsanelli E, Tadra-Sfeir M, Faoro H,
75. Maheshwari DK, Annapurna K. Endophytes:Crop Productivity and Protection. Berlin, Germany:Springer;2017. [CrossRef]
76. Yamada Y, Hoshino KI, Ishikawa T. The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA:The elevation of the subgenus
77. Stephan M, Oliveira M, Teixeira K, Martinez-Drets G, Döbereiner J. Physiology and dinitrogen fixation of
78. Döbereiner J. Isolation and identification of root associated diazotrophs. In:Skinner FA, Boddey RM, Fendrik I, editors. Nitrogen Fixation with Non-legumes. Dordrecht:Springer;1989. 103-8. [CrossRef]
79. Muthukumarasamy R, Revathi G, Lakshminarasimhan C. Influence of N fertilisation on the isolation of
80. James E, Reis V, Olivares F, Baldani J, Döbereiner J. Infection of sugar cane by the nitrogen-fixing bacterium
81. Dong Z, Heydrich M, Bernard K, McCully M. Further evidence that the N (inf2)-fixing endophytic bacterium from the intercellular spaces of sugarcane stems is
82. Fuentes-Ramirez LE, Jimenez-Salgado T, Abarca-Ocampo I, Caballero-Mellado J.
83. Muthukumarasamy R, Revathi G, Seshadri S, Lakshminarasimhan C.
84. Munoz-Rojas J, Fuentes-Ramírez LE, Caballero-Mellado J. Antagonism among
85. Piñón D, Casas M, Blanch Ma, Fontaniella B, Blanco Y, Vicente C,
86. Blanco Y, Arroyo M, Legaz M, Vicente C. Isolation from
87. Govindarajan M, Balandreau J, Muthukumarasamy R, Revathi G, Lakshminarasimhan C. Improved yield of micropropagated sugarcane following inoculation by endophytic
88. Boddey R, Urquiaga S, Reis V, Döbereiner J. Biological nitrogen fixation associated with sugar cane. Plant Soil 1991;137:111-7. [CrossRef]
89. Baldani J, Caruso L, Baldani VL, Goi SR, Döbereiner J. Recent advances in BNF with non-legume plants. Soil Biol Biochem 1997;29:911-22. [CrossRef]
90. Cojho EH, Reis VM, Schenberg AC, Döbereiner J. Interactions of
91. Alvarez B, Martínez-Drets G. Metabolic characterization of
92. Muthukumarasamy R, Cleenwerck I, Revathi G, Vadivelu M, Janssens D, Hoste B,
93. Rouws L, Meneses C, Guedes H, Vidal M, Baldani J, Schwab S. Monitoring the colonization of sugarcane and rice plants by the endophytic diazotrophic bacterium
94. Alquéres S, Meneses C, Rouws L, Rothballer M, Baldani I, Schmid M,
95. de Souza AR, De Souza S, De Oliveira M, Ferraz T, Figueiredo F, Da Silva N,
96. Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L. Genus?wide acid tolerance accounts for the biogeographical distribution of soil
97. Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P,
98. Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R,
99. Pérez?Pantoja D, Donoso R, AgullóL, Córdova M, Seeger M, Pieper DH,
100. Aizawa T, Ve NB, Nakajima M, Sunairi M.
101. Baldani V, Oliveira E, Balota E, Baldani J, Kirchhof G, Döbereiner J,
102. Sessitsch A, Coenye T, Sturz A, Vandamme P, Barka EA, Salles J,
103. Coenye T, Laevens S, Willems A, Ohlén M, Hannant W, Govan J,
104. Frommel MI, Nowak J, Lazarovits G. Growth enhancement and developmental modifications of
105. Barka EA, Belarbi A, Hachet C, Nowak J, Audran JC. Enhancement of
106. Nowak J, Asiedu S, Lazarovits G, Pillay V, Stewart A, Smith C,
107. Pillay V, Nowak J. Inoculum density, temperature, and genotype effects on
108. Barka EA, Gognies S, Nowak J, Audran JC, Belarbi A. Inhibitory effect of endophyte bacteria on
109. Luvizotto DM, Marcon J, Andreote FD, Dini-Andreote F, Neves AA, Araújo WL,
110. Compant S, Kaplan H, Sessitsch A, Nowak J, Ait Barka E, Clément C. Endophytic colonization of
111. Mitter B, Petric A, Shin MW, Chain PS, Hauberg-Lotte L, Reinhold-Hurek B,
112. Lowman S, Kim-Dura S, Mei C, Nowak J. Strategies for enhancement of switchgrass (
113. Baldani VD, Baldani JI, Döbereiner J. Inoculation of rice plants with the endophytic diazotrophs
114. Singh RK, Mishra RP, Jaiswal HK, Kumar V, Pandey SP, Rao SB,
115. Procópio R, Araújo W, Maccheroni W Jr., Azevedo J. Characterization of an endophytic bacterial community associated with
116. Van VT, Berge O, Ke SN, Balandreau J, Heulin T. Repeated beneficial effects of rice inoculation with a strain of
117. Mishra PK, Mishra S, Selvakumar G, Bisht J, Kundu S, Gupta HS. Coinoculation of
118. Wulff E, Mguni C, Mansfeld?Giese K, Fels J, Lübeck M, Hockenhull J. Biochemical and molecular characterization of
119. Rajendran L, Karthikeyan G, Raguchander T, Samiyappan R. Cloning and sequencing of novel endophytic
120. Chen Y, Gao X, Chen Y, Qin H, Huang L, Han Q. Inhibitory efficacy of endophytic
121. Wang H, Wen K, Zhao X, Wang X, Li A, Hong H. The inhibitory activity of endophytic
122. Liu M, Luo K, Wang Y, Zeng A, Zhou X, Luo F,
123. Khalifa AY, Almalki MA. Isolation and characterization of an endophytic bacterium,
124. Munjal V, Nadakkakath AV, Sheoran N, Kundu A, Venugopal V, Subaharan K,
125. Gupta C, Kumar B, Dubey R, Maheshwari D. Chitinase-mediated destructive antagonistic potential of
126. Chung BS, Aslam Z, Kim SW, Kim GG, Kang HS, Ahn JW,
127. Weyens N, Truyens S, Dupae J, Newman L, Taghavi S, Van Der Lelie D,
128. Aeron A, Dubey R, Maheshwari D, Pandey P, Bajpai VK, Kang SC. Multifarious activity of bioformulated
129. Pandey PK, Yadav SK, Singh A, Sarma BK, Mishra A, Singh HB. Cross?species alleviation of biotic and abiotic stresses by the endophyte
130. Gupta G, Panwar J, Jha PN. Natural occurrence of
131. Sun K, Liu J, Gao Y, Jin L, Gu Y, Wang W. Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium
132. Miller S, Mark G, Franks A, O'Gara F.
133. Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Ann Rev Microbiol 2009;63:541-56. [CrossRef]
134. Zhao LF, Xu YJ, Ma ZQ, Deng ZS, Shan CJ, Wei GH. Colonization and plant growth promoting characterization of endophytic
135. Tariq T, Shafiqe HA, Sultana V, Ehteshamul-Haque S. Management of root rot disease of wheat with endophytic plant growth promoting
136. Pham VT, Rediers H, Ghequire MG, Nguyen HH, De Mot R, Vanderleyden J,
137. Sandhya V, Shrivastava M, Ali SZ, Prasad VS. Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ Agric Sci 2017;43:22-34. [CrossRef]
138. Ma Y, Rajkumar M, Moreno A, Zhang C, Freitas H. Serpentine endophytic bacterium
139. Verma SK, Kingsley K, Irizarry I, Bergen M, Kharwar RN, White JF. Seed vectored endophytic bacteria modulate development of rice seedlings. J Appl Microbiol 2017;122:1680-91. [CrossRef]
140. Jha P, Kumar A. Characterization of novel plant growth promoting endophytic bacterium
141. Ladha JK, Reddy P. The Quest for Nitrogen Fixation in Rice. Los Baños, Philippines:International Rice Research Institute;2000.
