1. INTRODUCTION
The term probiotic was derived from ProG = favor, biosG = life [1]. In the early 1900s, Metchnikoff (1907) introduced probiotics in a scientific context to protect human microflora against pathogens. Henry Tissier also observed that a specific microbial concentration varies in stool samples of children with and/or without infection of diarrhea [2].
A required ecological balance of the gut flora is relevant to human health, as well as the probiotics and more than 100–1,000 microbial species in the gastrointestinal tract (GIT), which is already reported [3]. Half of the total weight of the colon material comprises bacterial cells whose numbers are exceed by the number of tissue cells of humans by 10-fold. Among these bacterial populations, only 103 different bacterial species are present in the stomach, whereas the total microbial population of the colon comprises about 1011–1012 cfu/ml of lyophilized bacterial culture [4]. These bacteria colonize in the gut since birth and they become stable at adulthood, depending on the various internal and external stresses [5].
Prebiotics enhance the growth and development of probiotics, which modify the gut microbial flora and are not easily digested by humans [6,7]. Some of the common known prebiotics includes bifidogenic properties like inulin, oligofructose, fructose-oligosaccharides, etc. [8,9]. The synergistic beneficial role of a synbiotic is promoted together with the probiotic and prebiotic in the living system, which beneficially enhances the gut health, including disease prevention [10,11].
The elite properties of probiotics are anti-pathogenicity, anti-obesity, anti-diabetic, anti-inflammatory, anti-allergic, anti-cancer activities, and others [12–14]. The adhesive property of probiotic is prerequisite for the colonization, which shows competitive advantage in this ecosystem that prevents enteropathogens from establishing an infection [14].
Inflammatory bowel disease (IBD) is a combination of Crohn’s disease and ulcerative colitis (UC) which have been associated with the risk of colon cancer [15,16]. UC is a large intestine disorder which is characterized by inflammation of the colonic lamina propria injury and disruption of the mucosal barrier [17]. The most specific clinical features of animal models which are very relevant to human IBD, which include weight loss, diarrhea, and sometimes mucus in the feces. Pathologically, human IBD shows ulceration of the mucosa and disappearance of the horizontal folds of the colon, whereas histologically it displays acute, subacute, and chronic inflammatory changes occur in the mucosa of the glandular epithelium [18].
Induction of colitis in animal models by chemicals includes carragenin [19], acetic acid [20], sodium hydroxide [21], TNBS (4, 6-trinitrobenzene sulfonic acid), and (dextran sodium sulfate) DSS [22].
Our study emphasized two segments, in one segment potential probiotic bacteria were isolated from different sources of human milk and in the other segment they were used for treating the gastrointestinal disease like human IBD.
2. MATERIALS AND METHODS
2.1. Collection of Samples
Human milk samples were collected from three different mothers in Burdwan town in the district of Purba Bardhaman, West Bengal, India. Safety measures were considered for surface sterilization by applying 70% ethanol on the upper surfaces of the breast and the sterile sucker milk samples were collected in a sterile tube for further analysis in the laboratory.
2.2. Isolation of LAB
Among three milk samples, only one milk sample was considered for further analysis; the other two were discarded due to some unwanted contamination. The selected milk sample was diluted up to 10−7 and aliquots were plated on nutrient agar media supplemented with Nystatin (50 µg/ml) to restrict the growth of fungus. After incubation at 37°C for 24 hours, total bacterial load was determined as cfu/mL and further isolated for pure culture formations. For pure culturing, the MRS (De Man, Rogosa, and Sharpe) agar media were used for selective isolation of LAB [23]. Bacterial colonies isolated from human milk samples were streaked across the MRS media and incubated at 37°C for 24 hours. After incubation, isolated single colonies were used for further identification purposes.
