1. INTRODUCTION
Endophytes are endosymbionts, generally bacterial or fungal microorganisms colonizing intercellularly or intracellularly a healthy plant material before completing their life cycle without generating signs of illness [1]. Endophytes are widespread, host-dependent cells that have been isolated from all plant species. Endophytes live inside the plants in a symbiotic relationship and use a number of strategies to respond to their environments [2]. Endophytes contain many compounds that stimulate plant growth and help them respond more to the environment to sustain a healthy symbiosis [3]. Endophytic bacteria support their host by developing phytohormones, providing protection from pathogens, and directly interacting with invaders [4,5]. Endophytes influence the growth of host plants and yield by suppressing pathogens, and they may also help in the removal of contamination, solubilize phosphate, and the provision of assimilable nitrogen to plants [6], they share mutualistic and antagonistic interaction with their host plant. Their relationship might be compulsory or optional hosts, and these endophytes utilize numerous ways to adapt progressively to their surroundings [7].
A biocontrol agent uses endophytic bacteria to produce secondary metabolites to protect its host against attack by fungi, insects, or mammals. In addition, because these endosymbionts are often also present in the target organism, they can be utilized to develop biocontrol agents [8,9].
Endophytes are known to provide nutrients to host plants by fixing atmospheric nitrogen and ion solubilization. This eventually improves the immune system of the host and protects from infection by plant pathogens [10,11]. Endophytic populations vary between plant to plant and species to species and certain plant species may have distinct populations of endophytes in various populations. As a result, the occurrence of endophytes is affected by temporal and climatic changes [12]. On the other hand, the presence of bacterial growth is typically deemed to be pollutants on an in vitro crop of plants that must be forbidden and eradicated [13,14]. The existence of endophytes must grab the attention in in vitro culture and micropropagated plants. The technique of plant tissue culture is a valuable approach and source for the recovery of beneficial microorganisms in certain species. Since endophytes live extensively in the plant, it is vital that surface contaminants are distinguished and removed from the plant tissue to recover endophytes effectively. Endophytes have an agronomic interest to enhance plant development and increase nutrition by fixing nitrogen, solubilizing phosphate, or iron chelation. Endophytic bacteria are plant beneficial bacteria that live throughout or portion of their life cycle in the plant tissue without inflicting any harm to their host plant [15]. They can directly benefit host plants with the improvement of plant nutrient uptake, modulating phytohormones associated with stress and improve plant health indirectly by using antibiotics to target pathogens and diseases, secreting lytic enzymes, and inducing plant resistance [16,17]. This work aimed to isolate and characterize the plant growth-promoting potential of the endophytic bacterial population found in the Musa acuminata plant.
2. MATERIALS AND METHODS
2.1. Isolation of Endophytic Bacteria
For the isolation of endophytic bacteria, samples were washed thoroughly under running tap water and then surface sterilized by sequentially dipping them into 70% ethanol for 60 s and 1% sodium hypochlorite for 30 s and then rinse with sterile distilled waters into a laminar airflow chamber [18]. The surface-sterilized samples were then cut of about 0.5 cm without the midrib under an aseptic environment, sterility tests of each sample were performed to assure the removal of surface microorganisms. Each piece of sample was then placed on a basic nutrient agar medium (HiMedia). Four pieces of samples per plate were inoculated at similar distances. The plates were incubated for 24–48 h in an incubator at 30 ± 1°C. Based on morphological characteristics and appearance, isolates were maintained on the NA medium, and then, subsequently samples were performed for Gram staining responses as described by Hans Christian Gram (1884).
2.2. Biochemical Characterization of Bacterial Isolates
The standard approach provided in Bergey’s Manual of Systematic Bacteriology identification of bacterial isolates, based on both morphological (shape, size, colony characteristics, gram staining) and biochemical (Oxidase, Catalase, Citrate, Urease, MR-VP, indole) characteristics Krieg et al. [19].
