1. INTRODUCTION
Recent investigations suggest that the illegal practice of species substitution and mislabeling is the most common fraud worldwide detected in the fish trade [1,2]. In the fish industry, intentional fraud is generally motivated by economic gain based on the substitution of one valuable species for another of lower value. Other motivations have also been highlighted such as the need to sell illegally caught species by hiding them under a specific false name on the label [1]. However, this type of fraud is feasible if the commercial product has undergone processing that has removed the morphological characteristics useful to recognize the species declared on the label. On the other hand, unintentional commercial fraud may be due to the difficulty of distinguishing species morphologically. An overview of fish fraud extracted from four databases [the European Union’s Rapid Alert System for Food and Feed, the Food Fraud Database, HorizonScan, and Nexis] was recently published [3]. Interestingly, the authors found that the scientific literature showed a higher level of species substitution than those found in the questioned databases. This result could indicate both a low level of attention by regulatory bodies to this important issue facing the seafood industry, but also the urgency of using, common guidelines to address the problem along the seafood production chain in a global market. In this regard, the need for greater coordination between research and policy actions has recently been claimed with the aim of minimizing mislabeling, decreasing its negative impacts, and improving transparency [4].
Polymerase chain reaction (PCR) is the most modern practical technology in diagnosing and compared with classical techniques [5,6]. It has been shown to be more rapid, with results obtained in a few hours, and also more reliable [7]. Moreover, PCR allows a faster identification directly from samples [8]. Genotyping, which is based on a more stable marker, DNA, is not dependent on gene expression [6,9]. In this context, DNA-based molecular techniques and in particular DNA barcoding offer an undoubted contribution to unraveling the fraudulent practice of species substitution in processed fish products [2]. More specifically, the standard region of approximately 650 bp of mitochondrial cytochrome oxidase I (COI) has been proposed as a reliable barcode gene for species discrimination [10] and could currently be considered the best barcode for animals. COI shows high interspecific and low intraspecific divergence (barcoding gap) and a very large number of sequences are available in public databases [that is, the Barcode of Life Database or BOLD, and GenBank accessible at the National Center for Biotechnology Information or NCBI]. On the other hand, besides being based on reliable molecular markers, a method for the authentication of processed products should also be easy to apply, quick, inexpensive, and easy to use in food control laboratories. Interestingly, the PCR-RFLP (Polymerase Chain Reaction–Restriction Fragment Length Polymorphism) method is based on the amplification of a DNA region and the subsequent digestion of the amplicon by restriction endonucleases that recognize restriction sites and cut the DNA sequence into fragments of different lengths [11,12]. The species-specific pattern of DNA fragments obtained has been widely used and validated for the genetic authentication of species in both fish and meat products using various nuclear and mitochondrial genes [13-18]. In particular, PCR-RFLP of the COI gene has been successfully used to identify tuna species in raw and cooked tuna products [13] and recently for the recognition and discrimination of 9 of 25 meat species [17]. However, it should be noted that in the studies just mentioned, at least two restriction enzymes are used simultaneously to increase the specific discriminatory power of the methodology. Therefore, the main advantage of PCR-RFLP, which is considered to be a cheap and fast method, is called into question.
Among the teleosts affected by the species substitution fraud is the family Sparidae which comprises 39 genera and 164 species currently listed in the catalog of fishes [19] and includes commercially and ecologically very important fishes. The ecomorphology of several sparid species has been studied based on the correlation between feeding behavior and body morphology [20]. Indeed, body shape provides relevant information on the biology and ecology of species, and geometric morphometric methods have proven to be an efficient way to explore various biological traits in fish species [21-23]. In particular, through a geometric morphometric approach conducted on 92 sparid species, it was found that several morphological traits show similarity between distinct species with respect to a common feeding and habitat utilization strategy [24]. As a result, many sparid species are difficult to recognize based on their morphological features, which increase the likelihood and frequency of misidentifications. Furthermore, the presence of hybridization between sparid species has been noted [25], which further confuses morphological identification.
Therefore, due to (i) the increase in the marketing of sparid species over the past decade, (ii) the high selling price of the most valuable species, and (iii) the morphological similarity between species, sparids are often subject to intentional or unintentional substitution fraud. In this regard, several investigations have recently been conducted focusing on the search for specific molecular markers and the development of specific and rapid tests for the identification of sparid species [26-28]. However, COI-DNA barcoding is still confirmed as a reliable molecular methodology for the identification of fish species [29-33] and for sparids in particular [26]. In this context, the aim of the present study was to identify sparid species in processed products using the COIBar-RFLP strategy, which exploits the specific discriminatory power of the COI gene (DNA barcode) and of the interspecific variation of the restriction enzymes digestion pattern. For this purpose, a single restriction enzyme will be selected in silico to simultaneously discriminate the sparid species examined in this study.