2.3 Identification of LAB
Seven isolated colonies were selected for further characterizations, on the basis of their Gram character. Gram positive bacterial isolates were selected in this case over gram negative bacterial isolates to avoid the pathogenicity (which may be conferred by O-antigen). Among the seven selected isolates, six were Gram positive coccus (named as PB1, PB2, PB3, PB4, PB5, and PB6) and one was a Gram positive rod (named as PB7). These seven isolates were further subjected to endospore staining, arginine hydrolase analysis test, urease test, oxidase test, catalase test, cellulase test, amylase test, carbohydrate utilizing test (using lactose as a reducing disaccharide, glucose as a monosaccharide, maltose as a reducing disaccharide, cellobiose as a reducing disaccharide, trehalose as a non-reducing disaccharide, dihydroyacetone as a keto-triose, erythrose as an aldo-tetrose, xylose as an aldo-pentose, and sorbitol as a sugar alcohol), and IMViC test [24].
2.3.1. Gram staining and endospore staining
All the seven isolates (PB1–PB7) were subjected to the Gram staining and endospore staining as described in Bergey’s Manual of Systematic Bacteriology [25] to identify and select the desirable group of bacteria.
2.3.2. Arginine hydrolase analysis test
The MRS (De Man, Rogosa and Sharpe) broth supplemented with 0.3% arginine and 02% sodium citrate was inoculated with seven isolates (PB1–PB7) for 24 hours at 37°C. After incubation, 100 µl of inoculated culture was mixed with 100- µl Nessler’s reagent. Bright orange color indicates a positive result.
2.3.3. Urease test
Sterilized urea agar media supplemented with phenol red prepared as slants were streaked with all the seven isolates (PB1–PB7) and incubated at 37°C for 24 hours to observe the change in the color.
2.3.4. Oxidase test
Sterile filter paper was soaked with 1% tetra methyl p-phenyldiamine dichloride and inoculated with all the seven isolates. The developing dark purple color within 10 seconds indicates the presence of potent cytochrome c oxidase producer.
2.3.5. Catalase test
The slants of sterilized trypticase soy agar were inoculated with all the seven isolates (PB1–PB7) and were incubated overnight at room temperature. After incubation, the slants were introduced to 3% H2O2 to observe the bubbles.
2.3.6. Cellulase test
Sterile Czapek mineral salt agar medium petri plates were inoculated with all the seven isolates (PB1–PB7) and incubated for 3–4 days at 37°C. After incubation, the petri plates were flooded with 1% aqueous solution of hexadecyl trimethyl ammonium bromide to observe the clear zones of the cellulose producers.
2.3.7. Amylase test
To identify the amylase producing bacteria, all the seven isolates were inoculated on a sterile starch agar medium and kept for 48 hours at 37°C. After incubation, the plates were flooded with Gram’s iodine for 30 seconds to observe the clear zones.
2.3.8. Carbohydrate utilizing test
To identify the lists of carbohydrates that can be utilized for all the seven isolates, a sterile medium was prepared containing trypticase, carbohydrates (lactose, glucose, maltose, cellobiose, trhalose, di-hydroxy acetone, erythrose, xylose, and sorbitol), NaCl, and phenol red (as indicators) with an inverted Durham’s tube in it. After 48 hours of incubation at 37°C, the isolates utilized the given carbohydrates and produced a yellow color acid or gas with bubbles, respectively.
2.4. Anti-Microbial Activity
The seven isolates were further analyzed for anti-microbial activity by the agar cup assay method. Anti-bacterial activity assessments were conducted against a clinically important human pathogen Vibrio cholerae (ATCC 14035) [26]. Suspensions of each of the seven selected isolates (106–107 cfu/mL ) were plated in Petri dishes to carry out a pour plate method, whereas the control used as uninoculated MRS broth. The final concentration of pathogenic bacteria (V. cholerae) in the liquid MRS agar medium is 104–105 cfu/ml. After the hardening of the agar wells (diameter of the well = 6 mm), 60 μl of selected seven isolates were pipetted in the wells of the plates and were incubated at 37ºC for 24 hours. Based on the diameter of the inhibition zone, anti-pathogen activities were divided into strong (diameter ≥ 20 mm), moderate (20 mm ≤ diameter ≥ 10 mm), and weak (diameter ≤ 10 mm) [27].
