1. INTRODUCTION
Dengue fever is recognized as the most significant arthropod-borne zoonotic disease [1], with an estimated 10,000 fatalities and roughly 100 million cases requiring hospitalization annually [2]. The incidence of dengue has been on a consistent rise over the years [3]. Concurrently, during the COVID-19 pandemic, a notable increase in dengue infections was observed in various American countries, including Argentina and Bolivia. In 2022, the reported cases reached 2.8 million [4]. However, in just the first half of 2023, there were reports of more than 3 million cases [5].
Dengue infection is caused by the dengue viruses from the Flaviviridae family. The four recognized serotypes of dengue viruses elicit complex immune responses in humans, posing significant challenges in the development of multivalent vaccines effective against all serotypes [6]. Clinical manifestations of dengue infection can range from asymptomatic to mild fever; however, severe complications can also arise. The most severe forms of the disease, dengue hemorrhagic fever (DHF) and dengue shock syndromes, are often the result of a secondary infection with a different serotype—for instance, an initial infection by dengue serotype 1 followed by a subsequent infection with serotype 2, 3, or 4 [1]. These complications have been linked to the phenomenon of antibody-dependent enhancement, wherein preexisting antibodies to an earlier serotype fail to neutralize a new infecting serotype, instead facilitating viral entry into host immune cells [7]. There is no specific treatment for dengue, and management is primarily supportive and symptomatic. While some vaccines, such as CYD-TDV (by Sanofi Pasteur), TV003 and TV005 (by NIAID), and TAK-003 (by Takeda), have either been licensed or are in phase 3 clinical trials, safety concerns, particularly in children, have limited their widespread use [8,9].
The dengue virus genome comprises a positive sense, single-stranded RNA that spans approximately 11 kb. It includes a single open-reading frame that encodes three structural proteins: the capsid (C), membrane (M), and envelope (E) glycoproteins, as well as seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) [1]. Research has intensely focused on the E and NS1 proteins, acknowledging their critical functions in vaccine development and diagnostic applications, respectively [10-12]. The E protein is integral to the virion’s architecture and facilitates the virus’s entry into host cells through receptor-mediated endocytosis [10-12]. On the other hand, NS1 is a multifaceted protein associated with the pathogenesis of dengue, influencing viral replication and contributing to the vascular leakage syndrome, a defining feature of severe dengue infection [12].
In its native state within the cell, NS1 is first synthesized as monomers, which subsequently associate into dimers in the rough endoplasmic reticulum. Beyond its intracellular role, NS1 is also secreted as hexameric lipo-particles into the bloodstream of infected individuals, where it can be detected in high concentrations during the acute phase of the disease [12,13]. The detection of NS1-specific IgG and IgM antibodies serves as an effective diagnostic tool. Notably, the ratio of these antibodies has been identified as a reliable biomarker for distinguishing between primary and secondary infections, which is pivotal for clinical prognosis and management [14].
The genetic diversity within each dengue virus serotype is profound, with further subdivisions such as genotypes, strain variations, and quasi-species compounding the complexity [15]. This immense variation presents a formidable challenge in the development of effective multivalent vaccines and the creation of universal diagnostic kits. The variability confounds the immune response and hinders the identification of viral markers necessary for consistent vaccine efficacy and diagnostic accuracy.
To address this issue, a common strategy involves the generation of consensus sequences for each dengue serotype or even a unified consensus across all serotypes. Consensus sequences are derived through various methods and analytical approaches, yet the underlying goal remains the same: to distill a genetic sequence that captures the core attributes shared across the vast diversity of dengue viruses. These consensus sequences aim to identify invariant or conserved regions that are less susceptible to genetic drift and are present in most viral variants [16-22]. By focusing on these conserved elements, researchers hope to formulate vaccines capable of eliciting a broad and protective immune response and develop widely applicable diagnostic tests that maintain their reliability despite the genetic variation among viral strains.
The pursuit of consensus sequences is not without challenges. It requires meticulous genetic analysis and the reconciliation of data from numerous viral isolates to ascertain the genetic motifs that are universally present. Despite these challenges, the creation of consensus sequences remains a cornerstone in the quest to mitigate the public health threat posed by dengue as it holds the promise of leading to vaccines and diagnostic tools that are both robust and broadly applicable, a critical need for a disease that affects millions globally. In this vein, we have developed a straightforward method to generate global consensus sequences for the NS1 protein of all four dengue serotypes. We codon optimized, synthesized, and expressed these genes in several Escherichia coli strains and identified the most robust system for the production of these proteins. We were able to purify the proteins to homogeneity. Our subsequent Western blot analysis utilizing antisera from outpatient dengue cases alongside those from hospitalized, severe acute dengue cases demonstrated the efficacy of our antigens in discriminating serotypes across the spectrum of disease severity.
2. MATERIALS AND METHODS
2.1. Strains and Culture Conditions
All cloning work was carried out using the E. coli TOP10 strain. For the expression of codon-optimized, dengue global consensus NS1 proteins, we utilized QIAGEN’s M15 strain, as well as Agilent’s BL21 CodonPlus-De3-RIPL strain. The latter contains additional copies of argU, ileY, proL, and leuW tRNA genes, which aid in the expression of heterologous proteins with rare codons. Additionally, Novagen’s Rosetta DE3 strain, a derivative of BL21 engineered for improved expression of eukaryotic genes, was used to evaluate the solubility of the target proteins.
