1. INTRODUCTION
The poultry industries largely satisfy the ever increasing animal protein demand globally that has arised due to population explosion [1]. People consume large quantity of chicken meat, consequently leads to the enormous generation of slaughterhouse waste [2]. Globally, 8.5 billion tons of poultry feather waste has been produced annually. In 2018, India produced 4.6 million tons of broiler chicken [3] and approximately 350 million tons of feather waste generated every year [4] and the feathers are indiscriminately dumped into the environment [5]. The accumulation of feathers in the environment leads to a variety of human ailments including chlorosis, fowl cholera, mycoplasmosis, and the outbreaks of H5N1 virus [6,7]. More than 90% of feathers are composed of protein and the major one is keratin. Extensive disulfide bonds and cross-linkages make keratin rigid and insoluble in water. Based on the sulfur content, keratins are classified as: hard keratins (feather, hair, hoof, and nail) and soft keratins (skin and callus) [8]. Sulfur content in keratin is high due to the presence of sulfur containing aminoacids such as cystine, cysteine, and methionine [9,10]. Keratin is remarkably resistant to degradation by regular proteolytic enzymes such as pepsin, papin, and trypsin. Nevertheless, feathers are considered as a good source of protein and essential amino acids if systematically recycled [11].
Various strategies have been adapted to manage the slaughterhouse waste generated in India. However, feather waste still remains a challenge [12,13].
Conventional methods of feather degradation such as acid/alkali hydrolysis and steam pressure cooking destroy the heat sensitive essential amino acids such as lysine, methionine, and tryptophan and generate non-nutritive amino acids apart from consuming huge amount of energy [14-16]. Land dumping and incineration methods are likely to result in environmental destruction and generates highly toxic emissions [17]. The conventional method of processing feather waste is cost and labor intensive that may reduce the nutritive value. Protease is used for hydrolysis of the peptide bonds that link amino acids together in a polypeptide chain [18,19]. The enzyme-catalyzed reaction is cost effective, highly efficient, and easily selective which usually requires mild reaction conditions and less energy which consequently improves the value of feathers [20]. A group of secreted proteolytic enzymes called as keratinases have the ability to hydrolyze insoluble keratins more efficiently than other proteases, are produced by microorganisms and some insects [21]. Hence, the alternative method of using microorganisms and their enzymes in feather processing technology is an environmental friendly approach. A myriad of actinomycetes, bacteria, fungi, and algae are known to produce keratinases to degrade keratin [22,23] and degradation of poultry feathers by microbial keratinases might afford a feasible alternative technique for bioconversion of keratin.
Keratinase is an extracellular enzyme produced in the presence of keratin at a broad range of pH and temperature which break the disulfide bond in keratin during degradation process. Soil holds a large number of phylogenetically diverse keratinophilic and keratinolytic microorganisms which produce enzymes with good stability at higher temperatures and alkaline conditions [24]. Microorganisms such as bacteria (Bacillus pumilus, Micrococcus sp., Microbacterium sp., and Pseudomonas sp.), actinomycetes (Streptomyces sp. and Thermoactinomyces candidus), and several fungi (Alternaria radicina, Aspergillus sp., Doratomyces microspores, and Rhizomucor sp.) have been reported to produce keratinase [25]. Recently, the production of keratinases using heterologous expression systems such as Escherichia coli, Bacillus sp. and Pichia pastoris in insect cells has attracted significant research interest [26]. Microbial keratinases are generally extracellular enzymes that are inducible in nature and some are cell bound and intracellular. Keratinases are classified into exoprotease and endoprotease based on their cleavage site as well as have many applications in leather, detergent, cosmetics, and pharmaceutical industries [27]. Hence, microbial keratinases have potential to conquer the distinctive position among the proteases which have an impact on tenvironment to bioprocess keratinous waste. Keratinolytic bacteria play an important role in the conversion of chicken feather keratin into amino acids and soluble protein. Consequently, there is a need for new process enabling ecological and economical valorization of this resistant chicken feather waste. Thus, the present study is to isolate and characterize keratinolytic bacteria and optimize their culture conditions for better degradation of chicken feather.
2. MATERIALS AND METHODS
2.1. Preparation of Chicken Feather Powder
Feather powder was prepared by the method of Larasati et al. [28] Chicken feathers were collected from a slaughterhouse at Virudhunagar, Tamil Nadu, India. The collected feathers were washed with detergent followed by water until the blood stains and dirt were removed and feathers were dried in sunlight. Then, the feathers were cut into small pieces and stored in an airtight plastic container.
2.2. Isolation and Identification of Keratinolytic Bacteria
The soil samples were collected from feather dumping site at Virudhunagar (9°35’ 13.9524’’ N Latitude and 77°57’ 5.1516’’ E Longitude), Tamil Nadu, India. Soil samples were collected from 3 to 4 cm depth using a sterile spatula and transferred to sterile zipper polythene bags and brought to the laboratory, Department of Zoology, Thiagarajar College, Madurai. The serial dilution was performed by following standard procedure [29] and diluted soil samples (10-3, 10-5, 10-7, and10-9) were inoculated on feather meal agar plates (the media composition consists of (g/l): 0.5 NH4Cl; 0.5 NaCl; 0.3 K2HPO4; 0.4 KH2PO4; 0.1 MgCl.6H2O; 10 feather powder; and 15 agar) and incubated at 37°C for 72 h. The well grown keratinolytic bacteria in the feather meal agar were isolated and maintained on agar slants. Further, all the bacteria that exerted better activities were identified and have taken for further studies. The isolated microorganisms were identified by morphological and physiological characteristics and they were acknowledged based on the prescriptions of Bergey’s Manual of Systematic Bacteriology [30].
