1. INTRODUCTION
Crop protection is increasingly reliant on pesticides to increase agricultural production. Easily accessible and effective, these chemical tools are being used for pest and disease control without much thought of their effects on the environment [1]. Unregulated use of pesticides leads to their accumulation in different ecosystems including agricultural soils [2]. The maximum residue levels of commonly used pesticides are between 0.2 parts per million (ppm) and 1.2 ppm; foods contaminated with pesticides above these levels may pose a health risk over time due to their bioaccumulation and biomagnifications [3,4]. The presence of pesticides above maximum residue limits has been reported by researchers worldwide in various foodstuffs [5-7].
Microbial bioremediation is an innovative and potentially safer and economically viable solution to clean up agricultural soils. Pesticide degrading enzymes present in these microorganisms enable them to break down the toxic pesticide molecules by cleaving their specific bonds such as P-O, P-F, P-S, and P-C bonds [8]. Based on experimental results and theoretical studies, multiple pathways have been identified for the degradation of organophosphorus pesticides in various microorganisms. These pathways involve one or the other pesticide degradative enzymes [9,10]. However, among all, organophosphate (OP) hydrolyzing enzymes are the most specific and extensively studied enzymes in terms of OP pesticide degradation [11]. Thakur et al., 2019 [12] list six types of OP hydrolyzing enzymes: OPDA, PTE, DFPase, SsoPox, OPAA, and PON 1 found in different bacteria and fungi.
Thousands of pesticides or agrochemicals are manufactured and being applied in agriculture worldwide. Each of these pesticides has a different chemical structure, toxicity, and environmental fate [4,13]. Remarkable research has been undertaken and reported over recent decades for the bioremediation of various pesticides [14-17]. However, studying each of them using conventional bioremediation techniques is next to impossible. The conventional bioremediation approach is cumbersome, time-consuming, and costly. Useful insights can be derived from the available genomes and transcriptome sequences in the databases.
In the last decade, many genes and enzymes involved in pesticide degradation have been identified and their sequences are now available in the public domain [18]. Here, a newly developed in silico bioremediation technology can be utilized for obtaining quick, low-cost, and timely information before proceeding with onsite remediation of toxic pollutants [19]. Protein modeling, molecular docking, and other high throughput techniques can be employed to predict the biodegradation potential of an organism. In silico bioremediation can address the research gaps and predict the customized degradative enzymes for instantaneous removal of toxic pesticides [20-23]. In silico analysis of pesticide degradation potential of some bacterial enzymes has been done, but studies with fungi (especially Trichoderma) are very few. Trichoderma, multicomponent plant growth-promoting fungi, is a key microbial partner in achieving sustainable agriculture. Since long it is being used as a biocontrol agent and biofertilizer in agriculture [24]. In recent years, its role in the bioremediation of toxic pollutants has also been established, but there is still a lot to explore. To date, only one or two research papers describe the probable molecular mechanism of OP pesticide degradation in Trichoderma harzianum. In a study by Sun et al., 2018 [25], a T. atroviride paraoxonase 1 like (TaPON1-like) protein was found in T. atroviride strain T23, which showed 27% query coverage and 34% sequence identity with HuPON1 protein. TaPON1-like is homologous to HuPON1, which is reported to be involved in the degradation of OP pesticides, esters, and lactones. Deletion of the TaPON1-like coding gene reduced P-O bond breakdown efficiency/OP pesticide degradation potential in T. atroviride.
The present study aims to investigate the mechanisms of pesticide degradation in a plant-friendly fungus T. harzianum using in silico protein modeling and molecular docking studies. The efficacies of binding of T. harzianum enzyme with the toxic pesticide ligands were calculated. In addition, a comparison of the results of both in silico, and in vitro seed germination studies was performed to check if the results are comparable. This study shed light on the probable molecular mechanism of OP degradation in T. harzianum and also demonstrates that in silico methods can be utilized to screen potential microorganisms before onsite bioremediation.
2. MATERIALS AND METHODS
2.1. Selection of Pesticides
Systematic literature search on OP pesticides using the online database PubMed (www.ncbi.nlm.nih.gov/pubmed) advanced search was performed using different combinations of appropriate search terms. Information on total registered pesticides, banned pesticides, and pesticide management bill 2020 were collected from government statistical databases and websites. Five-year compound annual growth rate (CAGR) (2019-2024) and market analysis reports were also considered to choose the pesticides that will be used more frequently in coming times.