2.3. Detection of Plant Growth-Promoting Activities
2.3.1. Quantitative estimation of indole-3-acetic acid (IAA)
According to Acun˜a et al. [20], quantitative estimates of IAA were carried out. The test culture in the Luria-Bertani (LB) broth, suspended at 0.1–0.2% L-tryptophan concentration and cultured at 28 ± 2°C for 3–5 days, were used for the assimilation of IAA-producing isolate. At the end of the incubation, the suspension cultures were centrifuged for 10 min at 10,000 rpm, and then, the supernatant collected 2 ml of culture filtrate allowed reactions at 28 ± 2°C for 30 min at 4 ml of Salkowski reagent (1 ml of 0.5 M FeCl3 in 49 ml of 37% perchloric acid). The development of pink color at the end of the incubation showed the existence of IAA. The optical density was measured at 530 nm using ultraviolet (UV)–visible spectrophotometer (UV 1800, Shimadzu). The amount of IAA produced was estimated using a standard curve with the known concentration of pure commercial IAA (HiMedia).
2.3.2. Screening for phosphate solubilization test
The phosphate solubilization activities of the isolates were evaluated in accordance with the method of Watanabe and Olsen [21]. Isolates were grown on the growth medium National Botanical Research Institute (NBRIP), a 10 g glucose, 5 g Ca3 (PO4)2, 5 g MgCl2, 0.25 g MgSO4, 0.2 g KCl, 0.1 g (NH4)2SO4, and 1.5 g agar for 100 ml of distilled water at pH 7. The endophytic bacterial colony was placed with a sterilized inoculation loop at the middle of the NBRIP medium agar plate and incubated for 7 days at 37°C. The phosphate solubilizers were screened based on halo zone development on agar plates. This is because organic acid synthesis in the surrounding medium experiment is carried out in triplicate for each bacteria. Phosphate solubilization was evaluated using the formula of Qureshi et al. [22] based on the halo zones around the colonies and measured by:
SE = Solubilization diameter/growth diameter × 100
2.3.3. Screening for siderophore production
The Chrome Azurol S (CAS) agar medium and the hexadecyltrimethylammonium bromide indicator were used to determine Siderophore formation, as reported by Schwyn and Neilands [23]. The addition of sterilized MM9 salt solution (850 ml) composed of piperazine-N, N-bis 2-ethane sulfonic acid (32.24 g), blue dye (100 ml), a 10% filter sterilized CAS amino acid solution (30 ml), and 20% glucose solution (10 ml). Isolates were inoculated on CAS agar plates and incubated at 28°C for 24–48 h, after incubation results recorded.
2.3.4. Screening for nitrogen fixation ability
Qualitative growth screening on solid N-free medium to assess the capability of the isolate to fix atmospheric nitrogen (1 g K2HPO4, 5 mg FeSO4.7H2O, 1 g CaCO3, 5 mg NaMoO4, and 10 g/l) used as the growth parameters and data were record after 4–10 days of inoculation following Santoyo et al., 2016 [6].
2.3.5. Ammonia production test
The capacity of isolated bacterial endophytic strains to produce ammonia was evaluated after the bacterial strains were grown in peptone water (peptone 10g/l, 5g/l NaCl) as proposed by Cappuccino and Sherman [24]. Freshly developed cultures were inoculated in 10 ml of peptone broth in each tube and incubated for 48–72 h at a temperature of 28 ± 2°C. After incubation adding Nessler’s reagent into each tube, the development of the faint yellow color indicated minimal ammonia production, while the deep yellow color to the brownish indicated the highest ammonia production.
2.3.6. Screening for hydrogen cyanide (HCN) production test
All endophytic bacterial isolates have been tested to produce HCN following the protocol stated by Lorck [25]. The isolates were screened on modified agar plates; the nutrient broth was changed with 4.4 g/l glycine. At the lid of the plate, Whatman filter paper No.1 was soaked in 0.5% picric acid solution with 2% sodium carbonate, was placed, and sealed with parafilm. The plates were incubated at 30°C for 4 days and the color changed from the orange to red of the filter paper indicated for HCN production, Bakker et al. [26].
2.4. Antifungal Activity of Isolated Cultures
Antifungal activity was examined using an overnight growth culture inoculated on both sides of the Petri plate containing the potato dextrose agar media as a single line streaked. Then, the fresh mycelia from Fusarium oxysporum (7693) and Macrophomina phaseolina (6630) obtained from Indian type culture collection, Delhi, were spot inoculated at the middle of the plates and incubated at 28°C for 6–7 days. The development of mycelium in the direction of bacterial colonies is taken into account to measure the antifungal activity [27,28]. The percentage of inhibition growth was recorded using the following formula:
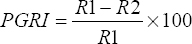 |
PGRI: Percentage of radial growth inhibition, R1: Radius of fungus, and R2: Radius of bacterial colony.