2.3. Molecular Identification of Bacteria
2.3.1. Bacterial genomic DNA isolation
The isolation of bacterial genomic DNA was carried out following Sankari and Khusro [31] method. Two milliliters broth culture of bacteria was centrifuged at 7000 rpm for 5 min and the supernatant was discarded. One milliliter UniFlex TM buffer 1 and 10μl RNAse were added and mixed well to the obtained pellet by pipetting and incubated for 30 min at 37°C in a water bath. Then, 1 ml phenol-chloroform mixture (1:1) was added and mixed well. Then, the sample was centrifuged for 15 min at 10,000 rpm a room temperature and separated the aqueous layer in a fresh vial (1.5 ml). Again 1 ml UniFlex TM buffer 2 was added to the aqueous layer and mixed well by pipetting. The mixture was centrifuged at 12,000 rpm for 15 min at room temperature, the supernatant was discarded and 500 μl ethanol (70%) was added to the pellet and centrifuged again at 10,000 rpm for 10 min at 4°C. Finally, the obtained pellet was air-dried until it completes the evaporation of ethanol. The pellet was re-suspended in 50–100 μl UniFlex TM elution buffer and the obtained DNA was stored at -20°C.
2.3.2. Amplification of 16S rRNA genes
The amplification of 16S rRNA gene fragment was carried out using universal primers, forward (5’-AGAGTTTGATCCTGGCTCAG-3’) and reverse (5’-GGTTACCTTGTTACGACT-3’) [32]. The PCR cycling conditions were as followed: denaturation for 5 min at 94°C, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The DNA amplification reaction mixture (30 μl) consisted of 2X Amplicon Red master mixes (amplicon®), with genomic DNA (10 ng total) and 10 pmol of forward and reverse primers were used. After amplification, the PCR products were electrophoresed in 1% agarose gel and stained with ethidium bromide. The stained gel was photographed using a gel documentation system. The amplified fragments were purified and sequenced by Eurofins Scientific (Bangalore). The sequences were compared with the GenBank database using National Center for Biotechnology Information (NCBI) BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for species level identification and phylogenetic trees were constructed using the neighbor-joining algorithm in Molecular Evolution Genetic Analysis version 8.0.
2.4. Effect of Different pH on Bacterial Growth
The effect of different pH on the growth of bacteria was studied. Overnight broth culture was inoculated into the feather minimal medium prepared at different pH values such as 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, 10, and 10.5 and incubated in shaking incubator for 120 h. The bacterial growth was estimated at every 24 h interval by reading the absorbance at 620 nm against a control blank without inoculums.
2.5. Effect of Different Temperature on Bacterial Growth
The impact of diverse temperatures on the growth of bacteria has also been studied. Overnight broth culture was inoculated into the feather minimal medium. The flasks were placed in a shaking incubator at 120 rpm and incubated for 120 h at different temperatures of 25, 30, 35, 40, and 45°C. The pH of the medium was adjusted before autoclaving. Growth was measured by reading the absorbance at 620 nm at every 24 h interval against a control blank without inoculums.
2.6. Effect of Different Feather Concentration and Various substrates on Bacterial Growth
Various concentrations of feather meal and different substrates were added in culture media to identify the better growth and higher degradation of bacteria. Different concentrations of feather powder such as 0.5, 1.0, 1.5, and 2.0 g with the bacterial inoculums were used for the determination of bacterial growth.
Minimal medium was supplemented with 1% each of bovine serum albumin and gelatin and peptone, respectively, to study the effect of various substrates on growth. Each flask was inoculated with the isolated bacteria and incubated at 37°C for 120 h at 120 rpm. The growth was estimated at every 24 h interval by reading the absorbance at 620 nm against a control blank without inoculums.
2.7. Protein Estimation
The protein content of the cell free supernatant was determined by standard protocol [33] with bovine serum albumin as standard. One milliliter sample was mixed with 5 ml freshly prepared alkaline copper sulfate and then incubated at room temperature for 10 min. Then added 0.5 ml Folin-ciocalteau’s reagent, incubated for 30 min and the absorbance were read at 660 nm against a reagent blank.
2.8. Estimation of Aminoacids
The concentration of free amino groups in the culture supernatant was determined by ninhydrin method [10] with glycine as standard.
2.9. Submerged Fermentation
Fermentation process was carried out following submerged fermentation method [34] using feather powder as substrate. The basal minimal medium consisted of 1% chicken feather powder as sole source of carbon and nitrogen. Further, fermentation tank was filled with 2 L of autoclaved medium. Fermentation was initiated by seeding the prepared inoculum and was carried out at 37°C, with pH 7.0 and 100 rpm. During the fermentation process, 10 ml media along with culture was siphoned from the tank at every 24 h interval for 120 h. The sample was centrifuged at 5000 rpm for 15 min, and the supernatant was used for the estimation of protein and amino acid. The biomass was calculated using the culture pellet. A thin smear of the feather hydrolysates was made on a clean glass slide and the changes in the architecture and shape of the feather were observed under the light microscope (LABOMAD LX 300) at 100× magnification.