2.2. Pesticide Degrading Enzyme Sequence Retrieval and Analysis
A list of OP pesticide degrading enzymes (OP hydrolases) was retrieved from various databases such as Uniprot, NCBI, BRENDA, ExPASY, KEGG, Swiss Prot, and literature survey. These enzymes were, further, searched in the T. harzianum genome using the NCBI blast server. The test sequence was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). Analysis of selected sequence was done by domain search and phylogenetic analysis using Pfam domain search and MEGA software, respectively. The subcellular localization of the target protein was predicted using the Target P-2.0 server (https://services.healthtech.dtu.dk/service.php?TargetP-2.0).
2.3. Homology Modeling and Validation
T. harzianum hypothetical protein was modeled using MODELLER version 10. MODELLER is used for homology or comparative modeling of protein three-dimensional structures [26]. The query sequence was BLAST against the PDB database to get a similar structure to the query and the template was downloaded from PDB (https://www.rcsb.org/search). Once the PDB files are obtained, the most matching template was selected, and the query was aligned with the template using the python script of MODELLER. The model for the query was generated using the MODELLER python script. Phyre2 tool was used to predict and analyze protein 3D structure (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The final model was evaluated based on DOPE profiles and the model with the lowest DOPE score was selected. Model validation was done by creating and analyzing the Ramachandran plot using the Ramachandran Plot Server of Zhiping Weng’s lab (https://zlab.umassmed.edu/bu/rama/).
2.4. Molecular Docking with Pesticide
Before docking the molecules, the binding site of the receptor protein was identified using an online tool called BiteNet (https://sites.skoltech.ru/imolecule/tools/bitenet/). Open babel version 2.4.1 was used for conversion of SDF file format of pesticide ligands to PDB format. Structures of the chemical compounds were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) [27]. The docking was performed using Autodock Vina [28]. The electrostatic potential calculation, model visualization, and image generation were performed using the PyMOL software (www.pymol.org). Discovery Studio Visualizer v20.1.0.19295 was used for visualization of a docked complex, 2D, 3D interaction, and other PDB files (https://discover.3ds.com/discovery-studio-visualizer-download). UCSF Chimera, candidate version 1.15, was also used for visualization of surface view of docked complex (https://www.cgl.ucsf.edu/chimera/).
2.5. Pesticide Tolerance and Seed Germination Restoration by T. harzianum