142. Egener T, Martin DE, Sarkar A, Reinhold-Hurek B. Role of a ferredoxin gene cotranscribed with the nif HDK operon in N2 fixation and nitrogenase “switch-off“of
143. Malik K, Bilal R, Mehnaz S, Rasul G, Mirza M, Ali S. Association of nitrogen-fixing, plant-growth-promoting rhizobacteria (PGPR) with kallar grass and rice. In:Ladha JK, de Bruijn FJ, Malik KA, editors. Opportunities for Biological Nitrogen Fixation in Rice and other Non-legumes. Dordrecht:Springer;1997. 37-44. [CrossRef]
144. Ahemad M, Khan MS. Effect of insecticide-tolerant and plant growth-promoting
145. Ramos PL, Van Trappen S, Thompson FL, Rocha RC, Barbosa HR, De Vos P,
146. Dutta D, Gachhui R. Novel nitrogen-fixing
147. Gillis M, Kersters K, Hoste B, Janssens D, Kroppenstedt RM, Stephan MP,
148. Caballero-Mellado J, Martínez-Aguilar L, Paredes-Valdez G, Santos PE-dl.
149. Zhu B, Zhou Q, Lin L, Hu C, Shen P, Yang L,
150. Lin L, Wei C, Chen M, Wang H, Li Y, Li Y,
151. Loganathan P, Nair S. Crop-specific endophytic colonization by a novel, salt-tolerant, N 2-fixing and phosphate-solubilizing
152. Tripathi AK, Verma SC, Chowdhury SP, Lebuhn M, Gattinger A, Schloter M.
153. Zhang GX, Peng GX, Wang ET, Yan H, Yuan QH, Zhang W,
154. Chaudhary HJ, Peng G, Hu M, He Y, Yang L, Luo Y,
155. Hardoim PR, Nazir R, Sessitsch A, ElhottováD, Korenblum E, van Overbeek LS,
156. Dreyfus B, Garcia JL, Gillis M. Characterization of
157. Glazebrook J, Walker GC. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by
158. Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA.
159. El?Hamdaoui A, Redondo?Nieto M, Rivilla R, Bonilla I, Bolanos L. Effects of boron and calcium nutrition on the establishment of the