3. RESULTS
3.1. Isolation and Identification of Keratinolytic Bacteria
In the present study, 14 bacteria displayed keratinolytic properties and three bacteria demonstrated better activities were chosen for further studies. Based on the morphological, physiological, and biochemical tests [Table 1] and in agreement with the characteristic features described in Bergey’s Manual of Systematic Bacteriology, the isolates were identified as Bacillus cereus, Bacillus licheniformis, Bacillus megaterium, Bacillus polymxa, B. pumilus, Bacillus subtilis, B. megaterium, Glutamicibacter arilaitensis, Micrococcus luteus, Micrococcus sp., Proteus vulgaris, Pseudomonas aeruginosa, Pseudomonas microphilus, and Serratia marcescens, respectively. Among them, three bacteria such as B. licheniformis, G. arilaitensis, and S. marcescens exerted better activity and they were considered for further studies. Those three bacteria were also confirmed by their 16S rRNA gene sequence and the sequences have been deposited in the NCBI and obtained GenBank accession numbers MH063522.1, MH063920.1, and MH064150.1 with high matches similarity [Figure 1] respectively.
Table 1: Biochemical characterization of the isolated keratinolytic bacteria.
| S. No | Biochemical Tests | Bacterial isolates | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | ||
| 1 | Gram staining | +R | +R | +R | +R | +R | +R | -R | -R | -R | +C | +R | -R | +C | +R |
| 2 | Methyl red | - | + | + | + | - | + | - | + | - | - | - | - | - | - |
| 3 | Voges–proskauer Test | + | + | + | + | - | + | - | - | + | - | + | - | - | - |
| 4 | Starch hydrolysis | + | + | + | - | + | + | + | NR | + | NR | + | NR | + | + |
| 5 | Urease test | - | NR | NR | NR | NR | - | + | + | + | NR | NR | - | + | - |
| 6 | Citrate utilization | + | + | + | - | + | + | + | + | + | + | - | + | - | NR |
| 7 | Indole test | - | NR | - | - | NR | - | + | - | NR | NR | - | + | - | |
| 8 | Casein hydrolysis | + | + | + | NR | + | + | NR | NR | + | + | + | NR | + | + |
| 9 | Oxidase test | NR | + | + | + | NR | NR | + | - | - | + | NR | NR | NR | - |
| 10 | Catalase test | + | + | + | + | + | + | NR | + | + | + | NR | NR | + | + |
| 11 | Mannitol salt agar test | NR | - | NR | NR | + | NR | NR | NR | + | NR | + | + | + | - |
Identified organisms: I= Bacillus subtilis, II= B. Cereus, III= B. licheniformis, IV= B. pumilus, V= B. megaterium, VI= B. thuriengiensis, VII= Pseudomonas microphilus, VIII= Proteus vulgaris, IX= Serratia marcescens, X= Micrococcus luteus, XI= Bacillus polymyxa, XII= Pseudomonas aeruginosa, XIII= Micrococcus SPe, XIV= Glutamicibacter arilaitensis+ R = Positive Rod; + C=Positive Cocci, NR=No Results.
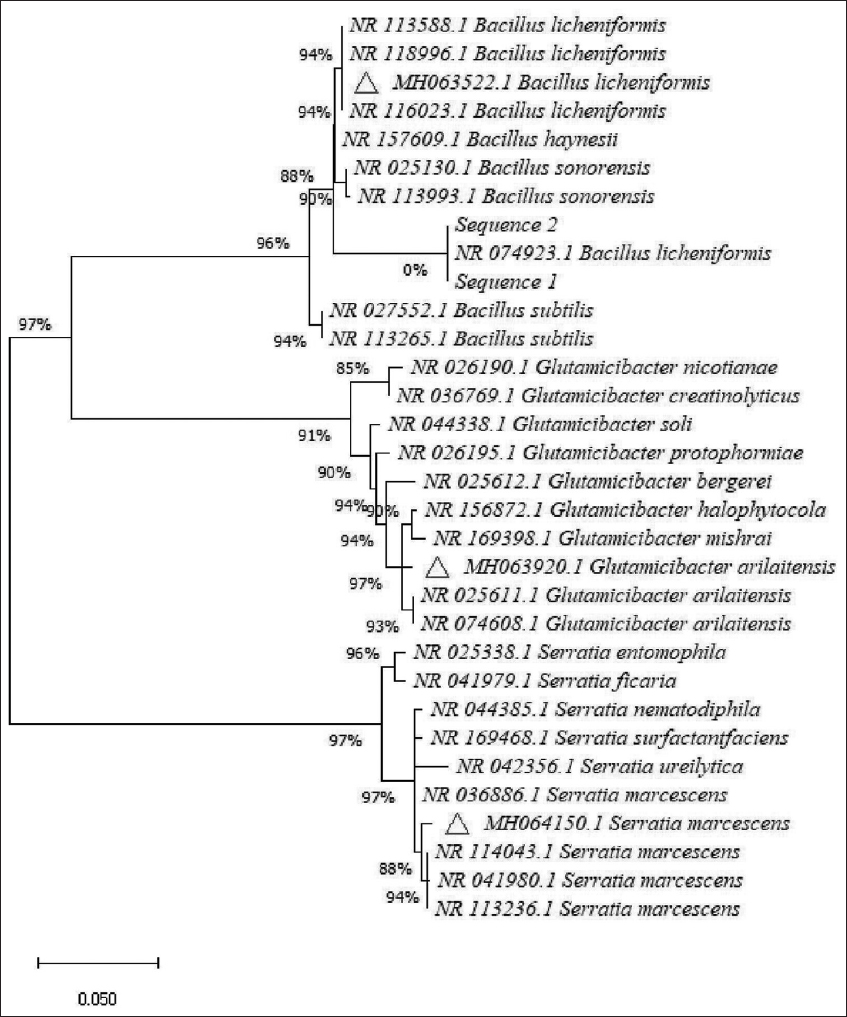 | Figure 1: Phylogenetic tree based on 16S rRNA sequence of isolated bacteria. The branching pattern was generated by neighbor-joining method. [Click here to view] |
3.2. Effect of Different pH and Temperature on Bacterial Growth
Feather powder was used as the substrates for pH and temperature optimization, various culture parameters have been optimized to attain maximum feather degradation by the bacterial isolates. As the temperature increased, growth of bacteria was also increased. The growth of G. arilaitensis was high at pH 8.5 and 40°C, respectively. Similarly, for B. licheniformis, it was 8.0 and 40°C and for Serratia marcesences, it was 8.5 and 40°C, respectively. The amount of protein produced by B. licheniformis was high at pH 8.5 and 40°C, respectively. G. arilaitensis, it was 8.0 and 40°C [Figures 2 and 3] and, for S. marcesences, it was 8.5 and 45°C, respectively, which suggest that keratinase activity depends on the growth of bacteria.