2.5. Anti-Oxidant Activity
2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity was evaluated using the method of Son and Lewis [28]. DPPH was used as a stable radical. A volume of 2 ml of DPPH in ethanol (500 mM) was added to 2 ml of the bacterial stock solution, mixed vigorously, and allowed to stand in the dark at room temperature for 30 minutes. The absorbance was measured at 517 nm. Ethanol was used as a blank, while DPPH solution in ethanol served as the control. The radical scavenging activity of the samples was expressed as % inhibition of DPPH absorbance:
where “Control” is the absorbance of the control sample (DPPH solution without sample) and “Test” is the absorbance of test sample (DPPH solution plus Sample) (Table 3)
2.6. Bile Salt Tolerance Assay
The selected seven isolates were suspended on MRS medium containing 0.2%–0.8% bile salt solution for examining their bile salt tolerance [29]. The medium was prepared following the method, where the aliquot (0.2 ml) from the suspension of bacteria was transferred to a 2 ml sterile eppendrof tube and mixed with 1 ml of 0.2%–0.8% bile salt solution in the MRS medium. This mixture was allowed to vortex for 10 seconds, followed by incubation at 37°C for 48 hours. After incubation, the result was recorded by determining the O.D. value at 620 nm [30].
2.7. Visualization under Scanning Electron Microscope (SEM)
Among the seven isolates the best anti-microbial activity, anti-oxidant activity, and bile salt tolerance were shown by PB7. Hence, this isolate was observed under SEM; as it is difficult to notice small changes in cell morphologies of bacteria under the light microscope, SEM was used in the present investigation to determine the size of the cells and also to review the changes in cell morphology (if any) due to environmental change from an in vivo to in vitro condition [31]. The bacterial cell was recovered by centrifugation at 6,000 rpm (revolutions per minutes) and the cells were washed twice with potassium phosphate buffer (50 mM, pH 7.0). Bacterial cell was then fixed by drowning in 2.5% glutaraldehyde in potassium phosphate buffer (50 mM, pH 7.0) for overnight at 4°C. Then, the specimen was washed twice with buffer and dehydrated by ethanol series (v/v) ranging from 30%, 40%, 50%, 60%, 70%, 80%, 90%, and 100% and stored in absolute ethanol. For SEM, the specimen was dried to critical point, coated with gold, and inspected with an S-200C scanning electron microscope with 1,000 KX magnifications.
2.8. Determination of Optimum pH and Temperature
Two sets of experiments were designed to determine the optimum pH and temperature of PB7. In Set I, PB7 was inoculated with three replica sets of MRS broth media and prepared for five different incubating temperatures, such as, 10°C, 20°C, 30°C, 40°C, and 50°C for 24 hours. In Set II, PB7 was inoculated with three replica sets of MRS media for each pH 2, 3, 4, 5, and 6 which was incubated at 37°C for 24 hours.
2.9. Molecular Identification by 16S ribosomal ribonucleic acid (rRNA) Sequencing
The determination of the 16S ribosomal ribonucleic acid (rRNA) gene sequencing acts as a tool to identify and confirm the bacterial strains from dairy samples, at KPC Medical College, Kolkata. The sequenced 16S rRNA gene was submitted in basic local alignment search tool for nucleotides (BLASTn) in the National Centre for Biotechnology Information (NCBI) server and then a phylogenetic tree was constructed in MEGA-X (a tool used for Molecular Evolutionary Genetic Analysis) with neighbor joining method by using the 14 most similar sequences to predict the species (Figure 4). The DNA of PB7 was isolated, purified, and amplified by the primers: 9F (“GAGTTTGATCCTGGCTCAG”) and 1492R (“GGTTACCTTGTTACGACTT”).
The GC contents of 16S rRNA gene of our isolates were utilized to find the specific species of the bacteria. By using SpeciesFinder Tools present in Center for Genome Epidemiology server (https://cge.cbs.dtu.dk/services/SpeciesFinder/) the species were identified and confirmed.