Maintenance of E. coli TOP10 strain was in Luria Broth (LB) and on LB agar plates. The BL21-Codonplus-De3-RIPL (hereon termed BL21 for convenience) was sustained in LB supplemented with chloramphenicol (CAM, 34 μg/ml), streptomycin (strep, 50 μg/ml), while the Rosetta De3 was maintained in LB with CAM (34 μg/ml). Selection of transformed strains was achieved by adding ampicillin (50 μg/ml) to their respective media. Agar plates were incubated at 37°C. Liquid cultures were grown at 200 rpm and 37°C in shaking incubators. Protein expression in the M15 strains was carried out at 37 and 20°C for 8 h, and in the BL21 strains was carried out at 15°C for 24 h. For the Rosetta strain, induction was conducted at both 37 and 20°C for 8 h to determine the optimal conditions for solubility.
2.2. Consensus Sequence Generation and Gene Optimization
The full polyprotein sequences for each dengue serotype were initially obtained from the UniProt database. These sequences served as the query for a BLASTp (protein BLAST) search to identify homologous protein sequences across the database. The retrieved sequences, in fasta format, were then screened to exclude any sequences with ambiguous amino acids, denoted by “X.” Subsequently, the sequences corresponding to each serotype were aligned using MUSCLE algorithm [23] from MEGA 11 program [24]. From these alignments, the NS1 protein sequences were isolated and preserved in fasta format for further analysis.
To visualize the consensus sequences, the alignments of NS1 proteins were uploaded to WebLogo (https://weblogo.berkeley.edu/logo.cgi). Concurrently, consensus sequences were generated using the EMBOSSCons tool (https://www.ebi.ac.uk/Tools/msa/emboss_cons/). The consensus sequences then served as the basis for additional BLAST searches against the NCBI database to confirm the presence of identical sequences.
For gene synthesis, the NS1 consensus sequences were subjected to codon optimization using the IDT Codon Optimization Tool (https://sg.idtdna.com/CodonOpt). This process yielded several gene variants for each NS1 protein. These variants were then analyzed for Codon Adaptation Index (CAI), mRNA secondary structure stability, GC content, and GC content at the third codon position utilizing the Visual Gene Developer software [25]. The selection criteria favored sequences with a high CAI and low mRNA folding energy [26]. Ultimately, one codon-optimized sequence for the NS1 gene from each serotype was chosen for synthesis. These genes are henceforth denoted as Denv1-NS1, Denv2-NS1, Denv3-NS1, and Denv4-NS1, respectively, and their corresponding protein products are referred to as DENV1-NS1, DENV2-NS1, DENV3-NS1, and DENV4-NS1, respectively.
To streamline the directional cloning process, restriction sites for BamHI (GGATTC) and SalI (GTCGAC) were engineered into the 5′ and 3′ ends of the synthetic genes. Gene synthesis, including cloning into pUC plasmids, was commissioned from PhusaGenomics (https://phusagenomics.com). Verification of the synthetic genes’ integrity was ensured by Sanger sequencing, confirming the absence of errors in the sequences.
2.3. Cloning and Expression Analysis
We excised all four synthetic genes from the pUC19 vector using BamHI/SalI enzymes (Thermo Fisher Scientific, FastDigest, Cat# FD0054 and FD0644) and purified them from the agarose gel with the Favorgen Gel/PCR Purification Kit. These genes were then cloned into three expression vectors: QIAGEN’s pQE30, Takara’s pColdII, and NovoPro Bioscience’s pGEX4-T1. Ligation was performed at 25°C for 1 h and then at 4°C for another hour using T4 DNA ligase (Thermo Fisher Scientific, EL0011). We transformed 10 µl of the ligation mixture into E. coli TOP10 cells and plated them on LB agar plates with ampicillin. Recombinant colonies were identified using one-step miniprep [27] and restriction enzyme digestion, followed by Sanger sequencing to confirm the absence of errors. These verified plasmids were then transformed into their respective expression hosts: pQE30 into M15, pColdII into BL21 and pGEX4-T1 into Rosetta, creating three expression systems: pQE30/M15 and pColdII/BL21 with a 6xHis tag, and pGEX4-T1/Rosetta with a GST tag. The NS1-His fusion protein has an approximate molecular weight of 40 kDa, while the NS1-GST fusion is around 65 kDa.
Protein expression was initiated as per the manufacturer’s protocols. A single colony was used to start a 5-ml LB culture with the appropriate antibiotics, incubated overnight at 37°C and 200 rpm. The next day, we inoculated 0.5 ml of this culture into 10 ml of fresh LB, again with antibiotics, and grew it to an OD600 of 0.5–0.7. Induction in pQE30/M15 and pGEX4-T1/Rosetta cultures was immediate upon reaching the desired OD600 at either 37 or 20°C for 8 h. For pColdII/BL21, a 30-min cold shock at 15°C preceded induction at the same temperature and the induction took 24 h. An initial IPTG concentration of 0.5 mM was used for induction. Expression of NS1 was verified by SDS-PAGE (12%), with the appearance of bands at the expected molecular weights indicating successful expression.