T. harzianum was exposed to Dimethoate (DM) and Monocrotophos (MCP) in the range of 0 to 2100 ppm and biomass was recorded to assess its tolerance to pesticide. Host plant (Sorghum bicolor) seeds were treated with two pesticides, namely, DM and MCP at the concentration of 300 ppm and the germination% was recorded after 48 h of incubation in the plant growth room. All the experiments were placed in triplicates. Seed germination restoration on T. harzianum treatment was calculated and compared [Figure 8]. Comparison of in silico docking scores and in vitro work was done to check the similarity of results.
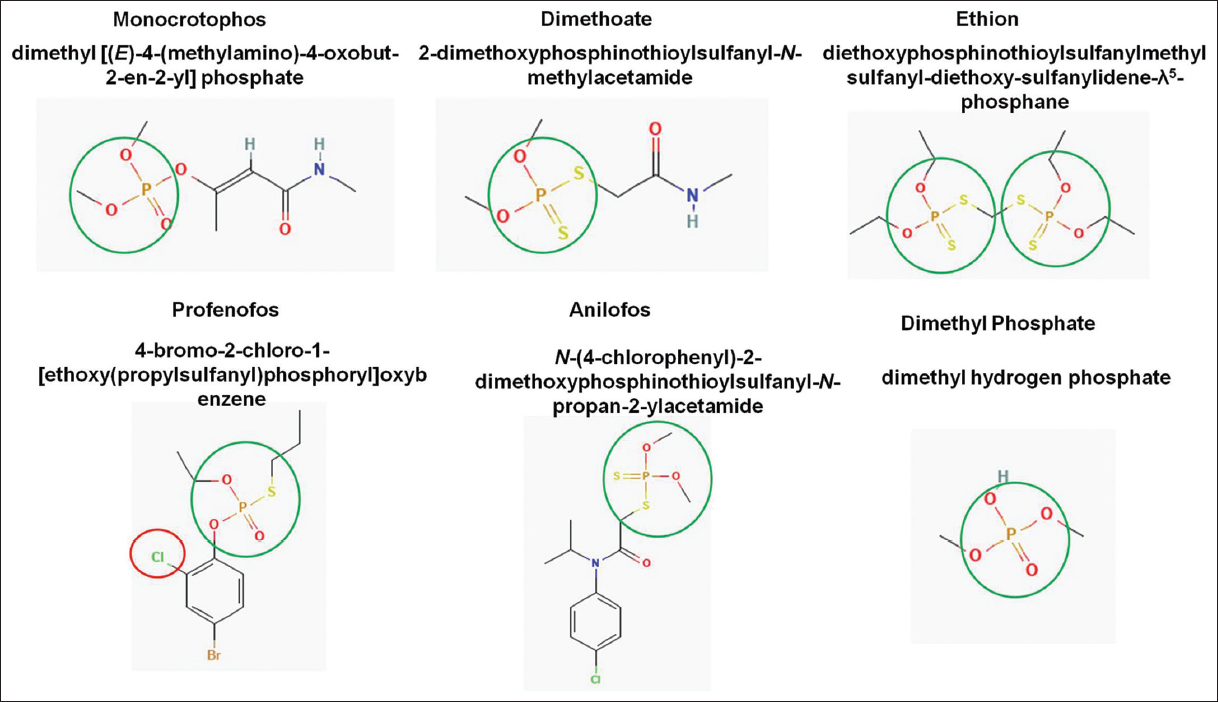 | Figure 1: Chemical structure of selected organophosphate pesticides. [Click here to view] |
 | Figure 2: Phylogenetic analysis of organophosphate hydrolyzing enzymes in different species. [Click here to view] |
 | Figure 3: Molecular mechanism of organophosphate pesticide degradation. [Click here to view] |
 | Figure 4: Sequence alignment of >PNP56599 with Serum paraoxonase/arylesterase 1. *Indicate the conserved amino acids; Sequence of Arylesterase domain was highly conserved. [Click here to view] |
 | Figure 5: Protein model construction and validation (a) modeled ThPON1-like, (b) DOPE profile (c) Ramachandran Plot [Click here to view] |
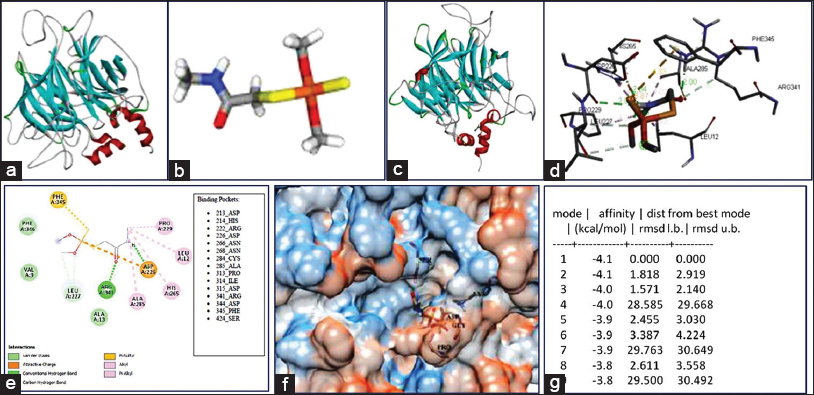 | Figure 6: Molecular docking studies with organophosphate weedicide dimethoate a. modeled ThPON1-like, b. dimethoate, c. docked complex, d. interaction between protein and ligand, e. 2D interaction of docked complex, and f. docked complex in surface view, g. Binding affinity (Kcal/mol) [Click here to view] |
 | Figure 7: Molecular docking studies with organophosphate insecticide monocrotophos. (a) modeled ThPON1-like, (b) monocrotophos, (c) docked complex, (d) interaction between protein and ligand, (e) 2D interaction of docked complex, and (f) docked complex in surface view, g. Binding affinity (Kcal/mol) [Click here to view] |
 | Figure 8: Comparative analysis of in silico and plant-based studies with selected pesticides. [Click here to view] |
3. RESULTS AND DISCUSSION
3.1. Selection of Pesticides
Total 30 OP pesticides were registered in India in 1968 for various agricultural purposes such as insecticide, weedicide, nematicide, and mosquito control [29]. Out of 30, 13 pesticides are banned and ten pesticides are used for other non-agricultural purposes. Acephate, Chlorpyrifos, Chlorpyriphos-methyl, Diazinon, Dichlorvos, Fenthion, Malathion, Parathion-methyl, Phorate, Phosphamidon, Quinalphos, Triazophos, and Trichlorfon are banned [30]. This study included Ethion, Profenofos, Anilofos, DM, MCP pesticides and one of the common pesticide degradation intermediate products, Dimethyl phosphate, based on the data collected from published literature, statistical reports, pesticide management bill 2020, CAGR, and market analysis reports. In addition, all the information about the application of these selected pesticides was collected and compared [Table 1]. Structural details such as bond structure and the presence of halogens were also studied to identify their potential environmental hazard [Figure 1]. Pesticides chosen for this study are readily available in the market and are frequently used by farmers.