2.10. Animal Handling
Seven specific pathogen-free, 8-week-old female BALB/c (an albino, immunodeficient inbred mouse strain) mice (Rita Animal House, Kolkata-52) were allocated to 4 groups for further proceeding of experiments and were maintained under a specific pathogen-free condition. The house was a filter-top cage in a specific pathogen-free environment of the animal facilities, under standard conditions (50%–60% humidity, 12-hours dark/12-hours light cycles), fed standard rat chow and saline water, which were placed in a feeding bottle.
2.11. Disease Induction
Mice were divided into two groups (n = 3) and one was kept as a control (R1), in which the disease was neither induced nor the treatment was applied. The disease (colitis) induction of the three mice of the first group (Group I) named as R2, R3, and R4 was done by Oxazolone, DSS, and TNBS (2,4,6 – Trinitrobenzenesulfonic acid), respectively. The 3% of these chemicals were gavaged two times a day which is mixed in saline water. Based on the result of Group I, only DSS was applied again on Group II named as R5, R6, and R7.
2.12. Disease Activation Index (DAI)
The weight was regularly noted as given below to determine the DAI. DAI was used to assess the clinical severity of colitis which was calculated on the daily basis, and measured as loss of body weight, stool consistency, and rectal bleeding.
The Loss of Body weight Index (LBI) was scored as follows: “0 - no weight loss”, “1 – weight loss of 0%–5%”, “2 – weight loss of 5%–10%”, “3 – weight loss of 10%–20%”, and “4 – weight loss >20%”. Stool consistency Index (SCI) was scored as follows: “0 – normally formed pellets”, “2 – pasty and semi-formed pellets”, and “4 – liquid stool”. Rectal bleeding Index (RBI) was scored as follows: “0 – no blood from the rectum” and “4 – gross bleeding from the rectum” [32]. After applying the chloroform to the animal models, the colons were removed from mice R1, R2, R3, and R4 after 60 days of disease induction and the same was done with R5, R6, and R7 after 20 days of treatment.
3. RESULTS AND DISCUSSION
3.1. Identification of LAB
From the study, it is observed that among the seven isolates, one isolate was non-spore former Gram positive rod, whereas the rest were spore former Gram positive coccus.
Following the characterization, the biochemical identification was carried out, which includes arginine hydrolase analysis, urease test, oxidase test, catalase test, cellulase test, amylase test, carbohydrate utilizing test (with lactose, glucose, and maltose), and IMViC (an acronym for the tests like, "I" is for indole test; "M" is for methyl red test; "V" is for Voges-Proskauer test, and "C" is for citrate test). From the result, it was also observed that PB1–PB7 showed positive result for arginine hydrolase analysis, cellulase test, and glucose utilizing test, but PB7 in addition to it also showed a positive result for lactose utilizing test, maltose utilizing test, and methyl red test of IMViC.
The result describes that among the seven isolates only PB7 is a Gram positive rod and has the ability to ferment glucose, lactose and maltose and can produce acid which lowers the pH of the medium. Besides this, PB7 posses arginine hydrolase enzyme and cellulase enzyme. And this result indicates the PB7 isolates towards a potent probiotic according to Bergeys Manual of Determinative Bacteriology (7th edition) (Table 1).
.png) | Table 1: The characterization and biochemical identification of the seven isolates (PB1–PB7), which includes staining, different enzyme analysis test, IMViC test, and carbohydrate utilization test. [Click here to view] |
3.2. Anti-Microbial Activity
After characterization another experiment which is equally important to prove the property of probiotic is its antimicrobial activity. In this study, among the seven isolates from human milk sample (namely PB1 – PB7), only PB7 showed a good antimicrobial activity against the potent human pathogen V. cholerae (ATCC 14035) [26]. Based on the diameter of the inhibition zone, anti-pathogen activities were divided into strong (diameter ≥ 20 mm), moderate (20 mm ≤ diameter ≥ 10 mm) and weak (diameter ≤ 10 mm) [27] (Table 2 and Figure 1).