To assess the solubility of the recombinant NS1 proteins, we subjected the induced cells to freeze–thaw cycles as described by Johnson and Hecht [28]. In brief, 3–5 ml of the induced cell culture was harvested, washed with distilled water, and resuspended in 80 µl of lysis buffer (50 mM NaH.PO., 300 mM NaCl, pH 8.0). This cell suspension was subjected to eight cycles of rapid freezing in –80°C alcohol for 2 min and subsequent thawing on ice at 0°C for 8 min, with a final 30 min rest on ice. The lysate was then centrifuged at 13,500 rpm for 10 min at 4°C to separate insoluble and soluble fractions. The insoluble fraction (pellet) was redissolved in denaturing lysis buffer with the same composition but with the addition of 8 M urea. Both fractions were then analyzed using SDS-PAGE and Western blot. For Western blot, we utilized mouse monoclonal antibodies against His Tag and GST (BioLegend, Cat# 362602 and 640820, respectively), and a goat anti-mouse IgG alkaline phosphatase-conjugated secondary antibody (Enzo, Cat# ADI-SAB-101-J).
2.4. Purification and Quantification
Purification of the target proteins was performed under denaturing conditions. For each gram of wet cell weight, 5 ml of denaturing lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 8 M urea, pH 8.0) was used. The cell lysis buffer mixture was stirred at room temperature for 1 h to ensure complete lysis, followed by disruption using an ultrasonicator (Qsonica, Q125). Post ultrasonication, the samples were centrifuged at 13,500 rpm and 4°C for 20 min to pellet cell debris. The clear supernatant was collected, and the pellet was discarded. The supernatant was then filtered using a 0.2-μm syringe filter (Millipore) to remove any remaining particulates.
The filtered supernatant underwent affinity purification on a 1 ml Histrap FF column (Cytiva) controlled by a peristaltic pump. The column was sequentially treated with denaturing lysis buffer containing increasing concentrations of imidazole: 10 mM for binding, 25 mM for washing, and 200 mM for elution. The yields of the purified proteins were quantified by comparing the band intensities on Western blots of the total lysate and the purified fractions using the ImageJ software for densitometry [29].
2.5. Protein Identification by Mass Spectrometry
Purified NS1 proteins were resolved on 12% SDS-PAGE and visualized with Coomassie Blue staining. Bands corresponding to the expected molecular weights were excised for analysis by liquid chromatography-mass spectrometry (LC-MS). The proteins were digested with trypsin and subjected to MALDI-TOF/TOF analysis. Peptide samples were combined with a matrix solution of 10 mg/ml R-cyano-4-hydroxycinnamic acid in 0.1% TFA/50% ACN and applied to a 384-well stainless steel MALDI target plate (Applied Biosystems, Foster City, CA). An ABI 4800 Proteomic Analyzer conducted the MS and MS/MS analyses. The resulting data were searched against the SwissProt and uniprotkb_organisms_Dengue_virus_2023_08_14 databases using GPS Explorer (Applied Biosystems version 3.6) linked to Mascot server (version 2.0, Matrix Science). A protein score above 70 for SwissProt and above 58 for uniprotkb_organisms_Dengue_ virus_2023_08_14 was considered statistically significant (p < 0.05).
2.6. Immunogenicity Assay
We assessed the specificity of our global consensus NS1 antigens in binding antibodies from individuals with dengue virus infections. The study utilized two distinct groups of sera: one from individuals who developed severe, acute dengue necessitating hospitalization, and another from individuals with dengue who exhibited milder symptoms. These sera were archived at the Tay Nguyen Institute of Hygiene and Epidemiology. The serotypes of the samples were determined previously by semi-nested PR-PCR [30]. Owing to the retrospective collection of these samples, they were categorized as exempt from further bioethics approval by local regulations. A total of 10 samples for each group were used. Before conducting the Western blot analysis, we utilized the Duo dengue Ag-IgG/IgM Rapid Test (CTK Biotech) to verify the presence of IgM, IgG, and NS1. In the Western blot assays, the human anti-sera acted as the primary antibodies against the purified NS1 proteins. We experimented with sera dilutions of 1:250, 1:500, and 1:750 to determine the ideal primary antibody titer. As secondary antibodies, we employed alkaline phosphatase-conjugated rabbit anti-human IgG and IgM polyclonal antibodies (Bethyl A80-199AP and A80-101AP, respectively) at a dilution of 1:5000. The procedure for Western blot was implemented as described in previous research [31].
3. RESULTS AND DISCUSSION
3.1. Consensus Sequence Generation and Optimization
Utilizing sequences retrieved from NCBI Genpept on June 26, 2020, we successfully created consensus sequences for the NS1 protein of all four dengue serotypes. Supplementary Table 1 summarizes the information of the consensus NS1s and the encoding genes.
Further analysis of the consensus NS1 sequences was conducted for linear epitopes using the IEDB database [https://www.iedb.org]. The experimentally verified linear epitopes of NS1 proteins for the four dengue serotypes showed high sequence identity to our consensus NS1 sequences, as presented in Supplementary Table 2. This finding suggests that our consensus NS1 has the potential for broad utility, extending beyond specific strains or substrains.