2. MATERIALS AND METHODS
2.1. Samples and DNA Extraction
Twenty-seven fish specimens belonging to the Sparidae family were purchased from several local markets (fish market, street fish market, and supermarket) in Southern Italy in 2021/2022. Among them, six fresh whole samples of the most commonly sold sparids were identified by morphological inspection using analytical keys [34] and the detected species were confirmed by molecular approach. The specimens were then used as reference samples for the remaining 21 fish products labeled with the names of the six reference species [Table 1]. To verify the correctness of the specific name given on the label, these samples, 11 fillets and 10 whole specimens, were identified only through the molecular approach [Table 2]. Genomic DNA was isolated from muscle tissue (25–30 mg) following the extraction protocol of the DNeasy tissue kit (Qiagen, Hilden, Germany).
Table 1: Reference sparid species identified on morphological characters.
| Sample Code | Scientific name | Common name | GenBank Accession Number | Matched GenBank Accession from BLAST | % identity with 100% coverage |
|---|---|---|---|---|---|
| S1 | Diplodus sargus | white seabream | OQ353062 | LC203132 | 99.10 |
| S2 | Diplodus vulgaris | common two-banded seabream | OQ353063 | LC195195 | 98.92 |
| S3 | Pagellus acarne | axillary seabream | OQ353065 | KJ012382 | 99.24 |
| S4 | Pagellus erythrinus | common pandora | OQ353066 | KX586210 | 99.54 |
| S5 | Pagrus pagrus | red porgy | OQ353067 | KJ012417 | 99.85 |
| S6 | Spondyliosoma cantharus | black seabream | OQ353069 | KJ012436 | 99.54 |
Table 2: Sparidae sampling in Sicilian fish market. In bold, misdescription cases.
| Code | Retail point | Retail format | Label Description | Scientific name of declared species | Identified species by DNA barcoding and BLAST search | GenBank Accession Number | Matched GenBank Accession from BLAST | % identity with 100% coverage |
|---|---|---|---|---|---|---|---|---|
| X1 | Street fish market | whole | white seabream | D. sargus | S. cantharus | OR268681 | KJ012436 | 99.39 |
| X3 | Street fish market | fillet | common pandora | P. erythrinus | P. pagrus | OR268682 | OQ865598 | 99.54 |
| X4 | supermarket | fillet | common pandora | P. erythrinus | P. erythrinus | OR268683 | KX586210 | 99.23 |
| X5 | fish market | fillet | red porgy | P. pagrus | P. pagrus | OR268684 | OQ865598 | 99.69 |
| X6 | supermarket | whole | red porgy | P. pagrus | P. pagrus | OQ359508 | KJ012417 | 99.85 |
| X7 | supermarket | fillet | white seabream | D. sargus | D. sargus | OR268685 | LC203132 | 98.80 |
| X8 | Street fish market | whole | common pandora | P. erythrinus | P. pagrus | OQ359510 | KJ012417 | 99.54 |
| X9 | Street fish market | whole | white seabream | D. sargus | D. sargus | OQ359511 | LC203131 | 99.85 |
| X10 | Street fish market | fillet | axillary seabream | P. acarne | P. acarne | OR268686 | KJ012382 | 98.93 |
| X12 | Street fish market | whole | common pandora | P. erythrinus | P. erythrinus | OQ359514 | KM538475 | 98.78 |
| X13 | Street fish market | whole | white seabream | D. sargus | S. cantharus | OQ359515 | K5J012439 | 99.54 |
| X14 | fish market | fillet | red porgy | P. pagrus | P. pagrus | OR268687 | OQ865598 | 99.54 |
| X15 | Street fish market | whole | common two-banded seabream | D. vulgaris | D. vulgaris | OQ359517 | LC195196 | 99.69 |
| X16 | Street fish market | fillet | common two-banded seabream | D. vulgaris | D. vulgaris | OR268688 | LC203527 | 99.06 |
| X17 | fish market | whole | red porgy | P. pagrus | P. pagrus | OQ359519 | KJ012417 | 99.24 |
| X18 | fish market | whole | common pandora | P. erythrinus | P. erythrinus | OQ359520 | KX586210 | 99.39 |
| X19 | supermarket | fillet | white seabream | D. sargus | D. sargus | OR268689 | LC203132 | 98.80 |
| X21 | Street fish market | whole | white seabream | D. sargus | S. cantharus | OR268690 | KJ012436 | 99.39 |
| X24 | Street fish market | Fillet | axillary seabream | P. acarne | P. acarne | OR268691 | KJ012382 | 98.71 |
| X26 | Street fish market | Fillet | axillary seabream | P. acarne | P. acarne | OQ359528 | KM538461 | 99.69 |
| X29 | fish market | fillet | red porgy | P. pagrus | P. pagrus | OQ359531 | KF461214 | 99.24 |
2.2. Barcode Amplification and Sequencing
The COI gene (~650 bp) was amplified using the universal primers VF2_t1 and FishR2_t1 [35]. A total of 25 μL of PCR amplification mix were prepared as described in [36], including negative controls in each run. All obtained amplicons were verified by 0.8% agarose gel electrophoresis and subsequently purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sequencing was performed by Eurofins Genomics using M13 forward and reverse primers [37]. All sequences were checked and edited using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and only those with a quality score above 20 were considered and deposited in the GenBank database [http://www.ncbi.nlm.nih.gov/genbank/] [Table 1]. The NCBI GenBank database was queried for the consensus sequences of each PCR product using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence similarity >98% and query coverage =100% were used as thresholds to identify species. ClustalX software [38] was used to align edited sequences. A COI reference library was constructed using six sparid species sequences obtained by us and six downloaded from GenBank [Table 1]. Subsequently, twenty-one unchecked (Xn) sequences were added to the reference COI sequences and a Maximum Likelihood (ML) tree was built by MEGA X software [39]. The bootstrap method was applied using 1000 non-parametric replicates [40].