.png) | Table 2: The zone of inhibition (mm) was tabulated against V. cholerae. [Click here to view] |
.png) | Figure 1: The anti-microbial effect of isolates (PB7, PB4, and control) against V. cholerae (ATCC 14035) by visible clear zone on the petriplate. [Click here to view] |
3.3. Anti-Oxidant Activity
Another most important property of a probiotic is its anti-oxidant activity. To determine the scavenging activity of the isolates besides being catalase negative, DPPH was used, which is shown in Figure 2.
From the given result, it was observed that PB7 has a significantly high scavenging activity. The standard taken here is the ascorbic acid which is considered to have maximum antioxidant activity, whereas PB7 also showed an enhanced DPPH activity as compared to the standard (Table 3).
3.4. Bile Salt Tolerance Assay
The concentration of the bile salt is highest in the GIT in comparison to the other internal organs of the human body. Thus, to establish an organism as a potent probiotic, its bile salt tolerance was examined.
The result depicts that among the seven isolates only one isolated sample, namely PB7, showed resistance against bile salt until 0.6% (w/v), but was unable to grow in and above 0.8% (w/v) (Table 4 and Figure 3)
.png) | Figure 2: The graphical representation of the enhanced DPPH activity of the PB7 in comparision with the other samples as well as the standard. [Click here to view] |
3.5. Visualization under SEM
From the result of characterization, biochemical identification, anti-microbial activity, anti-oxidant activity, and bile salt tolerance, it was determined that among the seven isolates (PB1 –PB7), only PB7 gives a significant good result which supports it toward a potent probiotic. Thus, only PB7 was visualized under SEM to confirm its physiological structural details. From the observation, it was concluded that it is in rod shape and can grow singly or in chains too (Figure 4).
3.6. Determination of Optimum pH and Temperature of PB7
From the experiment, it was concluded that the optimum temperature of PB7 was 40°C and the optimum pH was 3–4. Thus, we can conclude that PB7 can withstand acidic pH regions as well as normal body temperature, which supports its growth in the human gut (Tables 5 and 6, Figure 5A and B).
3.7. Species Identification and Phylogenetic Tree Determination
Based on the GC contents of the 16S rRNA gene, our isolates were analyzed using SpeciesFinder Tools 2.0, and the result showed that our querry or submitted sequence was highly similar with Lactobacillus fermentum; accession no. KC866340.
The 16S rRNA gene of PB7 was identified by using BLASTn tools available in NCBI database. The result from BLASTn represents the highly similar with L. fermentum FQ002 (97.73% identity), after the identification of the sample, the sequence of 16S rRNA gene of PB7 was submitted in NCBI through the Gene Bank Submission Tool which generated an accession number: MN121704 and is designated as L. fermentum (BURD PB7). The corresponding 14 similar sequences were also downloaded in fasta format and the phylogenetic tree was constructed, using MEGA-X. The phylogenetic tree revealed that (Figure 6) the 16S rRNA gene sequence of the L. fermentum BURD PB7 was highly similar with the same L. fermentum with different strains: TW20-4, TW21-6, FT41, FT58, CAU: 230, CAU: 235, and CAU: 237.
.png) | Table 3: The volume of the sample and DPPH solution used to determine its scavenging activity. [Click here to view] |
.png) | Table 4: The form of the O.D. value of seven isolates which are grown in MRS media with varying concentrations of bile salt, i.e., 0.2, 0.4, 0.6, and 0.8% (w/v). [Click here to view] |
.png) | Figure 3: The graphical representation of the comparative study of 7 isolates in different concentrations of bile salts, i.e., 0.2, 0. 4, 0.6, and 0.8 % (w/v). [Click here to view] |
3.8. Disease Activation Index
DAI was used to assess the clinical severity of colitis which is calculated on the basis of daily LBI, and also on the basis of SCI as well as RBI. And this LBI was divided into three sets as follows: SET-I denotes the result of R1, R2, R3, and R4 throughout 60 days of disease induction; SET-II denotes the results of R5, R6, and R7 throughout 60 days of disease induction; and SET-III denotes the result of R5, R6, and R7 throughout the 20 days of treatment.