To optimize NS1 expression in E. coli, we undertook codon optimization of the genes encoding NS1. Multiple gene variants for each serotype were produced, and selections were made favoring those with the highest CAI and the lowest mRNA folding energy for gene synthesis. Evidence suggests that codon optimization significantly enhances expression levels across various expression hosts, with a high CAI mitigating rare codon issues and low mRNA folding energy promoting ribosomal access and influencing translation speed [24,28,29].
A conventional method for producing recombinant antigens involves cloning and expressing genes from circulating pathogens specific to certain regions. While this approach is convenient, it is susceptible to sequence errors and may not account for genetic variation among strains, limiting the antigens’ applicability due to possible epitope mismatches. With dengue viruses, the substantial variation within serotypes discourages this method. Instead, a consensus approach is preferred, which has become popular for highly mutable viruses like dengue [16-21].
To minimize the potential for human errors in the sequencing and submission of dengue virus sequences, we intentionally selected only complete polyprotein sequences for all dengue serotypes when generating the consensus NS1 sequences. This stringent selection process reduces artifacts that could compromise the quality of our consensus NS1 sequences. By ensuring that each amino acid in the NS1 sequences is the most frequently occurring variant in the database, we infer that our NS1 sequences represent the most prevalent in the database at the time of analysis. Although our method for generating consensus NS1 sequences is relatively straightforward and does not consider the phylogenetic relationships or epitope reclassification as described by Sankaradoss et al. [17], it ensures that the NS1 sequences correspond to the strains most commonly reported worldwide, thereby ensuring the broad applicability of our NS1 proteins.
3.2. Cloning and Expression
We engineered all four genes to incorporate BamHI/SalI restriction sites at their 5′ and 3′ ends, respectively, to facilitate straightforward transfer between different expression plasmids. Initially, we examined NS1 expression in pQE30/M15 and pColdII/BL21 constructs. The pColdII/BL21 system was of particular interest due to its reported efficacy in producing high levels of soluble protein [32]. If soluble protein was not achieved, we proceeded to assess expression in the pGEX4-T1/Rosetta system. For each NS1 construct, four colonies were screened for the presence of a protein band around 40 kDa, the estimated molecular weight of NS1. All colonies exhibited protein bands at the expected molecular weight (data not shown), with one colony from each group being selected for further experiments.
During soluble expression analysis of the four recombinant E. coli strains—each expressing NS1 from a different dengue serotype—the pQE30/M15 constructs did not yield any soluble protein, even after lowering the induction temperature to 20°C [31,33] (data not shown). Likewise, the NS1 proteins remained insoluble in the pColdII/BL21 system [Figure 1]. This trend persisted with the pGEX4-T1/Rosetta constructs, in which the GST Tag was used, as depicted in Figure 2. Consequently, we deduced that NS1 proteins could not be expressed in a soluble form using these systems. This observation is consistent with prior studies using different expression systems, where dengue NS1 expression typically resulted in inclusion bodies, necessitating refolding [34,35]. Although the precise reasons for insolubility remain unclear, the high-level expression of NS1 may partly contribute to this outcome [36].
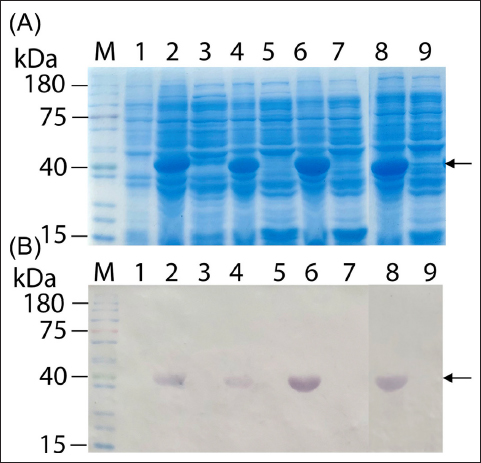 | Figure 1: Evaluation of soluble expression of the global consensus dengue NS1 proteins in pColdII/BL21: (A) SDS-PAGE analysis. Lane M: pre-stained protein ladder (Smobio PM5100); lane 1: pre-induction total protein; lanes 2, 4, 6, and 8: insoluble fractions of DENV1-NS1 to DENV4-NS1, respectively; lanes 3, 5, 7, and 9: soluble fractions of DENV1-NS1 to DENV4-NS1, respectively. (B) Western blot from a twin gel.