2.3. In silico Restriction Analysis and COIBar-RFLP
COI reference sequences of each investigated species were aligned using MEGA X [39] and then were uploaded to NEBcutter 3.0.15 (https://nc3.neb.com/NEBcutter/) to choose the most suitable restriction enzyme. The enzymes HinfI, AluI, MboI, and MspI (New England Biolabs, Inc.) were selected to perform COIBar-RFLP (Table 3). The fragments produced by the in silico digestion in each species are shown in Table 4. Only one enzyme was selected and used to digest the COI barcode amplicons obtained from the reference and unchecked samples. The digestion reaction and the visualization of the obtained fragments were carried out as performed by Ferrito et al. [41].
Table 3: List of the enzymes used in this study, their restriction sites (*), and their temperatures of use.
| Enzymes | Restriction sites | Temperature (°C)/time |
|---|---|---|
| HinfI | G*ANT*C | 37°C/1 h |
| AluI | AG*CT | 37°C/1 h |
| MboI | *GATC | 37°C/1 h |
| MspI | C*CGG | 37°C/1 h |
Table 4: Fragments greater than 100bp produced by the digestion of the selected enzymes (lengths in base pair) for each of the investigated species in this study.
| Diplodus sargus | Diplodus vulgaris | Pagellus acarne | Pagellus erythrinus | Spondilosoma cantharus | Pagrus pagrus | |
|---|---|---|---|---|---|---|
| HinfI | 376; 258 | ND | 564 | ND | 564 | 303; 282 |
| AluI | 252; 195; 165 | 187; 165; 158 | 244; 207; 159 | 252; 155; 144; 102 | 347; 187 | 182; 146; 102 |
| MboI | 376; 207 | 393; 170 | 244; 207; 204 | 421; 207 | 448; 207 | 484; 101 |
| MspI | 270; 153; 107 | 270; 246 | 210 | 338; 228 | 246; 234 | 293; 228 |
* ND, not digested.
3. RESULTS
3.1. COI Barcode
All COI gene fragments of 654 bp [Figure 1] resulted from functional mitochondrial sequences and not pseudogenes, not including insertions, deletions, or stop codons [42]. The COI sequences of the reference samples allowed to identify six sparid species confirming their morphological identification [Table 1]. The BLAST search in the GenBank database revealed 98.71 to 99.85% sequence identity for the PCR products to Pagellus erythrinus, Pagrus pagrus, Diplodus sargus, Spondyliosoma cantharus, Diplodus vulgaris, and Pagellus acarne [Table 2]. The COI sequences of the samples morphologically unidentified yielded the detection of the following six species by BLAST search in the GenBank database: P. erythrinus (n = 3), P. pagrus (n = 7), D. sargus (n = 3), S. cantharus (n = 3), D. vulgaris (n = 2), and P. acarne (n = 3) [Table 2]. Two cases of mislabeling were observed: P. pagrus in place of P. erythrinus and S. cantharus in place of D. sargus.
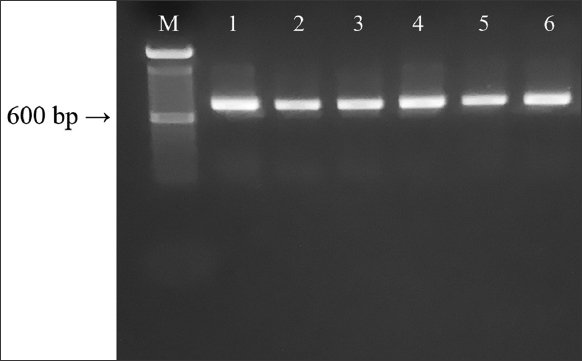 | Figure 1: Amplification products of the species examined in this study. M= 100bp ladder. Lane 1 = Diplodus sargus, Lane 2 = Diplodus vulgaris, Lane 3 = Pagellus acarne, Lane 4 = Pagellus erythrinus, Lane 5 = Spondyliosoma cantharus, Lane 6 = Pagrus pagrus. [Click here to view] |
To confirm the species revealed by BLAST, an ML tree was built using the 27 COI sequences of all aforementioned species and six additional sequences downloaded from GenBank. All species clustered into different groups corresponding to the six species matched from the BLAST search [Figure 2].