2.5. Molecular Identification of the Isolates
Bacterial isolates for DNA extraction and polymerase chain reaction (PCR) amplification were cultured in 100 ml of Erlenmeyer flask with 50 ml of LB medium (HiMedia) for overnight incubation on shaking incubator with 120 rpm at 37 ± 1°C. Centrifugations were used to collect the bacterial mass. DNA has been isolated from cultured bacteria using a DNA extraction kit (HiPurA™ MB505) as per the specifications of the manufacturer. Sequences of 16S rRNA gene sequences have been utilized for the molecular identification of isolated bacterial strains. The 16S rRNA gene was amplified with universal primers 10F (5`AGTTTGATCCTGGCTCAG3`) and 800R (3`TACCAGGGTATCTAATCC5`) [29]. Post-sequencing the isolate was further checked for homolog’s using BLAST tool available at https://blast.ncbi.nlm.nih.gov and phylogenetic tree analysis was done using MSA file generated through using Clustal W web tool available at https://www.genome.jp/tools-bin/clustalw and MEGAX software package.
2.6. Assessment of Lytic Enzyme Production
Screening of enzyme activity was performed using various enzyme assays. The carboxymethyl cellulose plate tests were done using 1% Congo red solution and NaCl to identify colonies exhibiting the area of inhibition [30]. A starch hydrolysis test was used to assess the amylolytic activity of the isolates, isolates are streaked onto the starch agar plates and incubated at 37 ± 2°C for 24–48 h, following the incubation, 1% iodine solution is flooded over a Petri plate, development of a clear zone around the colonies indicates the hydrolysis of starch [31]. The isolates have been single streaked to the middle of the plate for protease test confirmation using skimmed milk agar medium and are incubated at 37 ± 2°C during 24–48 h after incubation, formation of a clearing zone showing positive results [32]. The clear zone surrounding the inocula with tributyrin agar was discovered by streaking a single line followed by the incubation for 24–48 h at 37 ± 2°C for the detection of lipolytic activity [33]. The isolated strains were then inoculated using a 3% gelatin with nutrient agar for the determination of gelatinase activity followed by an incubation of 24–48 h at 37 ± 2°C. Isolates were maintained at 4°C for 15 min for gelatin liquefaction.
3. RESULTS
In this study, different parts of healthy M. acuminata plant samples including leaves, shoots, and root were collected from the various agricultural regions in Madhya Pradesh, India. Before isolation of microbes from plant, the epiphytic hosts were removed using surface sterilization method. Out of all the isolated cultures, 19 bacterial cultures were selected on the basis of biochemical tests such as catalase, oxidase, citrate, and urease based on their colony morphotypes, Gram staining mentioned in Table 1 and Figure 1. After the biochemical characterization test, the top five isolates were selected for 16S rRNA sequencing and phylogenetic analysis shown in Figures 3-8. The isolates were identified as Bacillus cereus, Enterobacter cloacae, and Enterobacter hormaechei and submitted to NCBI GenBank server.
Table 1: Morphological and biochemical characterization of isolated endophytic bacterial samples of Musa acuminata.
| Isolates | Morphological identification | Biochemical identification | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | Color | Shape | Gram | Motility | Catalase | Indole | Oxidase | Urease | Hydrogen sulfide | Citrate | |
| EMS 1 | Large | Yellow | Irregular | + | + | + | + | + | + | + | + |
| EMS 2 | Large | Yellow | Irregular | + | + | + | + | + | + | + | + |
| EMS 3 | Medium | White | Circular | − | + | + | + | + | + | + | + |
| EMS 4 | Large | Yellow | Irregular | + | − | + | + | + | + | + | + |
| EMS 5 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 6 | Large | Yellow | Irregular | + | − | + | + | + | + | + | + |
| EMS 7 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 8 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 9 | Medium | Brown | Circular | − | − | + | + | + | + | + | + |
| EMS 10 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 11 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 12 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 13 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 14 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 15 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 16 | Medium | White | Circular | − | − | + | + | + | + | + | + |
| EMS 18 | Medium | Brown | Circular | − | − | + | + | + | + | + | + |
| EMS 19 | Medium | White | Circular | − | − | + | + | + | + | + | + |
+: Present, −: Absent.