[Click here to view] |
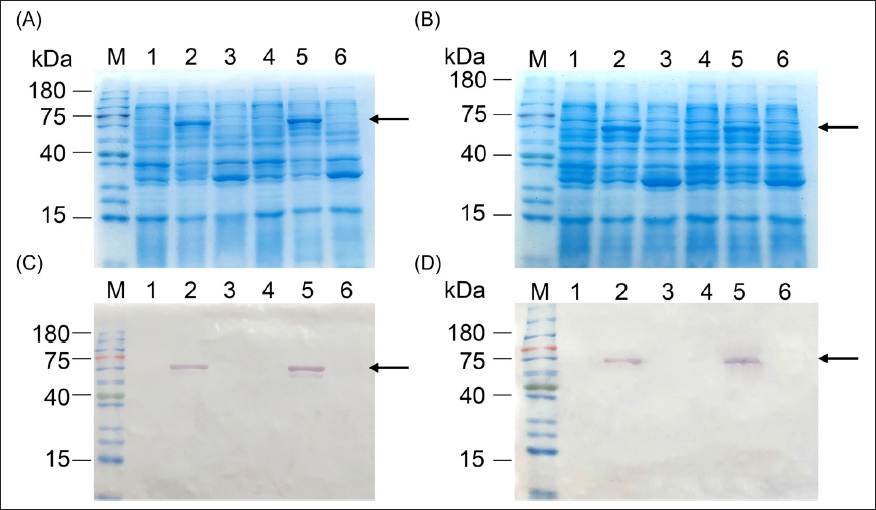 | Figure 2: Evaluation of the consensus dengue NS1 protein solubility using pGEX4-T1/Rosetta: Panels (A) and (B) show SDS-PAGE results while (C) and (D) show the Western blots from the twin gels. Lane M: pre-stained protein ladder (Smobio PM5100). Panels (A) and (C): lanes 1–3 are pre-induced total protein, insoluble fraction, and soluble fractions of induced DENV1-NS1, respectively; lanes 4–6 correspond to pre-induced total protein, insoluble fraction, and soluble fractions of induced DENV2-NS1, respectively. Panels (C) and (D) are of DENV3-NS1 and DENV4-NS1 with the exact same arrangement of pre-induced, insoluble, and soluble fraction.
[Click here to view] |
3.3. Purification and Biological Characterization
The four NS1 proteins were purified to near homogeneity, as demonstrated in Figure 3. Due to the lack of specific antibodies for each NS1 protein, we utilized semi-quantitative Western blot analysis to measure their expression, comparing band intensities to standards—purified NS1 proteins with known concentrations established by the Bradford assay. The expression levels of all NS1 proteins were estimated to be in the range of 40–45 mg/L, a figure that is likely conservative given the low specificity of the anti-His Tag antibody employed (data not shown).
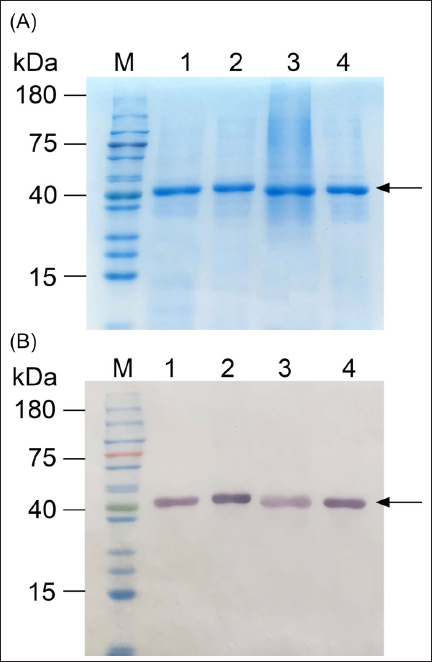 | Figure 3: Panels (A) and (B) SDS-PAGE and Western blot of purified NS1 from four dengue serotypes. Lane M: pre-stained protein ladder (Smobio); lanes 1–4 contain 200 ng of purified NS1 of DENV1 to DENV4, respectively.
[Click here to view] |
The identity of the recombinant NS1 proteins was confirmed using mass spectrometry. The molecular weights obtained were consistent with the theoretical weights, and peptides identified by MS/MS mapped to more than 34% of the NS1 sequences available in the database [Table 1], confirming the absence of frameshift mutations or translational errors.
Table 1: Summary of the identification of the dengue NS1 proteins using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS, MALDI-TOF/TOF).
| Name | Matching Results by Mascot Search | Molecular Weight by MASS Spectrometry (kDa) | Peptide Coverage on NS1 Protein (%) |
|---|
| DENV1-NS1 | Dengue virus 1 | 40.7 | 48 |
| DENV2-NS1 | Dengue virus 2 | 40.6 | 70 |
| DENV3-NS1 | Dengue virus 3 | 43.33 | 34 |
| DENV4-NS1 | Dengue virus 4 | 40.8 | 48 |
In investigating the diagnostic potential of our NS1 proteins within Vietnam’s clinical settings, we analyzed their ability to bind antibodies present in sera from dengue-infected patients. At sera dilutions of 1:250 and 1:500, robust signal detection was noted, with the 1:750 dilution yielding reduced reactivity (data not shown). The 1:250 dilution was found to be prone to substantial background noise, whereas the 1:500 dilution achieved an optimal signal-to-noise ratio. Consequently, we adopted the 1:500 dilution for further experiments.
We initiated our tests with sera from patients with mild dengue symptoms, encompassing a set of 10 samples. Distribution among the serotypes was as follows: 6 samples were identified as serotype 1, 3 as serotype 2, and 1 as serotype 4 [30]. The absence of serotype 3 in the recent epidemiological data [30,37,38] precluded the inclusion of this serotype in our analysis. In the IgG-based Western blot, serotype 1 samples demonstrated a strong affinity for the DENV1-NS1 antigen, with markedly lower cross-reactivity to other NS1 serotypes. Serotype 4 samples exhibited specific binding to DENV4-NS1. In contrast, serotype 2 samples showed equal reactivity to DENV1-NS1, DENV2-NS1, and DENV4-NS1, with no recognition of DENV3-NS1 [Figure 4].