160. Wang F, Wang ET, Wu LJ, Sui XH, Li Jr Y, Chen WX.
161. Sevilla M, Meletzus D, Teixeira K, Lee S, Nutakki A, Baldani I,
162. Black M, Moolhuijzen P, Chapman B, Barrero R, Howieson J, Hungria M,
163. Maillet F, Poinsot V, AndréO, Puech-Pagès V, Haouy A, Gueunier M,
164. Peng G, Wang H, Zhang G, Hou W, Liu Y, Wang ET,
165. Chen WM, James EK, Coenye T, Chou JH, Barrios E, De Faria SM,
166. Peng G, Zhang W, Luo H, Xie H, Lai W, Tan Z.
167. Gao JL, Lv FY, Wang XM, Yuan M, Li JW, Wu QY,
168. Dutta D, Gachhui R. Nitrogen-fixing and cellulose-producing
169. Valverde A, Velazquez E, Gutierrez C, Cervantes E, Ventosa A, Igual JM.
170. Xing K, Bian GK, Qin S, Klenk HP, Yuan B, Zhang YJ,
171. Román-Ponce B, Wang D, Vásquez-Murrieta MS, Chen WF, Estrada-de los Santos P, Sui XH,
172. Behera P, Ramana VV, Maharana B, Joseph N, Vaishampayan P, Singh NK,
173. Rozahon M, Ismayil N, Hamood B, Erkin R, Abdurahman M, Mamtimin H,
174. Guo GN, Zhou X, Zhao R, Chen XY, Chen ZL, Li XD,
175. Gao JL, Lv FY, Wang XM, Qiu TL, Yuan M, Li JW,
176. Madhaiyan M, Jin TY, Roy JJ, Kim SJ, Weon HY, Kwon SW,
177. Kesari V, Ramesh AM, Rangan L.
178. Lin DX, Wang ET, Tang H, Han TX, He YR, Guan SH,
179. Gisin J, Müller A, Pfänder Y, Leimkühler S, Narberhaus F, Masepohl B. A
180. Bhattacharyya P, Jha D. Plant growth-promoting rhizobacteria (PGPR):Emergence in agriculture. World J Microbiol Biotechnol 2012;28:1327-50. [CrossRef]
181. Loiret F, Ortega E, Kleiner D, Ortega?Rodés P, Rodes R, Dong Z. A putative new endophytic nitrogen?fixing bacterium
182. Defez R, Andreozzi A, Bianco C. The overproduction of indole-3-acetic acid (IAA) in endophytes upregulates nitrogen fixation in both bacterial cultures and inoculated rice plants. Microb Ecol 2017;74:441-52. [CrossRef]
183. Santi C, Bogusz D, Franche C. Biological nitrogen fixation in non-legume plants. Ann Bot 2013;111:743-67. [CrossRef]
184. Pan B, Vessey JK. Response of the endophytic diazotroph
185. Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and genetic analysis of the nifUSVWZM cluster from
186. Arnold W, Rump A, Klipp W, Priefer UB, Pühler A. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of
187. Merrick M, Edwards R. Nitrogen control in bacteria. Microbiol Mol Biol Rev 1995;59:604-22. [CrossRef]
188. Ureta A, Nordlund S. Evidence for conformational protection of nitrogenase against oxygen in
189. Vermeiren H, Willems A, Schoofs G, De Mot R, Keijers V, Hai W,
190. Egener T, Hurek T, Reinhold-Hurek B. Use of green fluorescent protein to detect expression of nlf genes of
191. Reis VM, Döbereiner J. Effect of high sugar concentration on nitrogenase activity of
192. Madhaiyan M, Poonguzhali S, Hari K, Saravanan V, Sa T. Influence of pesticides on the growth rate and plant-growth promoting traits of
193. Kennedy I. Integration of nitrogenase in cellular metabolism. In:Hardy RW, Bottomley R, Burns RC, editors. A Treatise of Dinitrogen Fixation:Inorganic and Physical Chemistry and Biochemistry. New York:Wiley;1979. 653-90.
194. Coppola D, Giordano D, Tinajero-Trejo M, Di Prisco G, Ascenzi P, Poole RK,
195. Glick BR. Plant growth-promoting bacteria:Mechanisms and applications. Scientifica 2012;2012:963401. [CrossRef]
196. Ronson CW, Primrose SB. Carbohydrate metabolism in
197. Thaweenut N, Hachisuka Y, Ando S, Yanagisawa S, Yoneyama T. Two seasons'study on nifH gene expression and nitrogen fixation by diazotrophic endophytes in sugarcane (
198. Roncato-Maccari LD, Ramos HJ, Pedrosa FO, Alquini Y, Chubatsu LS, Yates MG,
199. Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Cura JA. Plant growth promoting properties of a strain of
200. Hongrittipun P, Youpensuk S, Rerkasem B. Screening of Nitrogen Fixing Endophytic Bacteria in
201. Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003;255:571-86. [CrossRef]
202. Rana KL, Kour D, Sheikh I, Dhiman A, Yadav N, Yadav AN,
203. Rana KL, Kour D, Kaur T, Devi R, Negi C, Yadav AN,
204. Yadav AN, Singh J, Rastegari AA, Yadav N. Plant Microbiomes for Sustainable Agriculture. Cham:Springer;2020. [CrossRef]
205. Kour D, Rana KL, Kaur T, Yadav N, Yadav AN, Kumar M,
206. McAfee J. Potassium, a Key Nutrient for Plant Growth. Department of Soil and Crop Sciences;2008. Available from:http://jimmcafee tamu edu/files/potassium [Last accessed on 2022 Sep 29].