Table 1: Current status of selected organophosphate pesticides in India.
| Pesticide | Rate (Kg/ha) | Crops | Toxicity class | CAGR (2019-24) | Market availability | Production (2019-20) | Consumption (2019-20) | Environmental residues | Biodegradation |
|---|---|---|---|---|---|---|---|---|---|
| 1. Monocrotophos | 0.5-1.5 | Cotton, Maize, potato, fruits, sugar beet *Banned in vegetables | Class IB | −8.10% | Readily available | 5817 MT | 552 MT | Found>MRL | Bacillus, Pseudomonas, Streptomyces sp.Starkeya novella, Aspergillus, Macrophomina etc. |
| 2. Dimethoate | 0.2-2 | Horticultural crops, vegetables. cereals | Class II | 0.16% | Readily available | 1446 MT | 366 MT | Found>MRL | Paracoccus sp., Raoultella sp, Pseudomonas, Lactobacillus, Cyanobacteria |
| 3. Profenofos | 1.7 | Cotton and vegetables maize, potato, soybean, and sugar beet | Class II | 11.64% | Readily available | 12631 MT | 426 MT | Found>MRL | Pseudomonas putida Burkholderia Trichoderma |
| 4. Ethion | 0.25-1 | Tea, cotton, maize, cucurbits | Class III | 3.31% | Readily available | 2127 MT | 37 MT | Found>MRL | Pseudomonas and Azospirillum |
| 5. Anilofos | 0.4 | Transplanted paddy, soybean | Class II | Available | Weedicide (commonly given as weedicide) | 138 MT | Comparatively less reports on above MRL range | Rhodanobacter xiangquanii sp (Novel) |
MRL: Maximum residue limits, CAGR: Compound annual growth rate
3.2. Pesticide Degrading Enzyme Sequence Retrieval and Analysis
Varieties of microbial enzymes have evolved in nature with the capability of degrading OP compounds [31]. OP hydrolyzing enzymes have been extensively studied for their ability to degrade toxic OP pesticides [12,32,33]. OP hydrolyzing enzymes can be classified into six different types, each having its structure and origin [Table 2]. Comprehensive lists of OP pesticide degrading enzymes were prepared and their sequences were retrieved from various databases such as Uniprot, NCBI, BRENDA, ExPASY, KEGG, and Swiss Prot. These enzymes were, further, searched in the T. harzianum genome using NCBI blast. Among OP hydrolyzing enzymes, only PON1 type (TaPON1-like) of enzyme was found in T. harzianum (>PNP56599.1:1-437 hypothetical protein THARTR1_03295) and showed 65.23% identity with TaPON1-like enzymes of T. atroviride [25]. In the Pfam database, protein sequences are categorized into domains and families. Protein domains provide insight into the function of that protein. The probable biological function of a hypothetical protein can be deduced based on its domain structure [34]. Pfam domain search analysis revealed the presence of characteristic arylesterase (paraoxonases) domain in the test sequence that has a characteristic role in OP pesticide degradation. Subcellular localization using Target P server revealed peripheral likelihood = 6.74 (at 419). Moreover, multiple sequence alignment and phylogenetic analysis of various OP hydrolyzing enzymes revealed diversity in sequences. Bacterial OP hydrolase with PTE domain was clustered together, while those of higher organism (PON1 type) with arylesterase domain were clustered separately to them. The in silico protein modeling work done in this manuscript has the basis from pesticide degradative protein reported as TaPON1-like in T. atroviride [25]. The author of the manuscript proposes nomenclature as T. harzianum paraoxonase 1 like (ThPON1-like) as the work was conducted on T. harzianum. The TaPON1-like enzyme in T. harzianum (ThPON1-like) was closer to the protein of higher eukaryotic organisms [Figure 2]. A family of OP hydrolyzing enzymes hydrolyze and detoxifies a wide range of toxic OP pesticides by cleaving the various phosphorus-ester bonds (P-O, P-F, P-CN, and P-S) and converts them into less toxic intermediates and end products [12,31-33]. Figure 3 depicts a general mechanism of pesticide degradation pathway by OP hydrolyzing enzymes.