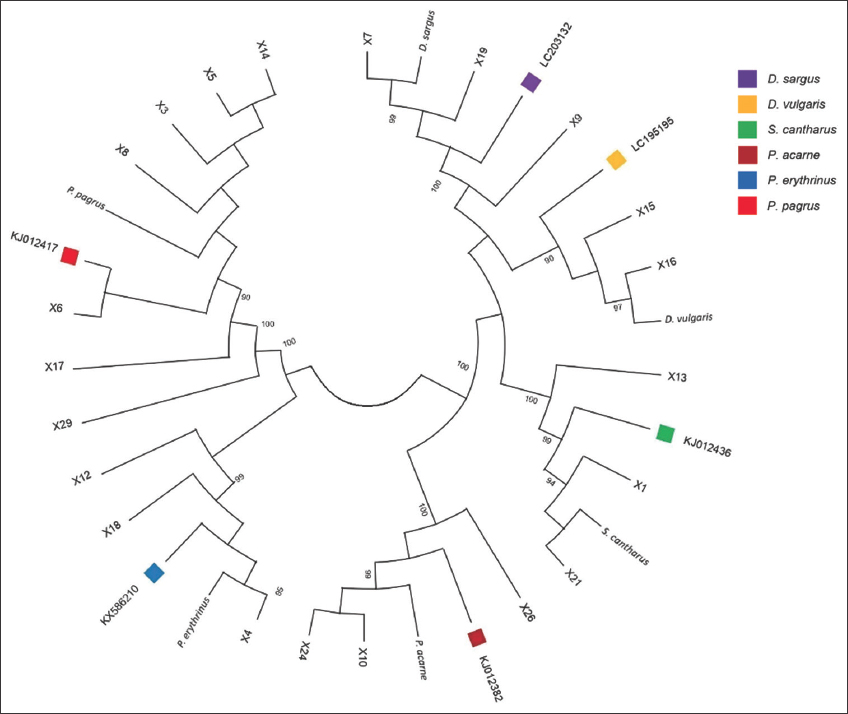 | Figure 2: Circular maximum likelihood (ML) tree constructed using COI sequences from sparid specimens and those downloaded from GenBank for each targeted species. Only bootstrap values greater than 70% are showed. [Click here to view] |
3.2. COIBar-RFLP
In silico analysis allowed to selection of the enzyme able to generate a differential restriction pattern for all tested sparid species: the fragments greater than 100 bp were reported in Table 4. In addition, Figure 3 shows the in silico MspI restriction patterns with all the virtual bands (greater and smaller than 100bp) visualized for each sparid species.
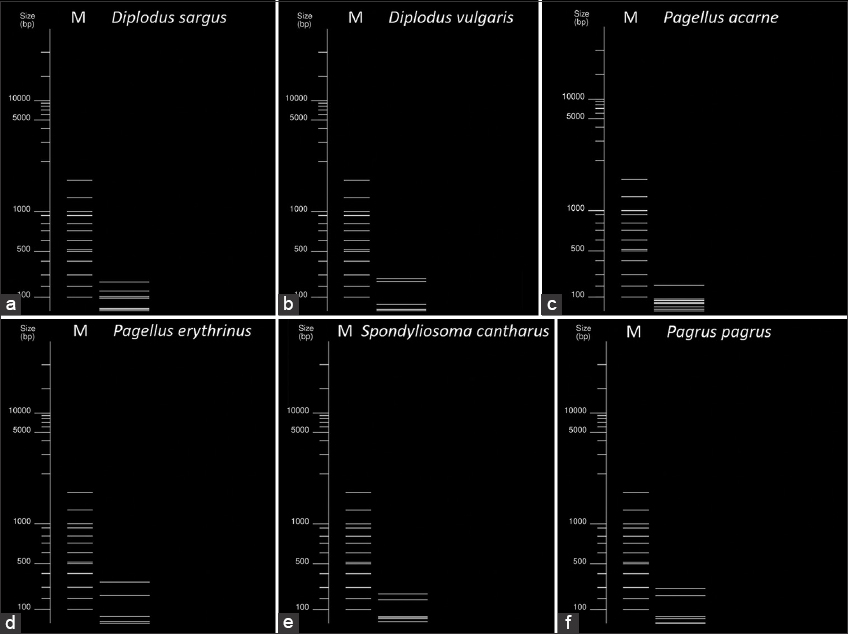 | Figure 3: In silico MspI restriction pattern of COI amplicons for sparid reference species in this study. “M” indicates the 100bp ladder used as a reference. Following are the sizes of each restriction band obtained in each species. (a) Diplodus sargus: 250,153,107,90,34 bp. (b) Diplodus vulgaris: 280,250, 74,50 bp. (c) Pagellus acarne: 210, 95, 85, 77, 70, 57, 40, 20 bp. (d) Pagellus erythrinus: 340, 220, 64, 30 bp. (e) Spondyliosoma cantharus: 260, 194, 70, 60, 50, 20 bp. (f) Pagrus pagrus: 290, 234, 70, 50, 10 bp. In each case, the sum of the individual band sizes is 654 bp. [Click here to view] |
The in silico patterns were confirmed by the in vitro digestion with MspI visualized in Figure 4. COI amplicon restriction yielded three fragments of 270, 153, and 107 bp in D. sargus. A single fragment of about 200 bp was observed in P. acarne, while in the remaining species, two fragments of different lengths were detected. In particular, P. erythrinus yielded a longer fragment of 340 bp and a shorter one of 220 bp while two fragments of 290 bp and 234 bp were produced by digestion of COI amplicon of P. pagrus. Finally, the two species D. vulgaris and S. cantharus shared the same fragment of 250 bp but the second fragment, which appears almost overlapping in the gel, was species-diagnostic being 280 bp for D. vulgaris and 220 bp for S. cantharus. The same specific pattern was obtained when COIBar-RFLP was applied in all samples identified only through a molecular approach.