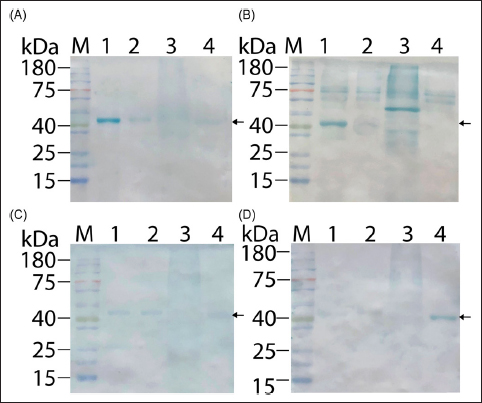 | Figure 4: Western blot using purified DENV1-NS1 to DENV4-NS1 as the target antigens to capture specific IgG antibodies using sera from milder dengue infection patients who did not require hospitalization (total of 10 samples). (A and B) The representative western results of six serotype 1 serum samples; (C) the representative western result of three serotype 2 serum samples; and (D) the western result of the serotype 4 serum. Due to the lack of DENV3 circulation in recent times in the region, there is no result for this serotype. Lane M: pre-stained protein ladder (Smobio); lanes 1–4 contain 100 ng of purified NS1 from serotype 1 to serotype 4, respectively.
[Click here to view] |
Sera from severe, acute dengue cases (10 samples), all corresponding to serotype 2, which predominated during the sampling period [39], exhibited heightened reactivity toward the corresponding antigens. The banding patterns observed were consistent with those from mild cases infected with the same serotype, namely serotype 2 [Figure 5]. Previous studies showed that the presence of NS1 is correlated with the severity of dengue infection [40] and the NS1 antibody titers are significantly higher in DHF patients [41], thus supporting our observation of heightened signals in severe dengue samples.
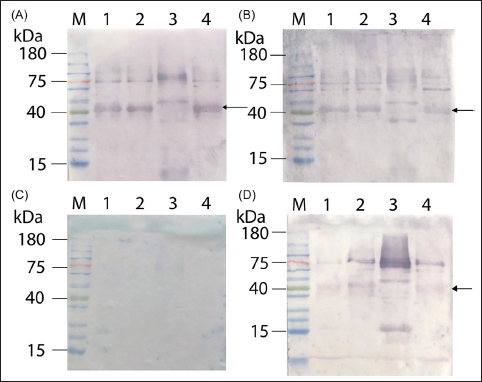 | Figure 5: Representative western results using DENV1-NS1 to DENV4-NS1 as the capture antigens to detect specific IgG antibodies from 10 severe, acute dengue infection patients who required hospitalization. All 10 samples are of serotype 2 as determined previously by semi-nested RT-PCR. Panels (A), (B), and (D) are the results from dengue-positive patients, while (C) is the result from dengue-negative patient. Lane M: pre-stained protein ladder (Smobio); lanes 1–4 contain 100 ng of purified NS1 from serotype 1 to serotype 4, respectively.
[Click here to view] |
When employing IgM secondary antibodies for the Western blot analysis, we observed divergent results. Samples from mild dengue-infection patients did not produce detectable signals, whereas sera from severe cases showed band patterns akin to those seen with IgG, albeit with reduced intensity [Figure 6]. This supports the established criterion that an IgG/IgM ratio greater than 1.1 is indicative of a secondary infection [14,40-42].
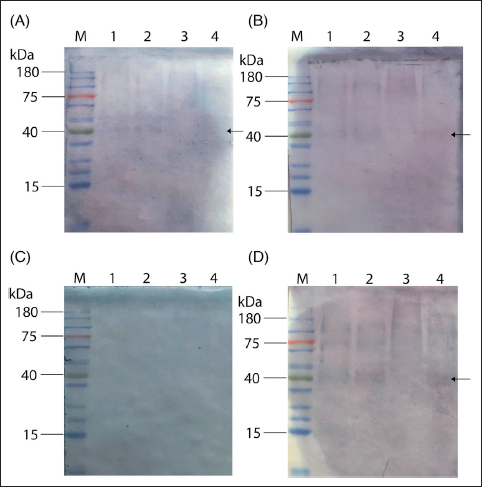 | Figure 6: Representative western results using purified DENV1-NS1 to DENV4-NS1 as the target antigens to capture specific IgM antibodies from severe, acute dengue infection patients. (A), (B), and (D) are from dengue-positive patients and (C) is the result from dengue-negative patient. The secondary antibodies used in the experiment are the AP conjugated, rabbit anti-human IgM (Bethyl A80-101AP). Lane M: pre-stained protein marker; lanes 1–4 contain 100 ng of purified NS1 of DENV1 to DENV4, respectively. The antisera were diluted at 1:500 in blocking buffer and used as the primary antibody. The data is a representative result of 10 samples.
[Click here to view] |
While the Western blot analysis is not commonly used for the serological examination of dengue-infected sera due to its limited scalability, it offers distinct advantages over ELISA [43], such as the ability to visually discern specific antigen–antibody interaction. Our global consensus NS1 antigens have demonstrated the ability to distinguish among serotypes, enhancing the specificity of the serological analysis. However, considering the limited number of samples, the absence of a comprehensive set of serum samples representing all four serotypes, and the specific geographical region they were taken from, further work involving a more diverse range of samples from different geographical locations is necessary to validate the findings.