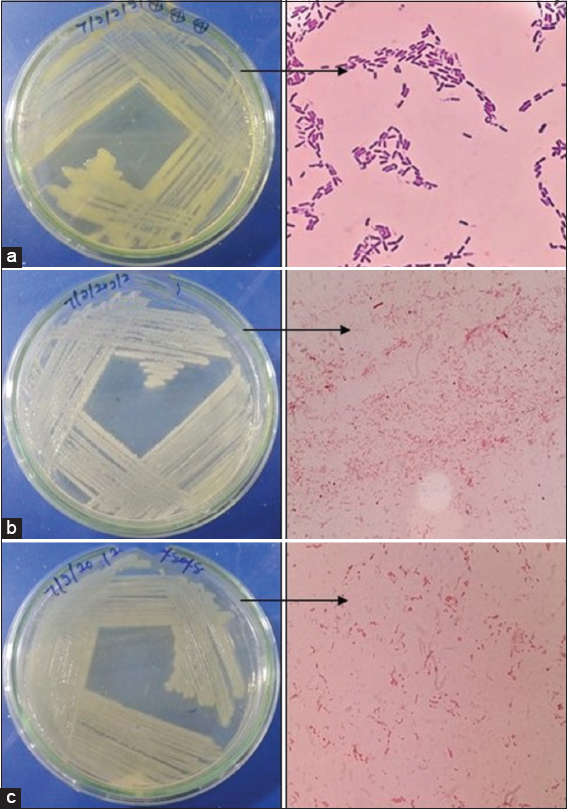 | Figure 1: Pure culture of endophytic bacterial isolates with morphological structure (a) Bacillus cereus (EMS1 and EMS4), (b) Enterobacter cloacae (EMS13, EMS14, and EMS16), (c) Enterobacter hormaechei (EMS18). [Click here to view] |
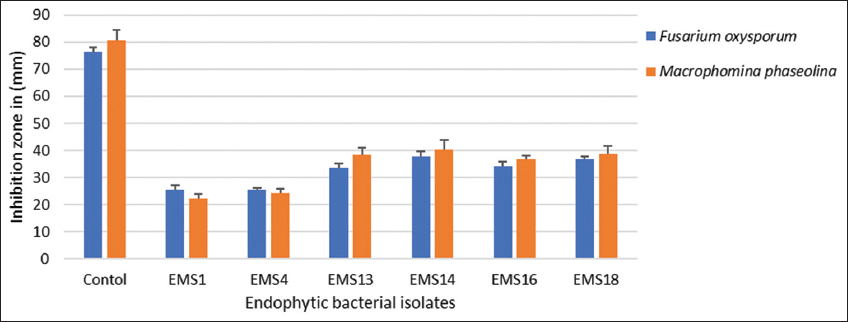 | Figure 2: Antagonistic activity by bacterial strains on fungal growth of Fusarium oxysporum and Macrophomina phaseolina. Plot mean with standard deviation error bars indicates significant differences (P<0.05). [Click here to view] |
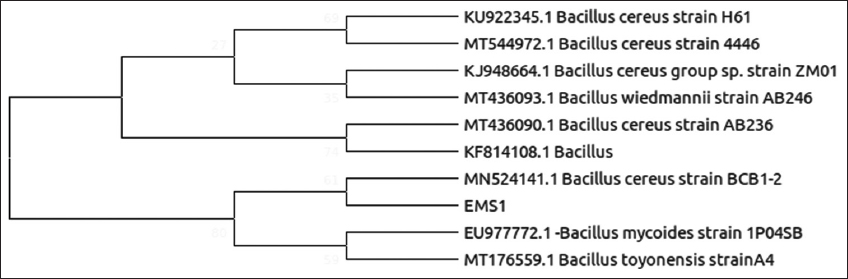 | Figure 3: Phylogenetic relationship of isolate EMS1 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
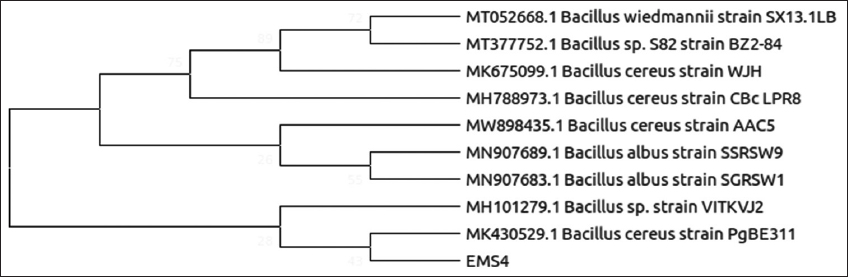 | Figure 4: Phylogenetic relationship of isolate EMS4 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
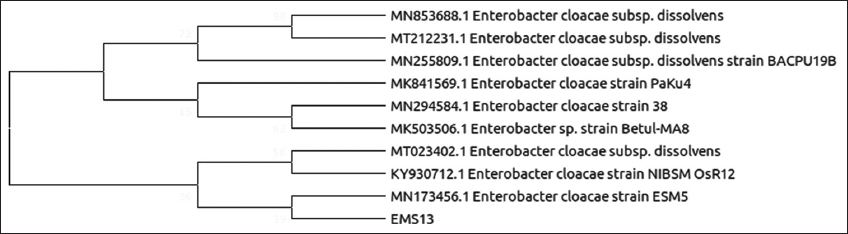 | Figure 5: Phylogenetic relationship of isolate EMS13 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
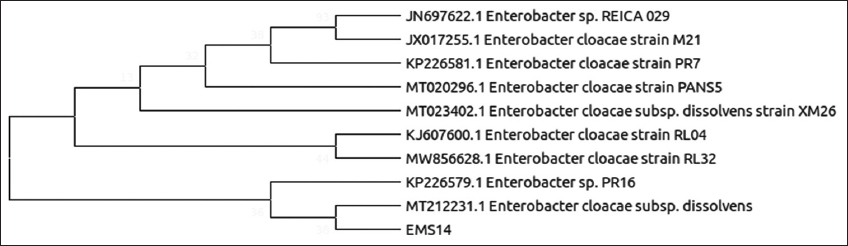 | Figure 6: Phylogenetic relationship of isolate EMS14 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