207. Rodr??guez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 1999;17:319-39. [CrossRef]
208. Kumar M, Tomar RS, Lade H, Paul D. Methylotrophic bacteria in sustainable agriculture. World J Microbiol Biotechnol 2016;32:120. [CrossRef]
209. Verma SC, Ladha JK, Tripathi AK. Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J Biotechnol 2001;91:127-41. [CrossRef]
210. Alori ET, Glick BR, Babalola OO. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol 2017;8:971. [CrossRef]
211. Sashidhar B, Podile AR. Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J Appl Microbiol 2010;109:1-12. [CrossRef]
212. Kapri A, Tewari L. Phosphate solubilization potential and phosphatase activity of rhizospheric
213. Martínez-Viveros O, Jorquera M, Crowley D, Gajardo G, Mora M. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr 2010;10:293-319. [CrossRef]
214. McGrath JW, Wisdom GB, McMullan G, Larkin MJ, Quinn JP. The purification and properties of phosphonoacetate hydrolase, a novel carbon?phosphorus bond?cleavage enzyme from Pseudomonas fluorescens 23F. Eur J Biochem 1995;234(1):225-30. [CrossRef]
215. Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion:A review. Ann Microbiol 2010;60:579-98. [CrossRef]
216. Otieno N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ,
217. You C, Zhou F. Non-nodular endorhizospheric nitrogen fixation in wetland rice. Can J Microbiol 1989;35:403-8. [CrossRef]
218. Oliveira Ad, Urquiaga S, Döbereiner J, Baldani J. The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 2002;242:205-15. [CrossRef]
219. Saubidet MI, Fatta N, Barneix AJ. The effect of inoculation with
220. Ji SH, Gururani MA, Chun SC. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol Res 2014;169:83-98. [CrossRef]
221. Bai Y, Zhou X, Smith DL. Enhanced soybean plant growth resulting from coinoculation of
222. James EK, Gyaneshwar P, Barraquio WL, Mathan N, Ladha JK. Endophytic diazotrophs associated with rice. In:Ladha JK, Reddy PM, editors. The Quest for Nitrogen Fixation in Rice. Makati City, Philippines:International Rice Research Institute;2000. 119-40.
223. Ladha J, Barraquio W, Watanabe I. Isolation and identification of nitrogen-fixing
224. Mirza MS, Ahmad W, Latif F, Haurat J, Bally R, Normand P,
225. Eskin N. Colonization of
226. Suman A, Shrivastava A, Gaur A, Singh P, Singh J, Yadav R. Nitrogen use efficiency of sugarcane in relation to its BNF potential and population of endophytic diazotrophs at different N levels. Plant Growth Regul 2008;54:1-11. [CrossRef]
227. Ladha J, Barraquio W, Revilla L. Isolation of endophytic diazotrophic bacteria from wetland rice. In:Ladha JK, de Bruijn FJ, Malik KA, editors. Opportunities for Biological Nitrogen Fixation in Rice and other Non-legumes. Dordrecht:Springer;1997. 15-24. [CrossRef]
228. Roesch LF, Camargo FA, Bento FM, Triplett EW. Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil 2008;302:91-104. [CrossRef]
229. Gyaneshwar P, James EK, Mathan N, Reddy PM, Reinhold-Hurek B, Ladha JK. Endophytic colonization of rice by a diazotrophic strain of
230. Puri A, Padda KP, Chanway CP. Can a diazotrophic endophyte originally isolated from lodgepole pine colonize an agricultural crop (corn) and promote its growth?Soil Biol Biochem 2015;89:210-6. [CrossRef]