The main aim of this study is to ensure that the consensus antigens are capable of capturing specific antibodies from patients’ sera. Despite the limitations aforementioned, we believe the findings demonstrate a proof of concept for the use of the antigens, especially in a resource-limited setting like Vietnam.
Interestingly, the Western blot analysis in this study seemed to exhibit enhanced sensitivity compared with the onsite rapid diagnostic test kits from CTK Biotech. While the rapid tests reliably identified NS1 antigens, they fell short in their ability to discern and capture IgG and/or IgM antibodies from the patients’ sera [Supplementary Figure 1]. This disparity underscores the potential of the Western blot technique to detect subtler immune responses, offering a more nuanced assessment of serological status compared to the more immediate, but less comprehensive, rapid tests. However, a more systematic assessment is necessary to validate the observation of banding patterns among different serotypes as well as the Western blot sensitivity compared to quick test kits.
We are also in the process of exploring more cost-effective and scalable methods like dot blot for broader application of the antigens. Additionally, as part of our efforts to enhance affordability and accessibility, we are optimizing the refolding of the NS1 proteins to assess their suitability for ELISA IgG/IgM capture assay, aiming to streamline the diagnostic process without compromising accuracy. Furthermore, we recognize the importance of cost-effectiveness in ensuring accessibility, particularly in resource-constrained settings. Therefore, we are working on optimizing the production process and reducing the costs associated with the diagnostic assays further.
In addition to the development of diagnostic tools, it is crucial to integrate these tools with existing surveillance systems for effective monitoring and control of dengue outbreaks. While molecular tests such as PCR are highly sensitive and specific, they may not capture the full extent of disease prevalence. By combining molecular testing with serological surveys, a broader spectrum of dengue cases can be captured [44,45], ultimately contributing to more effective disease management and control efforts.
REFERENCES
1. Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8(12 Suppl):S7–16. [CrossRef]
2. Messina JP, Brady OJ, Golding N, Kraemer MUG, Wint GRW, Ray SE, et al. The current and future global distribution and population at risk of dengue. Nat Microbiol. 2019;4(9):1508–15. [CrossRef]
3. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. [CrossRef]
4. Taylor L. Dengue and chikungunya cases surge as climate change spreads arboviral diseases to new regions. BMJ. 2023;380:717. [CrossRef]
5. Lenharo M. Dengue is breaking records in the Americas – what’s behind the surge? Nature. 2023. [CrossRef]
6. Katzelnick LC, Fonville JM, Gromowski GD, Bustos Arriaga J, Green A, James SL, et al. Dengue viruses cluster antigenically but not as discrete serotypes. Science. 2015;349(6254):1338–43. [CrossRef]
7. Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–32. [CrossRef]
8. Foucambert P, Esbrand FD, Zafar S, Panthangi V, Cyril Kurupp AR, Raju A, et al. Efficacy of dengue vaccines in the prevention of severe dengue in children: a systematic review. Cureus. 2022;14(9):e28916. [CrossRef]
9. Zainul R, Ansori ANM, Murtadlo AAA, Sucipto TH, Kharisma VD, Widyananda MH, et al. The recent development of dengue vaccine: a review. J Med Pharm Chem Res. 2024;6(4);362–82.
10. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–25. [CrossRef]
11. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–9. [CrossRef]
12. Glasner DR, Puerta-Guardo H, Beatty PR, Harris E. The good, the bad, and the shocking: the multiple roles of dengue virus nonstructural protein 1 in protection and pathogenesis. Annu Rev Virol. 2018;5(1):227-53. [CrossRef]
13. Puerta-Guardo H, Glasner DR, Harris E. Dengue virus NS1 disrupts the endothelial glycocalyx, leading to hyperpermeability. PLoS Pathog. 2016;12(7):e1005738. [CrossRef]
14. Kok BH, Lim HT, Lim CP, Lai NS, Leow CY, Leow CH. Dengue virus infection – a review of pathogenesis, vaccines, diagnosis and therapy. Virus Res. 2023;324:199018. [CrossRef]
15. Pierson TC and Diamond MS. Flaviviruses. In: Knipe DM, Howley PM editors. Fields virology. 6th ed. Philadelphia (PA): Lippincott Williams & Wilkins; 2013. vol 1. p. 757-8.