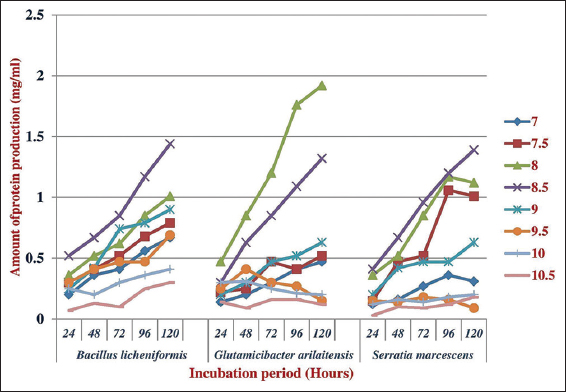 | Figure 2: Effect on pH on the production of protein by the isolated feather degrading bacteria. [Click here to view] |
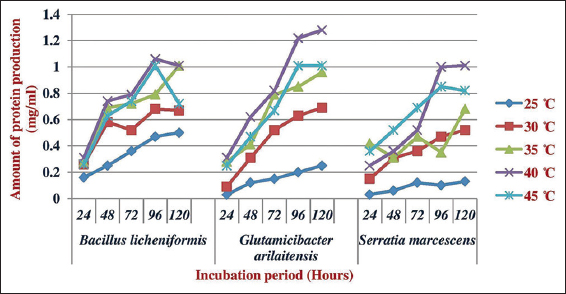 | Figure 3: Effect of temperature on the production of protein by the isolated feather degrading bacteria. [Click here to view] |
3.3. Effect of Different Feather Concentration and Various substrates on Bacterial growth
The optimum feather concentration for growth of all the tested bacteria was 1 g/100 ml and the obtained results are depicted in Figures 4 and 5. Different species of microbes releases different amount of protein production and growth using various growth substrates. Subsequently, for the growth and maximum protein, production of G. arilaitensis and S. marcescens utilized gelatin and bovine serum albumin exhibited highest protein production of B. lichenformis. Among the various substrates provided, gelatin was the least preferred by B. licheniformis and G. arilaitensis and for S. marcescens peptone was the least preferred substrate.
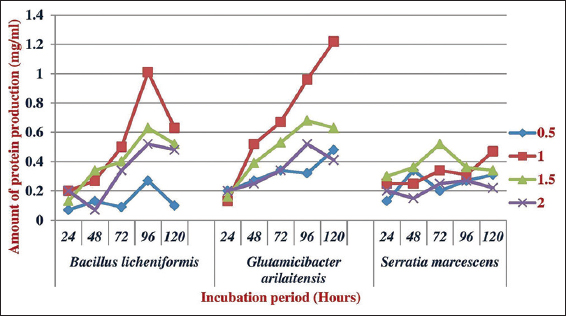 | Figure 4: Effect of concentration on the production of protein by the isolated feather degrading bacteria. [Click here to view] |
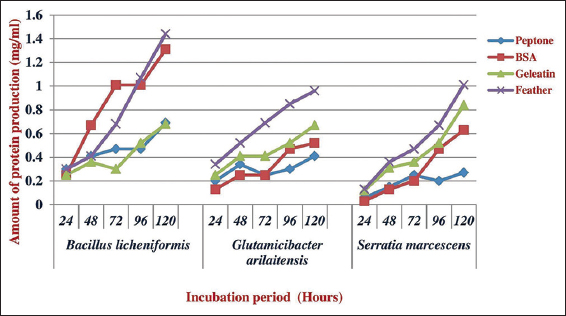 | Figure 5: Effect of various substrates on the production of protein by the isolated feather degrading bacteria. [Click here to view] |
3.4. Submerged Fermentation
The fermentation process was carried out using feather as the source of Carbon. The hydrolysate was tested at every 24 h interval for soluble amino acid, protein, and biomass and the results are tabulated [Tables 2-4]. The amount of amino acid and protein in the cell free supernatant was the best indicator for the progression of biodegradation. A thin smear of the feather hydrolysates was also prepared for every 24 h interval, examined under the microscope, and photographed. Feather rachea and barbs were ruptured into smaller fragments. Consequently, there was a lot of modification in the arrangement of barbs and rachis and architecture of the feather fragments [Figures 6-8]. Visual examination of unstructured feathers under the microscope at high magnification described the prominent variation in feather configuration at every 24 h interval. Considering the results, it has been concluded that, among the three bacteria, better degradation of feathers was exhibited by G. arilaitensis.