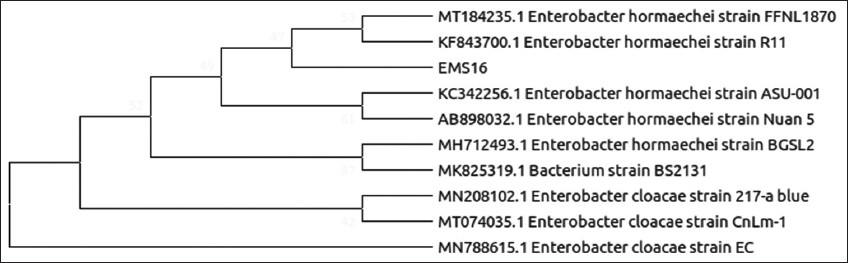 | Figure 7: Phylogenetic relationship of isolate EMS16 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
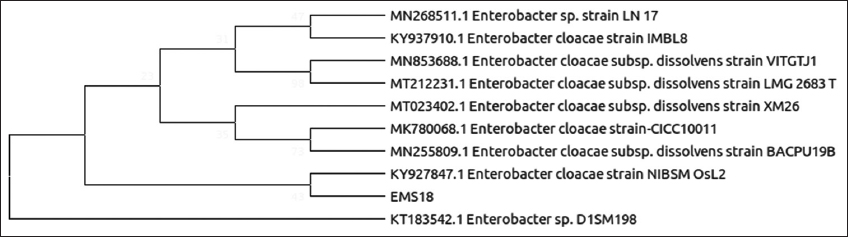 | Figure 8: Phylogenetic relationship of isolate EMS18 with the analyzed sequences by maximum likelihood method based on 16S rRNA gene sequences. [Click here to view] |
3.1. Screening of Endophytic Bacteria for Plant Growth-Promoting Properties
Endophytic bacterial isolates have been tested for their ability to solubilize phosphate, production of siderophore, HCN, IAA, and ammonia production [Table 2]. All 19 isolates were able to produce both ammonia and IAA. The isolates showed growth on N-free and suggesting their ability to fix nitrogen. After incubation observed, the formation of the visible halo zone on the NBRIP medium plate of phosphate solubilization activity and the development of an orange halo zone in a CAS agar plate indicated the presence of siderophore producing activity in all isolated strains. All isolates exhibiting negative results in the HCN production assay.