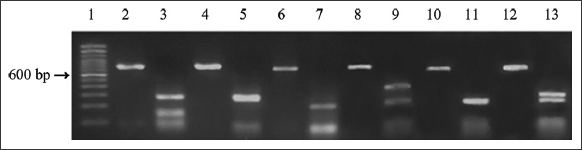 | Figure 4: In vitro MspI restriction pattern of COI amplicons for each sparid species in this study. Lane 1 = 100 bp ladder. Lane 2 = D. sargus (not digested amplicon, 654 bp). Lane 3 = D. sargus (digested amplicon, 270-153-107 bp). Lane 4 = D. vulgaris (not digested amplicon, 654bp). Lane 5 = D. vulgaris (digested amplicon, 280-250 bp). Lane 6 = P. acarne (not digested amplicon, 654 bp). Lane 7= P. acarne (digested amplicon, 210 bp). Lane 8 = P. erythrinus (not digested amplicon, 654 bp). Lane 9 = P. erythrinus (digested amplicon, 340-220 bp). Lane 10 = S. cantharus (not digested amplicon, 654 bp). Lane 11 = S. cantharus (digested amplicon, 250-220 bp). Lane 12 = P. pagrus (not digested amplicon, 654 bp). Line 13 = P. pagrus (digested amplicon, 290-250 bp). The fragments smaller than 100 bp were not taken into consideration for species identification. [Click here to view] |
4. DISCUSSION
The COI barcode sequencing of fresh whole samples of sparid species commonly sold in several local fish markets and supermarkets in Southern Italy, confirmed the detection of six species previously identified through morphological inspection using analytical keys. The COIBar-RFLP strategy applied to the COI amplicons of these reference samples allowed us to obtain different digestion patterns of the restriction enzyme MspI useful to discriminate all species simultaneously. In addition, the COI amplicons of the samples (11 fillets and 10 whole specimens), selected on the basis of the reference sparid species reported on the label, were analyzed by COIBar-RFLP using the MspI restriction enzyme that successfully discriminated all species. Considering that (i) no morphological identification was conducted for these specimens and (ii) the restriction pattern obtained confirmed that of the reference specimens, it can be stated that the advantage of the COIBar-RFLP strategy could be twofold: First, it is possible to quickly verify the correctness of the species declared on the label of the commercial products examined and Second, it is possible to achieve this by skipping the sequencing phase of the amplicons obtained, thus saving time and money. Five cases of mislabeling (24% of cases) were detected in which P. pagrus was found instead of P. erythrinus (two cases) and S. cantharus instead of D. sargus (three cases). The percentage of mislabeling we detected is slightly lower than the overall weighted rate of mislabeling (28.4%) in fish products sold on the Italian market (Giusti et al. 2023). Based on the sales prices of sparid species, we sampled (see Table S1 in Supplementary Materials) the substitution of S. cantharus in place of D. sargus, can be considered intentional, due to the obvious economic gain from the fraudulent replacement, while the substitution of P. pagrus for P. erythrinus can be considered unintentional since the two species were sold at the same price and are morphologically very similar. Moreover, as very similar sparid species are often distinguished on the basis of dentition alone [43], substitution fraud is hardly detectable by the consumer. This issue has been addressed in several studies that explored the efficacy of various molecular markers in discriminating sparid species. More specifically, the efficacy of DNA Inter-simple Sequence Repeat markers was tested in the identification of four species of Mediterranean sea bream and the Mediterranean common snapper [44]. Authentication of sparid fish species has also been achieved by sequencing the PCR products, Polymerase Chain Reaction–Single Strand Conformation Polymorphism (PCR-SSCP), and IsoElectric Focusing (IEF) [45]. More recently, the complete mitochondrial DNA of Dentex gibbosus [46], P. acarne [47], Dentex dentex [48], P. erythrinus [49], and Diplodus puntazzo [50] has been sequenced to provide useful genetic information for species identification. In particular, the comparison of the known sequences of the complete mitochondrial genome of sparid species [27,28,51] allowed to obtain new barcode genes to be used in place of the classic COI and Cytb which were considered less effective for the identification of sparid species. However, it should be emphasized that new barcode genes would have to be tested on a large number of species before being used in the forensic field. In contrast, the efficacy of COI barcode sequences in discriminating 75 sparid species was demonstrated by Armani et al. [26] in a comprehensive study aimed at testing the complete COI barcode for the identification of sparid species, also highlighting the effectiveness of the COI mini barcode for discriminating species in highly processed commercial products.