Table 2: The impact of Bacillus licheniformis on fermentation.
| Hours | Dry feather weight (g) | Biomass (g) | Protein (mg/ml) | Amino acids (µg/ml) |
|---|---|---|---|---|
| 24 | 1.24 | 0.002 | 0.57±0.11 | 0.063±0.011 |
| 48 | 0.67 | 0.016 | 0.55±0.08 | 0.10±0.028 |
| 72 | 1.01 | 0.103 | 0.41±0.03 | 0.125±0.037 |
| 96 | 0.44 | 0.012 | 1.45±0.03 | 0.218±0.019 |
| 120 | 0.62 | 0.008 | 0.58±0.10 | 0.180±0.018 |
Data represents mean±standard deviation of independent triplicate.
Table 3: The impact of Glutamicibacter arilaitensis on fermentation.
| Hours | Dry feather weight (g) | Biomass (g) | Protein (mg/ml) | Aminoacids (µg/ml) |
|---|---|---|---|---|
| 24 | 1.33 | 0.03 | 0.81±0.11 | 0.10±0.028 |
| 48 | 0.99 | 0.016 | 0.90±0.10 | 0.025±0.018 |
| 72 | 0.064 | 0.012 | 1.37±0.03 | 0.317±0.011 |
| 96 | 0.018 | 0.018 | 1.42±0.03 | 0.386±0.049 |
| 120 | 0.012 | 0.004 | 2.15±0.04 | 0.498±0.019 |
Data represents mean±standard deviation of independent triplicate.
Table 4: The impact of Serratia marcescens on fermentation.
| Hours | Dry feather weight (g) | Biomass (g) | Protein (mg/ml) | Amino acids (µg/ml) |
|---|---|---|---|---|
| 24 | 0.092 | 0.02 | 0.92±0.08 | 0.205±0.011 |
| 48 | 0.083 | 0.04 | 1.24±0.06 | 0.262±0.101 |
| 72 | 0.092 | 0.05 | 1.49±0.10 | 0.305±0.011 |
| 96 | 0.062 | 0.06 | 1.42±0.03 | 0.448±0.011 |
| 120 | 0.018 | 0.02 | 1.66±0.06 | 0.504±0.028 |
Data represents mean±standard deviation of independent triplicate.
 | Figure 6: (a-e) Microscopic view of degraded feather during fermentation by Bacillus licheniformis. [Click here to view] |
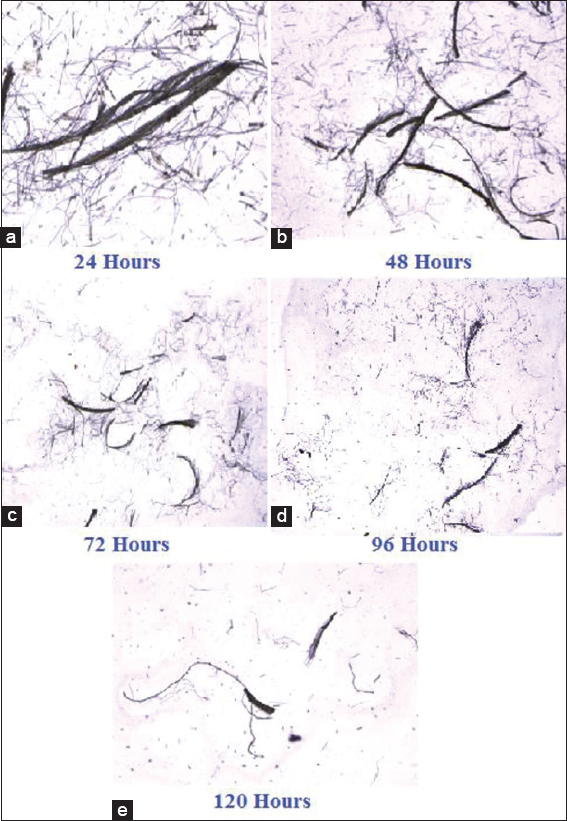 | Figure 7: (a-e) Microscopic view of degraded feather during fermentation by Glutamicibacter arilaitensis. [Click here to view] |
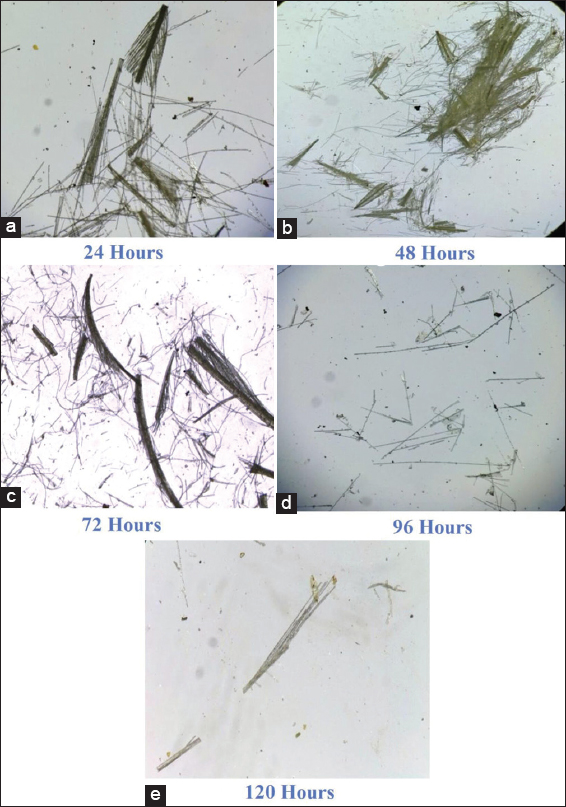 | Figure 8: (a-e) Microscopic view of degraded feather during fermentation by Serratia marcescens. [Click here to view] |
4. DISCUSSION
Several million tons of feathers are generated every year by the poultry sectors across the globe which leads to troublesome environmental pollution and subsequent wastage of protein [10]. In this study, 14 keratinolytic bacteria were isolated from soil based on the growth, biomass, and protein content in the medium. The three better strains such as B. licheniformis, G. arilaitensis, and S. marcescens were chosen for further studies. Microorganisms utilize feathers as a source of nutrient for their growth by hydrolyzing keratin through keratinolysis, sulfitolysis, and proteolysis [35]. Optimization is notably an important criterion in the development of microbial process. Cost-effective optimization of culture conditions for keratinolytic bacteria is a challenge, and only few significant results were achieved for large scale process [36]. Furthermore, increasing in temperature causes the denaturation of enzyme and also microorganisms could not survive at high temperatures. This indicates that the isolates might be mesophiles and not favorable for enzyme production at high temperatures. These results are in accordance with results described by Sahoo et al. [37] who obtained optimum enzyme production at 40°C with Bacillus weihenstephanensis.