231. Singh D, Rajawat MV, Kaushik R, Prasanna R, Saxena AK. Beneficial role of endophytes in biofortification of Zn in wheat genotypes varying in nutrient use efficiency grown in soils sufficient and deficient in Zn. Plant Soil 2017;416:107-16. [CrossRef]
232. Bayliss W, Starling EH. Croonian lecture:The chemical regulation of the secretory process. Proc R Soc Lond 1904;73:310-22. [CrossRef]
233. Li JH, Wang ET, Chen WF, Chen WX. Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang Province of China. Soil Biol Biochem 2008;40:238-46. [CrossRef]
234. Ambawade M, Pathade G. Production of gibberellic acid by
235. Yoon V, Tian G, Vessey JK, Macfie SM, Dangi OP, Kumer AK,
236. Rodrigues EP, Soares CP, Galvão PG, Imada EL, Simões-Araújo JL, Rouws LF,
237. Bhutani N, Maheshwari R, Negi M, Suneja P. Optimization of IAA production by endophytic
238. Crowley DE. Microbial siderophores in the plant rhizosphere. In:Barton LL, Abadia J, editors. Iron Nutrition in Plants and Rhizospheric Microorganisms. Dordrecht:Springer Netherlands;2006. 169-98. [CrossRef]
239. Bultreys A. Siderotyping, a tool to characterize, classify and identify fluorescent pseudomonads. In:Varma A, Chincholkar SB, editors. Microbial Siderophores. Berlin, Heidelberg:Springer;2007. 67-89. [CrossRef]
240. Loaces I, Ferrando L, Scavino AF. Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb Ecol 2011;61:606-18. [CrossRef]
241. Abbamondi GR, Tommonaro G, Weyens N, Thijs S, Sillen W, Gkorezis P,
242. Kong Z, Deng Z, Glick BR, Wei G, Chou M. A nodule endophytic plant growth-promoting
243. Dolphen R, Thiravetyan P. Reducing arsenic in rice grains by leonardite and arsenic-resistant endophytic bacteria. Chemosphere 2019;223:448-54. [CrossRef]
244. Abeles FB, Morgan P, Saltveit Jr M. Ethylene in Plant Biology. San Diego:Academic Press;1992.
245. Glick BR. The enhancement of plant growth by free-living bacteria. Can J Microbiol 1995;41:109-17. [CrossRef]
246. Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase?containing plant growth?promoting rhizobacteria. Physiol Plant 2003;118:10-5. [CrossRef]
247. Glick BR. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 2005;251:1-7. [CrossRef]
248. Shaharoona B, Arshad M, Zahir ZA, Khalid A. Performance of
249. Nadeem SM, Zahir ZA, Naveed M, Arshad M. Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can J Microbiol 2007;53:1141-9. [CrossRef]
250. Rothballer M, Eckert B, Schmid M, Fekete A, Schloter M, Lehner A,
251. Karthikeyan B, Joe MM, Islam MR, Sa T. ACC deaminase containing diazotrophic endophytic bacteria ameliorate salt stress in
252. Vargas L, de Carvalho TL, Ferreira PC, Baldani VL, Baldani JI, Hemerly AS. Early responses of rice (
253. Nascimento FX, Brígido C, Glick BR, Oliveira S. ACC deaminase genes are conserved among
254. Ahmad E, Khan MS, Zaidi A. ACC deaminase producing
255. Mahanty T, Bhattacharjee S, Goswami M, Bhattacharyya P, Das B, Ghosh A,
256. Yadav AN, Rastegari AA, Yadav N. Microbiomes of Extreme Environments, Volume 2:Biotechnological Applications in Agriculture, Environment and Industry. Vol. 2. Boca Raton, USA:CRC Press, Taylor and Francis Group;2020. [CrossRef]
257. Kumar P, Dubey RC, Maheshwari DK, Bajpai VK. ACC deaminase producing
258. Win KT, Tanaka F, Okazaki K, Ohwaki Y. The ACC deaminase expressing endophyte