Table 2: Plant growth-promoting potential of isolated bacterial strains.
| Sample number | Strain | IAA production test (µg ml−1) | Phosphate solubilization (µg ml−1) | Nitrogen fixation test | Siderophore production test | HCN production test | Ammonia production test |
|---|---|---|---|---|---|---|---|
| EMS 1 | Bacillus cereus | + | + | + | + | + | + |
| EMS 4 | Bacillus cereus | + | + | + | + | + | + |
| EMS 13 | Enterobacter cloacae | + | + | + | + | − | + |
| EMS 14 | Enterobacter cloacae | + | + | + | + | − | − |
| EMS 16 | Enterobacter hormaechei | + | + | + | + | + | + |
| EMS 18 | Enterobacter cloacae | + | + | + | + | − | + |
+: Present, −: Absent, HCN: Hydrogen cyanide, IAA: Indole-3-acetic acid.
3.2. Antifungal Activity of the Isolates
Endophytic bacterial isolates were examined for the inhibitory impact on the isolates shown in Figure 2. In antagonistic activity against fungal pathogens such as F. oxysporum (7693) and M. phaseolina (6630), the bacterial isolates exhibited significant antagonism. The results indicate that EMS1 and EMS4 inhibited pathogenic growth significantly, with a mean inhibition diameter of 25 ± 0.02 mm and 24 ± 0.08 mm, inhibit F. oxysporum with other strains, such as EMS13 (28 ± 0.1 mm), EMS14 (23 ± 0.2 mm), EMS16 (32 ± 0.01 mm), and EMS18 (32 ± 0.1 mm) and of 25 ± 0.02 mm and 24 ± 0.08 mm, similarly against M. phaseolina EMS1 and EMS4 showing 22 ± 0.01and 21 ± 0.02 and other strains such as EMS13 (30 ± 0.1 mm), EMS14 (29 ± 0.2 mm), EMS16 (28 ± 0.01 mm), and EMS18 (28 ± 0.1 mm).
3.3. Enzymatic Activity
The results of the isolates for enzymatic production activity are displayed in Table 3. On the basis of the screening of their extracellular enzymes production activity, all isolates such as cellulase, lipase, pectinase, amylase, protease, and gelatinase were tested. The maximum number of enzyme activities is present among all isolated bacterial strains especially Bacillus spp. (EMS 1 and EMS 4) and Enterobacter spp. (EMS 16 and 18). Seven isolated strains showed good results. There were 12 isolates showing negative results in the gelatinase test, while 19 isolates showed a negative result in the amylase test.
Table 3: Screening for lytic enzyme production of endophytic bacterial isolates.
| Serial number | Isolates | Cellulase | Amylase | Protease | Lipase | Gelatinase | Pectinase |
|---|---|---|---|---|---|---|---|
| 1 | EMS 1 | + | − | + | + | + | + |
| 2 | EMS 2 | + | − | + | + | + | + |
| 3 | EMS 3 | + | − | + | + | − | + |
| 4 | EMS 4 | + | − | + | + | + | + |
| 5 | EMS 5 | − | − | + | + | + | + |
| 6 | EMS 6 | + | − | + | + | + | + |
| 7 | EMS 7 | + | − | + | + | − | − |
| 8 | EMS 8 | + | − | + | + | − | + |
| 9 | EMS 9 | + | − | + | + | − | + |
| 10 | EMS 10 | + | − | + | + | − | + |
| 11 | EMS 11 | + | − | + | + | + | − |
| 12 | EMS 12 | + | − | + | + | − | + |
| 13 | EMS 13 | + | − | + | + | − | + |
| 14 | EMS 14 | + | − | + | + | − | + |
| 15 | EMS 15 | + | − | + | + | − | + |
| 16 | EMS 16 | + | − | + | + | + | + |
| 17 | EMS 17 | + | − | + | + | − | + |
| 18 | EMS 18 | + | − | + | + | + | + |
| 19 | EMS 19 | + | − | + | + | − | − |
+: Present, −: Absent.