Chaisemsaeng et al. [15] reported that the best temperature was between 40 ºC and 45ºC for the production of keratinolytic enzyme by bacilli (B. licheniformis and Brevibacillus brevis). B. licheniformis ALW 1 produced keratinase that exhibited good stability over a range of pH (7–9) and temperature (50–60°C) and was able to degrade native feathers up to 63% in redox free system [38]. Similarly, Mosavi et al. [39] also reported maximum keratinase activity by B. subtilis isolated from poultry waste at pH 11 and 40ºC on the 6th day of culture. A novel keratin degrading protease, keratinase BsKER71 from B. subtilis S1-4, was cloned and expressed under high temperature regime of 50–55ºC in a wide range of pH 9 and 10 [40]. Furthermore, B. licheniformis optimally produced the enzyme at pH 10 and temperature 40ºC and Geobacillus stearothermophilus produced keratinase at 55 ºC and pH 7.0–8.0 [41].
The pH changes not only affect nature of the medium, substrates, and also affect growth and enzyme production by affecting enzymes transport across to the substrate so that growth in unsuitable conditions limits the microbial growth and enzyme production. The rate of microbial growth may be lower or higher than optimum due to the changes in three dimensional structure of proteins, affect the ionization of R groups of amino acids in active site or in other parts of the enzyme so that the enzyme changes shape and it decreases enzyme catalytic ability. Hence, change in pH is one of the significant characteristics of keratin degradation and the present research used the pH of the basal medium at 7–9 for better bacterial growth and stimulation of the enzyme. In this study, G. arilaitensis shows good degradation by producing more keratinase. Barman et al. [32] reported a synonymous strain Arthrobacter sp. which was a good candidate for keratinase production; however, reports are scarce on the feather degradation ability of G. arilaitensis. Most of the keratinases are released extracellular and the enzyme was stable at pH 6–8 for Bacillus strains [42,43] and the keratinases produced by the Bacillus species might be active in neutral to basic pH conditions. B. licheniformis BBE11-1 and Stenotrophomonas maltophilia BBE11-1efficiently degraded 50 g/L of chicken feather waste and produced large amounts of amino acids and antioxidant substances at a conversion rate of 70.0% [44].
Different mechanisms were proposed for microbial decomposition of keratin, in which cleavage of disulfide bonds before proteolytic breakdown is inherent [45]. The protease produced by Arthrobacter arilaitensis Re117 and Serratia sp. has high stability and purified with ion exchange chromatography technique [46-48]. Biodegradation of keratin under culture conditions is a process dependent on the microbial strain and type of keratinase. Keratinase shows variable keratinolytic activity toward different substrates. Moreover, the efficiency degradation rate linked with type of substrates and amount of cysteine of the substrates. Aeromonas hydrophila K12, Chryseobacterium indologenes A22, and S. marcescens P3 produced high amount of soluble proteins and formed thiol groups during keratinase production [37]. Khardenavis et al. [49] isolated Serratia HPC 1383 to enhance the degradative capacity to further shorten the time for digestion of feather waste to protein-rich animal feed by optimizing the kinetic contents of keratinase. Aarti et al. [50] reported that G. arilaitensis ALA4 produced amylase at 4°C with goat dung as substrate; these results indicate that G. arilaitensis has higher ability to degrade keratin from feathers. The present study is in agreement with their findings, wherein under optimized culture conditions keratinase production was high and degraded feathers in a better way.