3.8.1. Loss of body weight (SET-I)
From Table 7 and Figure 7A, it can be concluded that due to normal healthy diet R1, the control gained its body mass throughout the 60 days of experimental period. But in comparison to that, the animal models R2, R3, and R4 had a severe loss of body mass throughout the disease induction period (60 days). Following the comparative study among R2, R3, and R4, it was evident that R3 with the highest LBI score as well as loss of body mass in which the disease was induced by DSS. Thus, in the next set of experiments, the disease was induced only by DSS to all three animal models (namely R5, R6, and R7). The days of the experiments are similar as before. The loss of body mass is shown in Figure 8 and the score is tabulated in Tables 7 and 8.
.png) | Figure 4: The visualization of PB7 under SEM. [Click here to view] |
.png) | Table 5: The form of O.D. of MRS containing PB7 after incubation at different temperatures (10, 20, 30, 40, and 50°C). [Click here to view] |
.png) | Table 6: The O.D. values of the growth of PB7 at different pH. [Click here to view] |
.png) | Figure 5: (A & B). The comparative study of growth of PB7 at different temperatures. [Click here to view] |
.png) | Figure 6: The phylogenetic tree of PB7 with the bacteria which has maximum similarity with the sequence of the strains that have been submitted and assigned an acession no. by NCBI. [Click here to view] |
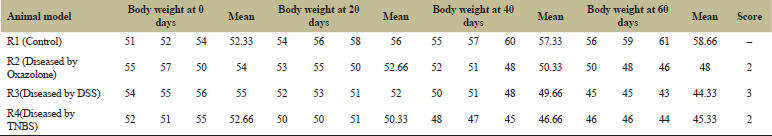 | Table 7: The loss of body weight of R, R2, R3, and R4 in the course of 60 days were tabulated and designated with proper scoring as per references. [Click here to view] |
.png) | Figure 7: (A). A comparative study of the body weight throughout the course of disease induction by three different agents: DSS, Oxazolone, and TNBS. (B) The animal models R5, R6, and R7 in which colitis was induced by DSS showed a permanent loss of body mass throughout the experiments. (C) The gain of body mass by the colitis-induced mice R5, R6, and R7 after treatment by probiotic PB7 and Vitamin C either independently or in combination. [Click here to view] |
3.8.2. Loss of body weight (SET-II & SET III)
From Tables 8 and 9 and Figure 7B and C for LBI – SET III and LBI – SET III experiments, it was observed that the colitis-induced mice (by DSS) on treating with synbiotic (PB7 and Vitamin C) rather that treating either with prebiotic (Vitamin C) or with only probiotic (PB7) showed a significant enhancement in their body mass. But in a comparative study between the R5, R6, and R7, R5 received the treatment of synbiotic and showed highest increase in the body mass. Although the gain in body mass was observed in all the three mice, the treatment with synbiotics showed significantly good results.
.png) | Figure 8: (A & B) The SCI and RBI are represented from 0 to 80 days of all the animal models. [Click here to view] |
.png) | Table 8: The loss of body mass throughout the period of disease induction (60 days) and the LBI score was tabulated. [Click here to view] |
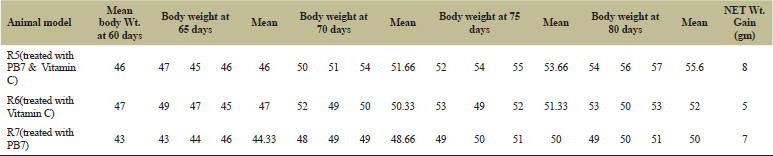 | Table 9: The DSS-induced colitis on R5, R6, and R7 showed a net gain in body mass after treatment with PB7 and vitamin C. [Click here to view] |
3.8.3. Stool consistency index and RBI
According to the SCI and RBI mentioned in the previous section, the tabulated form is represented in Table 10. From Figure 8A and B, it is clearly evident that probiotic is a good therapeutic agent for the treatment of colitis in animal model, but synbiotic is far better. The SCI and RBI was decreased to zero after treating with synbiotic within 10 days, but in case of treating with probiotic and prebiotic separately it took 15–20 days. Thus, for the treatment of colitis, it is preferred to combine probiotic and prebiotic to get best results.