4. DISCUSSION
All the bacterial endophytes isolated from M. acuminata belong to different taxonomical categories Proteobacteria and Firmicutes, respectively. Enterobacter spp. and Bacillus spp. were difficult to identify because of high sequence similarity at subspecies level due to that isolates were subjected to PCR and 16S rRNA sequencing for molecular characterization at least 2 times for assurance [34]. In the previously mentioned research, these bacterial isolates were able to promote plant growth at hormone level such as IAA, siderophore, phosphate solubilization test, and other test [35]. Among the tested isolates, we found that the highest IAA was produced by isolates EMS 1 and EMS 4 of Bacillus spp. These isolates were able to grow on a nitrogen-free substrate, only two EMS 1 and EMS 2 Bacillus spp. have capability to grow in nitrogen-free medium, as shown in Table 2. Interestingly, none of the selected isolates were able to produce a significant amount of HCN. However, Enterobacter spp. was previously reported in the case of PGPR from soils that were able to produce HCN [33]. The result shows that Bacillus spp. (EMS 1 and EMS 4) were more effective in the production of antibiotics compared to Enterobacter spp. (EMS13, EMS14, EMS16, and EMS18) which were tested against F. oxysporum and M. phaseolina, as shown in Figure 2. Enterobacter spp. has been previously reported against M. phaseolina and other pathogenic fungi. The results of antifungal activity on the plates show clear zone of minimum 25 mm approx. which is much more in comparison to previously reported work [36]. Plant growth bacteria have applications such as biocontrol agents because they are capable of resisting plant diseases and directly or indirectly stimulating plant growth by producing antimicrobial compounds such as HCN [33,37]. Hence, further studies should be conducted to understand the relation of gene producing HCN with respect to antifungal activity at transcriptomic level.
5. CONCLUSION
The present study suggested that endophytic bacterial strains found in M. acuminata have ability to produce plant growth-promoting activities such as IAA production, phosphate solubilization, and ammonia production. Hence, these endophytes are responsible for plant survival against several fungal pathogens and their applications should be implemented in field study also. Additional research is needed to evaluate the ability of these microorganisms to stimulate plant development in banana trees by introducing these isolates under in vitro conditions.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This work doesn’t include harm to any type of animal during this work.
10. DATA AVAILABILITY
All the data included in this work submitted to NCBI.
1. EMS-1 - MZ377101.1
2. EMS-4 - MZ377142.1
3. EMS-13 - MZ612853.1
4. EMS-14 - MZ612852.1
5. EMS-16 - MZ377118.1
6. EMS-18 - MZ377140.1.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. ACKNOWLEDGMENT
The authors are grateful to AKS University, Satna, for extending all facilities to carry out this work.
REFERENCES
1. Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol Ecol 2008;65:193-201. [CrossRef]
2. Dudeja SS, Giri R, Saini R, Suneja-Madan P, Kothe E. Interaction of endophytic microbes with legumes. J Basic Microbiol 2012;52:248-60. [CrossRef]
3. Das A, Varma A. Symbiosis:The art of living. In:Varma A, Kharkwal AC, editors Symbiotic Fungi Principles and Practice. Berlin, Germany:Springer;2009. 1-28. [CrossRef]
4. Iniguez AL, Dong Y, Carter HD, Ahmer BM, Stone JM, Triplett EW. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact 2005;18:169-78. [CrossRef]
5. Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 2006;19:827-37. [CrossRef]
6. Santoyo G, Moreno-Hagelsieb G, del Carmen Orozco-Mosqueda M, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol Res 2016;183:92-9. [CrossRef]
7. Ngamau CN, Matiru VN, Tani A, Muthuri CW. Isolation and identification of endophytic bacteria of bananas (Musa spp.) in Kenya and their potential as biofertilizers for sustainable banana production. Afr J Microbiol Res 2012;6:6414-22. [CrossRef]
8. Compant S, Duffy B, Nowak J, Clément C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases:Principles, mechanisms of action, and future prospects. Appl Environ Microbiol 2005;71:4951-9. [CrossRef]
9. Monnanda SN, Harischandra SP, Mysore VT. Bioactive potentials of novel molecules from the endophytes of medicinal plants. In:Egamberdieva D, Tiezzi A, editors. Medically Important Plant Biomes. Ch. 13. Singapore:Springer;2020. 293-355. doi:10.1007/978-981-13-9566-6. [CrossRef]
10. Doty SL. Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol 2008;179:318-33. [CrossRef]
11. Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, et al. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 2004;22:583-8. [CrossRef]
12. Nair DN, Padmavathy S. Impact of endophytic microorganisms on plants, environment and humans. ScientificWorldJournal 2014;2014:250693. [CrossRef]
13. George EF, Hall MA, De-Klerk GJ. Plant Propagation by Tissue Culture. 3rd ed. Dordrecht:Springer;2008. 1-28. [CrossRef]
14. Abreu-Tarazi MF, Navarrete AA, Andreote FD, Almeida CV, Tsai SM, Almeida M. Endophytic bacteria in long-term in vitro cultivated “axenic“pineapple microplants revealed by PCR-DGGE. World J Microbiol Biotechnol 2010;26:555-60. [CrossRef]
15. Liaqat F, Eltem R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 2016;6:120. [CrossRef]
16. Thomas P, Soly TA. Endophytic bacteria associated with growing shoot tips of banana (Musa sp.) cv. Grand Naine and the affinity of endophytes to the host. Microb Ecol 2009;58:952-64. [CrossRef]
17. Pinski A, Betekhtin A, Hupert-Kocurek K, Mur L, Hasterok R. Defining the genetic basis of plant-endophytic bacteria interactions. Int J Mol Sci 2019;20:8. [CrossRef]
18. Araújo WL, Marcon J, Maccheroni W Jr., Van Elsas JD, Van Vuurde JW, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 2002;68:4906-14. [CrossRef]
19. Krieg NR, Holt JG. Bergey's Manual of Determinative Bacteriology. 9th ed. Baltimore, London:Williams and Wilkins;1994.