The simplicity and ease of use of PCR-RFLP together with its low cost are certainly the most advantageous aspects of this technique which is widely used in food control laboratories [52]. Indeed, a high degree of specialization or experience in the application of the molecular biology technique is not required to perform it, nor the use of expensive and sophisticated instruments. However, the disadvantages of this strategy cannot be ignored either, which lie in the choice of the specific restriction enzyme and processing conditions, the poor performance of incorrectly stored enzymes, and the need to use more than one enzyme in the same reaction to increase the discriminating power for all the species studied. This particular problem increases the processing times and costs of the methodology [53]. For these reasons, the criterion we used for our COIBar-RFLP was to select a single restriction enzyme, which simultaneously and successfully discriminated all target species both in the present study and in similar studies we had previously conducted [54-58].
5. CONCLUSION
The need to protect consumers from the adulteration of seafood products is explicitly stated in Annex II of the European Commission Recommendation (12.3.2015), according to which in each Member states “Competent authorities should carry out official controls in order to establish whether fish species found in unprocessed or processed fishery and aquaculture products complies with the species that is declared on the label or in other means of information accompanying the food product.”
In this context, our contribution focused on the COIBar-RFLP method as a reliable molecular strategy to identify fish species in processed products. Six sparid species were recognized using the differential digestion pattern produced by the restriction enzyme MspI.
The proposed methodology could be used in food control laboratories to combat the widespread habit of fraudulent species substitution in the fishing industry. At least two important implications derive from this problem, the first is purely economic to the detriment of consumers and the second concerns consumer health due to the risk of allergic reactions as demonstrated by the existence of cases of monosensitivity to single species of sparids [59].
6. ACKNOWLEDGMENT
A special thanks to Dr. Samira Vinjau, Mr. Andrea Amore, and the fish market “Il Delfino” for providing the reference specimens.
7. AUTHOR CONTRIBUITIONS
Venera Ferrito: Conceptualization, writing original draft, reviewing, editing, and funding acquisition. Marta Giuga: methodology, analyzing data, reviewing and editing. Giada Santa Calogero: methodology and analyzing data. Anna Maria Pappalardo: conceptualization, methodology, analyzing data, writing original draft, reviewing, editing, and supervision.
8. FUNDING
This research was funded by the University of Catania: PIACERI linea 2, 2022.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
In this study, no in vivo experiments on animals were performed. Tissues used for DNA extraction were pieces of processed fish products (fillets or whole specimens) purchased at fish markets and supermarkets. Therefore, approval from the Ethical Committee is not required.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Donlan CJ, Luque GM. Exploring the causes of seafood fraud:A meta-analysis on mislabeling and price. Mar Policy 2019;100:258-64. [CrossRef]
2. Cermakova E, Lencova S, Mukherjee S, Horka P, Vobruba S, Demnerova K, et al. Identification of fish species and targeted genetic modifications based on DNA analysis:State of the art. Foods 2023;12:228. [CrossRef]
3. Lawrence S, Elliott C, Huisman W, Dean M, van Ruth S. The 11 sins of seafood:Assessing a decade of food fraud reports in the global supply chain. Compr Rev Food Sci Food Saf 2022;21:3746-69. [CrossRef]
4. Kroetz K, Luque GM, Gephart JA, Jardine SL, Lee P, Chicojay Moore K, et al. Consequences of seafood mislabeling for marine populations and fisheries management. Proc Natl Acad Sci U S A 2020;117:30318-23. [CrossRef]
5. Mohammadabadi MR, Shaikhaev GO, Sulimova GE, Rahman O, Mozafari MR. Detection of bovine leukemia virus proviral DNA in Yaroslavsl, Mongolian and black pied cattle by PCR. Cell Mol Biol Lett 2004;9:766-8.
6. Shahdadnejad N, Mohammadabadi MR, Shamsadini M. Typing of Clostridium perfringens isolated from broiler chickens using multiplex PCR. Genet Third Millennium 2016;14:4368-74.
7. Mohammadabadi MR, Soflaei M, Mostafavi H, Honarmand M. Using PCR for early diagnosis of bovine leukemia virus infection in some native cattle. Genet Mol Res 2011;10:2658-63. [CrossRef]
8. Ahsani MR, Mohammadabadi MR, Shamsaddini MB. Clostridium perfringens isolate typing by multiplex PCR. J Venom Anim Toxins Incl Trop Dis 2010;16:573-8. [CrossRef]
9. Mohammadabadi MR. Inter-simple sequence repeat loci associations with predicted breeding values of body weight in kermani sheep. Genet Third Millennium 2016;14:4383-90.
10. Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life:Cytochrome c oxidase subunit 1 divergence, among closely related species. Proc Biol Sci 2003;270:S96-9.
11. Mohammadabadi MR, Torabi A, Tahmourespoor M, Baghizadeh A, Esmailizadeh K, Mohammadi A. Analysis of bovine growth hormone gene polymorphism of local and Holstein cattle breeds in Kerman province of Iran using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). Afr J Biotechnol 2010;9:6848-52.