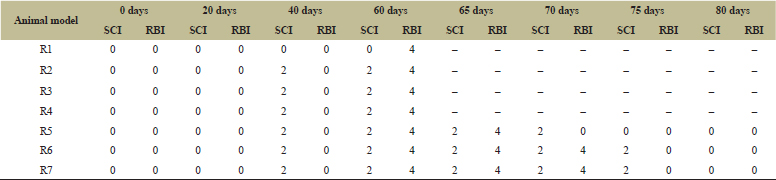 | Table 10: The SCI and RBI of seven animal models throughout disease induction and treatment. [Click here to view] |
4. CONCLUSION
The result of the present study supports the efficacy of the use of synbiotics (probiotic + prebiotic) as a therapeutic tool for the treatment of the intestinal colitis. From the seven isolates (isolated from human milk samples), only PB7 was selected for the treatment of the colitis induced by DSS, due to its superior antimicrobial property, antioxidant activity, and bile salt tolerance activity. The accession number of PB7 isolate is MN121704 provided by Gene Bank of NCBI which was designated as L. fermentum (BURD PB7). The addition of Vitamin C (Prebiotic) along with PB7 (Probiotic) gave better results on colitis-induced mice than the application of probiotic or prebiotic alone. Future studies should focus on the histological and immunological aspects on colitis and its treatment with probiotic.
ACKNOWLEDGMENTS
This study was supported by the laboratory of Oriental Institute of Science and Technology, Burdwan, and the genome sequencing was done by KPC Medical College, Kolkata. This study received no research grant from any funding agency like government, public, commercial, or non-profitable sectors.
CONTRIBUTION
Bidyut Bandyopadhyay and Madhumita Mitra helped in designing the experiments, analyzing data, development of methods, and material preparation. Experiments were carried out by Atrayee Roy and supervised by the other two authors mentioned prior. Atrayee Roy wrote the manuscript with contributions from Bidyut Bandyopadhyay and Madhumita Mitra. All authors approved the manuscript before submission.
CONFLICT OF INTEREST
Atrayee Roy, Madhumita Mitra, and Bidyut Bandyopadhyay confirm that this article content has no conflicts of interest.
FUNDING
The entire funding of the experiment was supported by the OIST laboratory and self-finance was also invested in some cases.
7. REFERENCES
1. Veera Jothi V, Anandapandian KTK, Shankar T. Bacteriocin production by probiotic bacteria from curd and its field application to poultry. Arch Appl Sci Res 2012;4(1):336–47.
2. Tissier H, Tritement des infections intestinales par la methode de translormation de la flore bacterienne de lintestin. C R Soc Biol 1906;60:359–61.
3. Aziz Q, Dore J, Emmanuel A, Guarners F, Quigley EMM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neuro Gastroenterol Motil 2013;25:4–15. CrossRef
4. Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients 2013;5:1417–35. CrossRef
5. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med 2016;8:1–11. CrossRef
6. Rastall RA, Gibson GR. Recent developments in prebiotics to selectively impact beneficial microbes and promote intestinal health. Curr Opinion Biotechnol 2015;32:42–6. CrossRef
7. Thomas LV. Probiotics-the journey continues. Int J Dairy Tech 2016;69:1–12. CrossRef
8. Hutkins RW, Krumbeck JA, Bindels LB, Cani PD, Fahey G, Goh YJ. Prebiotics: why definitions matter. Curr Opin Biotechnol 2016;37:1–13. CrossRef
9. Tanaka R, Takayama H, Morotomi M, Kuroshima T, Ueyama S, Matsumoto K. Effects of administration of TOS and Bifidobacterium breve 4006 on the human fecal flora. Bifidobact Microflora 1983;2:17–24. CrossRef
10. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 1995;125:1401–12. CrossRef
11. Tufarelli V, Laudadio V. An overview on the functional food concept: prospectives and applied researches in probiotics, prebiotics and synbiotics. J Exp Biol Agric Sci 2016;4:274–8.
12. Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC. Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis 2009;15:300–10. CrossRef
13. Sherman PM, Ossa JC, Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr Clin Pract 2009;24:10–4. CrossRef
14. Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 2008;72:728–64. CrossRef
15. Langholz E, Munkholm P, Davidsen M, Binder V. Colorectal cancer risk and mortality in patients with ulcerative colitis. Gastroenterology 1992;103:1444–51. CrossRef
16. Gillen CD, Walmsley RS, Prior P, Andrews HA, Allan RN. Ulcerative colitis and Crohn’s disease: a comparison of the colorectal cancer risk in extensive colitis. Gut 1994;35:1590–2. CrossRef
17. Karp LC, Targan SR, Ogra PL, Mestecky J, Lamm ME, Strober W. et al. Evidence for an updated hypothesis of disease pathogenesis in Ulcerative colitis. Mucosal Immunology. Academic Press, San Diego, CA, pp 1047–53, 1999.
18. Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models, Nat Rev Microbiol 2010;8:564–77. CrossRef
19. Moyana TN, Lalonde JM. Carrageenan-induced intestinal injury in the rat model for inflammatory bowel disease. Ann Clin Lab Sci 1990;20:420–6.
20. MacPherson BR, Pfeiffer CJ. Experimental production of diffuse colitis in rats. Digestion 1978;17:135–50. CrossRef
21. Kocak E, Koklu S, Akbal E, Tas A, Karaca G, Astarci MH, et al. NaOH-induced Crohn’s colitis in rats: a novel experimental model. Dig Dis Sci 2011;56(10):2833–7. CrossRef
22. Alex P, Zachos NC, Nguyen T, Gonzales L, Chen TE, Conklin, LS, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis 2009;15:41–52. CrossRef
23. De Man JC, Rogosa M. Elisabeth Sharpe, ME. A medium for the cultivation of Lactobacilli. J Appl Bact 1960;23:130–5. CrossRef
24. Buchanan RE, Gibbons NE. Bergey’s manual of determinative bacteriology. 8th edition, The Williams and Wilkins Company, Baltimore, MD, 1974.
25. Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey’s manual of determinative bacterial. 9th edition, Williams and Wilkins, London, UK, p 787, 1994.
26. Haghshenas B, Nami Y, Abdullah N, Radiah D, Rosli R, Khosroushahi AY. Anti-proliferative effects of Enterococcus strains isolated from fermented dairy products on different cancer cell lines. J Func Foods 2014;11:363–74. CrossRef
27. Lim YS. Probiotic characteristics of bacteriocinogenic Lactobacillus plantarum strains isolated from malaysian food, biotechnology and biomolecular science. PhD thesis, University Putra Malaysia (UPM), Kuala Lumpur, Malaysia, 2010.
28. Son S, Lewis BA. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure-activity relationship. J Agric Food Chem 2002;50:468–72. CrossRef
29. Kumar AM, Murugalatha N. Isolation of Lactobacillus plantarum from cow milk and screening for the presence of sugar alcohol producing gene. J Microbiol Antimicrob 2012;4(1):16–22. CrossRef
30. Bassyouni RH, Abdel-all WS, Fadl MG, Abdel-all S, Kamel Z. Characterization of lactic acid bacteria isolated from diary products in Egypt as a probiotic. Life Sci J 2012;9:2924–30.
31. Raouf T, Marion B, Marielle G, Jean-Paul V. In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J Med Microbiol 2013;62:637–49. CrossRef
32. Murano M, Maemura K, Hirata I, Toshina K, Nishikawa T, Hamamota N, et al. Therapeutic effect ofb intracolonically administered nuclear factor κB (p65) antisense oligonucleotide on mouse dextran sulphate sodium (DSS) – induced colitis. Clin Exp Imunol 2000;120:51–8. CrossRef