20. Acun˜a JJ, Jorquera MA, Mart?'nez OA, Menezes-Blackburn D, Ferna'ndez MT, Marschner P, et al. Indole acetic acid and phytase activity produced by rhizosphere bacilli as affected by pH and metals. J Soil Sci Plant Nutr 2011;11:1-12.
21. Watanabe FS, Olsen SR. Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soil. Soil Sci Soc Am Proc 1965;29:677-8. [CrossRef]
22. Qureshi M, Ahmad Z, Akhtar N. Role of phosphate solubilizing bacteria (PSB) in enhancing P availability and promoting cotton growth. J Animal Plant Sci 2012;22:204-10.
23. Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 1987;160:47-56. [CrossRef]
24. Cappuccino, J.C., Sherman, N. In:Microbiology:A Laboratory Manual, third ed. Benjamin/cummings Pub. Co., New York, pp.1992:125-179.
25. Lorck H. Production of hydrocyanic acid by bacteria. Physiol Plantarum 1948;1:142-6. [CrossRef]
26. Bakker AW, Bakker P, Schippers B. Deleterious cyanide producing rhizosphere pseudomonads as a factor limiting potato root growth and tuber yield in high frequency potato cropping soil. In:Effects of Crop Rotation on Potato Production in the Temperate Zones. Dordrecht:Kluwer Academic Publishers;1989. 153-62. [CrossRef]
27. Sessitsch A, Reiter B, Berg G. Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can J Microbiol 2004;50:239-49. [CrossRef]
28. Jimtha JC, Smitha PV, Anisha C. Isolation of endophytic bacteria from embryogenic suspension culture of banana and assessment of their plant growth promoting properties. Plant Cell Tiss Organ Cult 2014;118:57-66. [CrossRef]
29. Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004;17:840-62. [CrossRef]
30. Gupta P, Samant K, Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol 2012;?578925:1687-918X. [CrossRef]
31. Silva Filho GN, Vidor C. Phosphate solubilization by microorganisms in the presence of different carbon sources 2000;24:311-9.
32. Smibert RM, Krieg NR. Phenotypic characterization. In:Gerhardt P, Murray RG, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology. Washington, DC:American Society of Microbiology;1994. 607-54.
33. Deepa CK, Dastager SG, Pandey A. Isolation and characterization of plant growth promoting bacteria from non-rhizospheric soil and their effect on cowpea (Vigna unguiculata (L.) Walp.) seedling growth. World J Microbiol Biotechnol 2010;26:1233-40. [CrossRef]
34. Jovel J, Patterson J, Wang W, Hotte N, O'Keefe S, Mitchel T, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 2016;7:459. [CrossRef]
35. Sobral JK, Araujo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of soybean-associated bacteria and their potential forplant growth promotion. Environ Microbiol 2004;6:1244-51. [CrossRef]
36. Rani MU, Reddy G. Bacillus cereus and Enterobacter cancerogenus screened for their efficient plant growth promoting traits rhizobacteria (PGPR) and antagonistic traits among sixteen bacterial isolates from rhizospheric soils of Pigeon Pea. Afr J Microbiol Res 2011;5:209-4. [CrossRef]
37. Ahemad M, Khan MS. Plant growth promoting activities of phosphate-solubilizing Enterobacter asburiaeas influenced by fungicides. Eur Asia J Biosci 2014;4:88-95. [CrossRef]