Filamentous fungi and bacteria degrade feather by keratinase through sulfitolytic cleavage of cystine [18] and accumulation of reduced thiols due to direct reduction with specific reductase-like enzymes [51]. Verea et al. [52] concluded that keratinase production and feather degrading activity in vitro are not only the properties of bacteria but also depend on other factors including keratin substrate, sterilization method, and culture conditions. High feather powder concentration above 1% may lead to increase in medium viscosity which results in oxygen limitation for bacterial growth. Cheng et al. [53] reported that 1% feather powder resulted in the highest keratinase activity for B. licheniformis PWD-1. B. subtilis S1 with 1% feather powder yielded better degradation activity [54]. Recently, the products of feather degradation have been used as a component of fertilizers which augment seed germination and growth rate [55]. The treatment of non-digestible biomass by these microorganisms may offer an industrial opportunity along with degradation of keratin [56]. The thermal alkaline microwave method, enzymatic method [57], and green technology based methods are most significant in the production of keratin hydrolysates which have enormous biotechnological significance [17].
5. CONCLUSION
G. arilaitensis degrades feather waste under optimized culture conditions in a better, safe, and ecofriendly way. Further, validation of this research has a huge potential to employ this strain in the large scale degradation of chicken feather waste. Further studies are required to possibly convert the recalcitrant chicken feather waste into utilizable products for sustainable development.
6. ACKNOWLEDGMENT
We would like to thank the management of Thiagarajar College, Madurai, for providing the required facilities to carry out the study. In addition, we would like to thank the MHRD National Centre for Excellence, Thiagarajar College for permitting us to make use of the fermenter facility. The fund received from University Grants Commission, Government of India through a major research project (MRPMAJOR-ZOOL-2013-24210 [943-573/2014(SR)]) to the corresponding author is gratefully acknowledged.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included in this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Brandelli A, Sala L, Kalil SJ. Microbial enzymes for bioconversion of poultry waste into added-value products. Food Res Int 2015;73:3-12. [CrossRef]
2. Mazotto AM, Couri S, Damaso MC, Vermelho AB. Degradation of feather waste by
3. Prabhavathi TG, Ramani R, Rao VA, Babu RN, Ramesh J, Vanathi A,
4. Samuel P, Maheswari M, Vijayakumar J, Selvarathinam T, Amirtharaj K, Deenathayalan R. Bio-prospecting of marine-derived fungi with special reference to production of ecovaluable enzyme keratinase-a need-based optimization study. J App Biol Biotech 2018;6:35-41.
5. Gurav GR, Tang J, Jadhav JP. Sulfitolytic and keratinolytic potential of
6. Agrahari S, Wadhwa N. Degradation of chicken feather a poultry waste product by keratinolytic bacteria isolated from dumping site at Ghazipur poultry processing plant. Int J Poult Sci 2010;9:482-9. [CrossRef]
7. Haq I, Akram F, Jabbar Z. Keratinolytic enzyme-mediated biodegradation of recalcitrant poultry feathers waste by newly isolated
8. Subugade S, Gupta SG, Mokashe S. Isolation screening of keratinase producing bacteria from chicken feather dumping site. Int J Chem Tech Res 2017;10:900-5.
9. Manirujjman M, Amin R, Nahid AA, Alam MS. Isolation and characterization of feather degrading bacteria from poultry waste. Afr J Bacteriol Res 2016;8:14-21.
10. Laba W, ?arowska B, Chorazyk D, Pud?o A, Piegza M, Kancelista A,
11. Kshetri P, Ningthoujam DS. Keratinolytic activities of alkaliphilic
12. Jayathilakan M, Sultana K, Radhakrishna K, Bawa AS. Utilization of byproducts and waste materials from meat, poultry and fish processing industries:A review. J Food Sci Technol 2012;49:278-93. [CrossRef]
13. Bhange K, Chaturvedi V, Bhatt R. Feather degradation potential of
14. Chhimpa S, Yadav CS, John PJ. Isolation and identification of keratin degrading (keratinolytic) bacteria from poultry feather dumping sites. J Biodiversity Environ Sci 2016;8:109-19.
15. Chaisemsaeng P, Sabu A, Ansanan S, Sirisan, S. Feather degradation and keratinase production by
16. Tamilkani P, Karan M, Kanimozhi K, Panneerselvam A. Screening of keratinolytic bacteria from keratin waste dumped soil in Thanjavur (Dt), Tamil Nadu, India. Int J Pharm Pharma Res 2017;8:25-32.
17. Prajapati S, Koirala S, Anal AK. Bioutilization of chicken feather waste by newly isolated keratinolytic bacteria and conversion into protein hydrolysates with improved functionalities. Appl Biochem Biotechnol 2021;193:2497-515. [CrossRef]
18. Godbole S, Pattan J, Gaikwad S, Jha, T. Isolation, identification and characterization of keratin degrading microorganisms from poultry soil and their feather degradation potential. Int J Environ Agric Biotechnol 2017;2:2060-8. [CrossRef]
19. Li ZW, Liang S, Ke Y, Deng JJ, Zhang MS, Lu DL,
20. Suharti S, Riesmi MT, Hidayati A, Zuhriyah UF, Wonorahardjo S, Susanti E. Enzymatic dehairing of goat skin using keratinase from
21. Khodayari S, Kafilzadeh F. Separating keratinase producer bacteria from the soil of poultry farms and optimization of the conditions for maximum enzyme production. Eur Exp Biol 2018;8:35. [CrossRef]
22. Tamreihao K, Mukherjee S, Khunjamayum R, Devi LJ, Asem RS, Ningthoujam DS. Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J Basic Microbiol 2019;59:4-13. [CrossRef]
23. Moonnee YA, Foysal MJ, Hashem A, Miah MF. Keratinolytic protease from
24. Shah M, Vaidya RB. A novel feather degrading
25. Kiruthika R, Devakumar J, Rajam JS, Vaishali S, Pavithra KS. Isolation and identification of chicken feather degrading organisms from soil sample. Int J Progress Res Sci Eng 2021;2:124-32.