12. Rohallah A, Mohammadreza MA, Shahin MB. Kappa-casein gene study in Iranian Sistani cattle breed (Bos indicus) using PCR-RFLP. Pak J Biol Sci 2007;10:4291-4. [CrossRef]
13. Aranishi F. Rapid PCR-RFLP method for discrimination of imported and domestic mackerel. Mar Biotechnol (NY) 2005;7:571-5. [CrossRef]
14. Mata W, Chanmalee T, Punyasuk N, Thitamadee S. Simple PCR-RFLP detection method for genus- and species-authentication of four types of tuna used in canned tuna industry. Food Control 2020;108:106842. [CrossRef]
15. Rahat MA, Haris M, Ullah Z, Ayaz SG, Nouman N, Rasool A, et al. Domestic animals'identification using PCR-RFLP analysis of cytochrome B gene. Adv Life Sci 2020;7:113-6.
16. Yao L, Lu J, Qu M, Jiang Y, Li F, Guo Y, et al. Methodology and application of PCR-RFLP for species identification in tuna sashimi. Food Sci Nutr 2020;8:3138-46. [CrossRef]
17. Khalid A, Imran M, Ali A, Muzammil S, Badar M, Hayat S, et al. Molecular marker (PCR-RFLP) assisted identification of meat species by mitochondrial cytochrome C oxidase subunit I (COI) gene. J Anim Plant Sci 2022;32:1724-30.
18. Giusti A, Malloggi C, Tosi F, Boldini P, Larrain Barth MA, Araneda C, et al. Mislabeling assessment and species identification by PCR-RFLP of mussel-based products (Mytilus spp.) sold on the Italian market. Food Control 2022;134:108692. [CrossRef]
19. Fricke R, Eschmeyer WN, Fong JD. Eschmeyer's Catalog of Fishes. California:Caliornia Academy of Science;2023.
20. Russo T, Costa C, Cataudella S. Correspondence between shape and feeding habit changes throughout ontogeny of gilthead sea bream Sparus aurata L., 1758. J Fish Biol 2007;71:629-56. [CrossRef]
21. Fruciano C, Tigano C, Ferrito V. Body shape variation and colour change during growth in a protogynous fish. Environ Biol Fishes 2012;94:615-22. [CrossRef]
22. Franchini P, Fruciano C, Spreitzer ML, Jones JC, Elmer KR, Henning F, et al. Genomic architecture of ecologically divergent body shape in a pair of sympatric crater lake cichlid fishes. Mol Ecol 2014;23:1828-45. [CrossRef]
23. Moreira C, Froufe E, Vaz-Pires P. Triay-Portella R, Correia AT. Landmark-based geometric morphometrics analysis of body shape variation among populations of the blue jack mackerel, Trachurus picturatus, from the North-East Atlantic. J Sea Res 2020;163:101926. [CrossRef]
24. Antonucci F, Costa C, Aguzzi J, Cataudella S. Ecomorphology of morpho-functional relationships in the family of Sparidae:A quantitative statistic approach. J Morphol 2009;270:843-55. [CrossRef]
25. Seyoum S, Adams DH, Matheson RE, Whittington JA, Alvarez AC, Sheridan NE, et al. Genetic relationships and hybridization among three western atlantic sparid species:Sheepshead (Archosargus probatocephalus), sea bream (A. rhomboidalis) and Pinfish (Lagodon rhomboides). Conserv Genet 2020;21:161-73. [CrossRef]
26. Armani A, Guardone L, Castigliego L, D'Amico P, Messina A, Malandra R, et al. DNA and Mini-DNA barcoding for the identification of porgies species (Family Sparidae) of commercial interest on the international market. Food Control 2015;50:589-96. [CrossRef]
27. Ceruso M, Mascolo C, Anastasio A, Pepe T, Sordino P. Frauds and fish species authentication:Study of the complete mitochondrial genome of some Sparidae to provide specific barcode markers. Food Control 2019;103:36-47. [CrossRef]
28. Ceruso M, Mascolo C, De Luca P, Venuti I, Smaldone G, Biffali E, et al. Arapid method for the identification of fresh and processed Pagellus erythrinus species against frauds. Foods 2020;9:1397. [CrossRef]
29. Pappalardo AM, Ferrito V. DNA barcoding species identification unveils mislabeling of processed flatfish products in Southern Italy Markets. Fish Res 2015;164:153-8. [CrossRef]
30. Pappalardo AM, Cuttitta A, Sardella A, Musco M, Maggio T, Patti B, et al. DNA barcoding and COI sequence variation in Mediterranean lanternfishes larvae. Hydrobiologia 2015;749:155-67. [CrossRef]
31. Pappalardo AM, Copat C, Ferrito V, Grasso A, Ferrante M. Heavy metal content and molecular species identification in canned tuna:Insights into human food safety. Mol Med Rep 2017;15:3430-7. [CrossRef]
32. Pappalardo AM, Copat C, Raffa A, Rossitto L, Grasso A, Fiore M, et al. Fish-based baby food concern-from species authentication to exposure risk assessment. Molecules 2020;25:3961. [CrossRef]
33. Pappalardo AM, Raffa A, Calogero GS, Ferrito V. Geographic pattern of sushi product misdescription in Italy-a crosstalk between citizen science and DNA barcoding. Foods 2021;10:756. [CrossRef]
34. Fischer W, Bauchot ML, Schneider M. Fiches FAO D'identification des Espèces Pour les Besoins de la Pêche. (Révision 1). Méditerranée et mer Noire. Zone de Pêche 37. Vertébrés. Publication Préparée par la FAO, Résultat d'un Accord Entre la FAO et la Commission des Communautés Européennes (Projet GCP/INT/422/EEC) Financée Conjointement par Ces Deux Organisations. Vol. 2. Rome:FAO;1987. 761-1530.
35. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. DNA barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci 2005;360:1847-57. [CrossRef]
36. Pappalardo AM, Federico C, Sabella G, Saccone S, Ferrito V. A COI nonsynonymous mutation as diagnostic tool for intraspecific discrimination in the european anchovy Engraulis encrasicolus (Linnaeus). PLoS One 2015;10:e0143297.
37. Messing J. New M13 vectors for cloning. Methods Enzymol 1983;101:20-78. [CrossRef]
38. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface:Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876-82. [CrossRef]
39. Stecher G, Tamura K, Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 2020;37:1237-9. [CrossRef]
40. Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution 1985;39:783-91. [CrossRef] [CrossRef]
41. Ferrito V, Bertolino V, Pappalardo AM. White fish authentication by COIBar-RFLP:toward a common strategy for the rapid identification of species in convenience seafood. Food Control 2016;70:130-7. [CrossRef]
42. Zhang DX, Hewitt GM. Nuclear integrations:challenges for mitochondrial DNA markers. Trends Ecol Evol 1996;11:247-51. [CrossRef]
43. Parenti P. An annotated checklist of the fishes of the family Sparidae. J Fish Taxonomy 2019;4:47-98.
44. Casu M, Lai T, Curini-Galletti M, Ruiu A, Pais A. Identification of Mediterranean Diplodus spp. and dentex dentex (Sparidae) by means of dna inter-simple sequence repeat (ISSR) markers. J Exp Mar Biol Ecol 2009;368:147-52. [CrossRef]
45. Schiefenhövel K, Rehbein H. Differentiation of Sparidae species by DNA sequence analysis, PCR-SSCP and IEF of sarcoplasmic proteins. Food Chem 2013;138:154-60. [CrossRef]
46. Mascolo C, Ceruso M, Palma G, Anastasio A, Pepe T, Sordino P. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes:Sparidae). Mitochondrial DNA B Resour 2018;3:525-6. [CrossRef]
47. Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes:Sparidae). Mitochondrial DNA B Resour 2018;3:434-5. [CrossRef]
48. Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes:Sparidae). Mitochondrial DNA B Resour 2018;3:391-2. [CrossRef]
49. Ceruso M, Mascolo C, Lowe EK, Palma G, Anastasio A, Sordino P, et al. The complete mitochondrial genome of the common pandora Pagellus erythrinus (Perciformes:Sparidae). Mitochondrial DNA B Resour 2018;3:624-5. [CrossRef]
50. Ceruso M, Venuti I, Osca D, Caputi L, Anastasio A, Crocetta F, et al. The complete mitochondrial genome of the sharpsnout seabream Diplodus puntazzo (Perciformes:Sparidae). Mitochondrial DNA B Resour 2020;5:2379-81. [CrossRef]
51. Ceruso M, Mascolo C, De Luca P, Venuti I, Biffali E, Ambrosio RL, et al. Dentex dentex Frauds:Establishment of a new DNA barcoding Marker. Foods 2021;10:580. [CrossRef]
52. Griffith AM, Sotelo CG, Mendes R, Perez Martin RI, Schroder U, Shorten M, et al. Current methods for seafood authenticity testing in Europe:Is there a need for harmonisation?Food Control 2016;45:95-100.
53. Hashim HO, Al-Shuhaib MB. Exploring the potential and limitations of PCR-RFLP and PCR-SSCP for SNP detection:A review. J Appl Biotechnol Rep 2019;6:137-44. [CrossRef]
54. Pappalardo AM, Ferrito V. A COIBar-RFLP strategy for the rapid detection of Engraulis encrasicolus in processed anchovy products. Food Control 2015;57:385-92. [CrossRef]
55. Pappalardo AM, Federico C, Saccone S, Ferrito V. Differential flatfish species detection by COIBar-RFLP in processed seafood products. Eur Food Res Technol 2018;244:2191-201. [CrossRef]
56. Pappalardo AM, Petraccioli A, Capriglione T, Ferrito V. From fish eggs to fish name:Caviar species discrimination by COIBar-RFLP, an efficient molecular approach to detect fraudulent caviar trade. Molecules 2019;24:2468. [CrossRef]
57. Ferrito V, Raffa A, Rossitto L, Federico C, Saccone S, Pappalardo AM. Swordfish or shark slice?A rapid response by COIBar-RFLP. Foods 2019;8:537. [CrossRef]
58. Pappalardo AM, Giuga M, Raffa A, Nania M, Rossitto L, Calogero GS, et al. COIBar-RFLP molecular strategy discriminates species and unveils commercial frauds in fishery products. Foods 2022;11:1569. [CrossRef]
59. Taylor SL, Kabourek JL, Hefle SL. Fish allergy:Fish and products thereof. J Food Sci 2004;69:R175-80.