16. Huang HJ, Yang M, Chen HW, Wang S, Chang CP, Ho TS, et al. A novel chimeric dengue vaccine candidate composed of consensus envelope protein domain III fused to C-terminal-modified NS1 protein. Vaccine. 2022;40(15):2299-310. [CrossRef]
17. Sankaradoss A, Jagtap S, Nazir J, Moula SE, Modak A, Fialho J, et al. Immune profile and responses of a novel dengue DNA vaccine encoding an EDIII-NS1 consensus design based on Indo-African sequences. Mol Ther. 2022;30(5):2058-77. [CrossRef]
18. Park C, Kim WB, Cho SY, Oh EJ, Lee H, Kang K, et al. A simple method for the design and development of flavivirus NS1 recombinant proteins using an in silico approach. Biomed Res Int. 2020;2020:3865707. [CrossRef]
19. Hussain M, Idrees M, Afzal S. Development of global consensus of dengue virus envelope glycoprotein for epitopes based vaccine design. Curr Comput Aided Drug Des. 2015;11(1):84-97. [CrossRef]
20. Ayub A, Ashfaq UA, Idrees S, Haque A. Global consensus sequence development and analysis of dengue NS3 conserved domains. Biores Open Access. 2013;2(5):392-6. [CrossRef]
21. Danecek P, Lu W, Schein CH. PCP consensus sequences of flaviviruses: correlating variance with vector competence and disease phenotype. J Mol Biol. 2010;396(3):550-63. [CrossRef]
22. Leng CH, Liu SJ, Tsai JP, Li YS, Chen MY, Liu HH, et al. A novel dengue vaccine candidate that induces cross-neutralizing antibodies and memory immunity. Microbes Infect. 2009;11(2):288–95. [CrossRef]
23. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [CrossRef]
24. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–7. [CrossRef]
25. Jung SK, McDonald K. Visual gene developer: a fully programmable bioinformatics software for synthetic gene optimization. BMC Bioinform. 2011;12:340. [CrossRef]
26. Webster GR, Teh AY, Ma JK. Synthetic gene design-The rationale for codon optimization and implications for molecular pharming in plants. Biotechnol Bioeng. 2017;114(3):492–502. [CrossRef]
27. Lezin G, Kosaka Y, Yost HJ, Kuehn MR, Brunelli L. A one-step miniprep for the isolation of plasmid DNA and lambda phage particles. PLoS One. 2011;6(8):e23457. [CrossRef]
28. Johnson BH, Hecht MH. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Biotechnology (NY). 1994;12(13):1357–60. [CrossRef]
29. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. [CrossRef]
30. Nguyen TTM, Le VP, Le VT. Epidemiological characteristics and circulation of dengue virus serotypes causing dengue fever in Dak Lak province in 2020. Can Tho J Med Pharm. 2023;63:63–9.
31. Nguyen TNT, Huynh TTH, Nguyen PT, Duong TAT, Cao TR, Le THT, et al. Expression of a synthetic gene encoding the enhanced green fluorescent protein in various Escherichia coli strains. Vietnam J Biotechnol. 2022;20(2):359–68.
32. Qing G, Ma LC, Khorchid A, Swapna GV, Mal TK, Takayama MM, et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol. 2004;22(7):877–82. [CrossRef]
33. Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. [CrossRef]
34. Das D, Mongkolaungkoon S, Suresh MR. Super induction of dengue virus NS1 protein in E. coli. Protein Expr Purif. 2009;66(1):66–72. [CrossRef]
35. Allonso D, da Silva Rosa M, Coelho DR, da Costa SM, Nogueira RM, Bozza FA, et al. Polyclonal antibodies against properly folded Dengue virus NS1 protein expressed in E. coli enable sensitive and early dengue diagnosis. J Virol Methods. 2011;175(1):109–16. [CrossRef]
36. Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb Cell Fact. 2015;14:41. [CrossRef]
37. Vien CC, Pham NT, Nguyen LMH, Ly TTT, Vu SN. Dengue fever in central highlands region, 2000-2020. Vietnam J Prevent Med. 2023;32(Suppl. 2):46–52.
38. Nguyen TTV, Phan TTN, Le VT, Nguyen VT, Le DMQ, Vien CC. Circulation of Dengue virus serotypes causing dengue fever in the Central Highlands Region, 2003-2020. Vietnam J Prevent Med. 2022;32(Suppl. 2):64–9.
39. Nabeshima T, Ngwe Tun MM, Thuy NTT, Hang NLK, Mai LTQ, Hasebe F, et al. An outbreak of a novel lineage of dengue virus 2 in Vietnam in 2022. J Med Virol. 2023;95(11):e29255. [CrossRef]
40. Baihaki I, Dewi BI, Kharisma VD, Murtadlo AA, Taman MB, Purnamasari D, et al. Correlation of the presence of non-structural-1 (NS1) antigen dengue virus with severity of dengue infection. Pharmacognosy J. 2022;14(6);813–6.
41. Jayathilaka D, Gomes L, Jeewandara C, Jayarathna GSB, Herath D, Perera PA, et al. Role of NS1 antibodies in the pathogenesis of acute secondary dengue infection. Nat Commun. 2018;9(1):5242. [CrossRef]
42. Changal KH, Raina AH, Raina A, Raina M, Bashir R, Latief M, et al. Differentiating secondary from primary dengue using IgG to IgM ratio in early dengue: an observational hospital based clinico-serological study from North India. BMC Infect Dis. 2016;16(1):715. [CrossRef]
43. Kuno G, Vorndam AV, Gubler DJ, Gómez I. Study of anti-dengue NS1 antibody by western blot. J Med Virol. 1990;32(2):102–8. [CrossRef]
44. Haselbeck AH, Im J, Prifti K, Marks F, Holm M, Zellweger RM. Serology as a tool to assess infectious disease landscapes and guide public health policy. Pathogens. 2022;11(7):732. [CrossRef]
45. Wilson SE, Deeks SL, Hatchette TF, Crowcroft NS. The role of seroepidemiology in the comprehensive surveillance of vaccine-preventable diseases. CMAJ. 2012;184(1):E70–6. [CrossRef]