26. Huang M, Chen R, Ren G. Secretory expression and purification of
27. Tork SE, Shahein YE, Hakim AE, Abdel-Aty AM, Aly MM. Production and characterization of thermostable metallo-keratinase from newly isolated
28. Larasati D, Tsurayya N, Koentjoro MP, Prasetyo EN. Keratinase from Newly Isolated Strain of Thermophilic
29. Sarkar P, Dutta E, Sen P, Banerjee R. Isolation and Molecular characterization of extracellular keratinase from
30. Bernner DJ, Kriej NR, Staley, JT. Bergey's Manual of Systematic Bacteriology 2nd. ed. New York:Springer;2004. 323-58.
31. Sankari D, Khusro A. Biochemical, molecular characterization and sequence analysis of keratinase producing novel strain of
32. Barman NC, Zohora FT, Das KC, Mowla MG, Banu NA, Salimullah M,
33. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem 1951;193:265-75. [CrossRef]
34. Mazotto AM, de Melo CA, Macrae A, Rosado AS, Peixoto R, Cedrola SM,
35. Sharma R, Devi S. Versatility and commercial status of microbial keratinases:A review. Rev Environ Sci Biotechnol 2017;19:19-45. [CrossRef]
36. Nnolim NE, Okoh AI, Nwodo UU. Proteolytic bacteria isolated from agro-waste dumpsites produced keratinolytic enzymes. Biotechnol Rep (Amst) 2020;27:e00483. [CrossRef]
37. Sahoo DK, Das A, Thatoi H, Mondal KC, Mohapatra PK. Keratinase production and biodegradation of whole chicken feather keratin by a newly isolated bacterium under submerged fermentation. Appl Biochem Biotechnol 2012;167:1040-51. [CrossRef]
38. Fattah AM, El-Gamel MS, Ismail SA, Emran MA, Hashem AM. Biodegradation of feather waste by keratinase produced from newly isolated
39. Mosavi S, Salouti M, Shapoury R, Heidari Z. Optimization of keratinase production for feather degradation by
40. Yong B, Fei X, Shao H, Xu P, Hu Y, Ni W,
41. Alahyaribeik S, Sharifi SD, Tabandeh F, Honarbakhsh S, Ghazanfari S. Bioconversion of chicken feather wastes by keratinolytic bacteria. Process Safe Environ Protect 2020;135:171-8. [CrossRef]
42. Poopathi S, Thirugnanasambantham K, Mani C, Lakshmi PV, Ragul K. Purification and characterization of keratinase from feather degrading bacterium useful for mosquito control--a new report. Trop Biomed 2014;31:97-109.
43. Jin M, Chen C, He X, Zeng R. Characterization of an extreme alkaline-stable keratinase from the draft genome of feather-degrading
44. Peng Z, Mao X, Zhang J, Du G, Chen J. Effective biodegradation of chicken feather waste by cocultivation of keratinase producing strains. Microb Cell Fact 2019;18:84. [CrossRef]
45. Nnolim NE, Okoh AI, Nwodo UU.
46. Siala R, Hammemi I, Sellimi S, Vallaeys T, Kamoun AS, Nasri M.
47. Bach E, Daroit DJ, Correa AP, Brandelli A. Production and properties of keratinolytic proteases from three novel gram-negative feather-degrading bacteria isolated from Brazilian soils. Biodegradation 2011;22:1191-201. [CrossRef]
48. Murthy VN, Murulidhara VN, Mariswamy M. Production and purification of keratinase enzyme from
49. Khardenavis A, Kapley A, Purohit HJ. Processing of poultry feathers by alkaline keratin hydrolyzing enzyme from
50. Aarti C, Khusro A, Agastian P. Carboxymethyl cellulase production optimization from
51. Laba W, Choinska A, Rodziewicz A, Piegza M. Keratinolytic abilities of
52. Verea C, Vitelli-Flores J, Dorta B, Isturiz T, Solorzano A, Rodríguez-Lemoine V,
53. Cheng SW, Hu HM, Shen SW, Takagi H, Asano M, Tsai YC. Production and characterization of keratinase of a feather-degrading
54. Singh S, Masih H, Jeyakumar GE, Lawrence R, Ramteke PW. Optimization of fermentative production of keratinase by
55. Koentjoro MP, Prasetyo EN. Advances in use of keratinase from feather wastes for feedstock modification. Appl Food Biotechnol 2021;8:19-30.
56. Cheong CW, Ahmad SA, Ooi PT, Phang LY. Treatments of chicken feather waste. Pertanika J Scholarly Res Rev 2017;3:32-41.
57. Cheong CW, Lee YS, Ahmed SA, Ooi PT, Phang LY. Chicken feather valorization by thermal alkaline pretreatment followed by enzymatic hydrolysis for protein-rich hydrolysate production. Waste Manag 2018;79:658-66. https://doi.org/10.1016/j.wasman.2018.08.029 PMid:30343798