Table 2: Enzymes involved in pesticide biodegradation.
| Enzymes | Enzyme (kDa) | Gene | Organism | Source |
|---|---|---|---|---|
| 1. Alkaline phosphatase | 86 kDa | Alpl/phoA | Bacteria, Fungi | NCBI UNIPROT PDB KEGG BRENDA |
| 2. Carboxyl esterase | 56.5 kDa | CE/carE1 | Bacteria, Fungi | |
| 3. OP hydrolyzing enzymes | ||||
| OPH | 72 kDa | opd | Bacteria | |
| PTE | 19 kDa | hocA | Bacteria | |
| DFPase | 35.21 kDa | dfpase | Sea squid | |
| TaPON1-like | 43 kDa | tapon1 | T. atroviride | |
| SsoPox | 144 kDa | php (S) | Archaea | |
| 178.28 kDa | opaa | Alteromonas Proteobacteria | ||
| 4. Monooxygenase | 41 kDa | Cyp450 | Bacteria, Fungi |
3.2.1. Arylesterase domain - (NDLFAESPTSFFVTNDHYYTEGFM RAVEDLLPRATWTNVLH)
The query sequence was BLAST against the PDB database to find the closest structure. The best template was Serum paraoxonase/arylesterase 1 (PDB ID: 1V04). The hypothetical protein >PNP56599 when aligned with arylesterases 1 shared 21.0% identity and 33% query coverage with 0.48E-09 E value. MODELLER guidelines suggest that even around 25% identity indicates a potential target unless it is <100 residues and this query sequence is more than 467 residues and it has 33% query coverage to the template. Dutta et al., 2013 [35], constructed a similar protein model for docking studies when query template identity was around 25%, but the domain responsible for the function was conserved. Protein domains are the fundamental units of proteins, which can fold, function, and evolve independently. The domains of proteins fold separately from the rest of the protein and serve a specific function. Protein domains with the same functions might be found in diverse groups of organisms [36]. In this study, also the comparative sequence alignment of query and template displayed conserved amino acids. The target sequence had maximum conserved residue in the coverage of the arylesterase domain that is specifically required for interaction with OP pesticide [Figure 4]. Arylesterase domain is mainly found in serum paraoxonases/arylesterases; some reports also mention their presence in Trichoderma atroviride [25]. The entire arylesterase domain was present both in the template and query proteins and is highly conserved. Even though there is less sequence identity between query and template sequences, a sound model was made, because the arylesterase domain was retained. As per Sun et al., 2019 [25], despite the low identity (34%) between PON1 and TaPON1-like, due to the presence of conserved motifs belonging to the same six-blade β-propeller hydrolase subfamily, PON1 protein is a valid template for modeling the structure of TaPON1-like. In another study, the query template identity was around 25%, but a sound model was built as the PDZ1 domain of the protease chain was conserved [35].
3.3. Homology Modeling and Validation
Homology modeling is one of the prerequisites for in silico bioremediation as molecular docking studies relies on the protein model and its specific three-dimensional structural properties (α-helix, beta-sheet, loop, etc.) [19]. The comparative modeling of >PNP56599-ThPON1 was performed using a restrained-based approach implemented in MODELLER10. A set of five models for the target protein was constructed. The resulting three-dimensional models of ThPON1-like were sorted according to scores calculated from the discrete optimized protein energy (DOPE) scoring function. The final model that shared the lowest Root Mean Square Deviation (RMSD), relative to the trace (Ca atoms) of the crystal structure was selected. The plotted DOPE score profile (below) shows regions of relatively high energy for the long active site loop between residues 90 and 200. Ramachandran plot is a plot to visualize energetically allowed regions for a polypeptide backbone torsion angles psi (y) against (phi) f of amino acid residues present in a protein structure. In our model, most of the amino acids fall on the highly preferred region (>93%), 4% in preferred except for a few (2%) indicates a good model. A graphical illustration of protein’s three-dimensional structure, DOPE score profile, and Ramachandran plot is given in Figure 5. Dutta et al., 2013 [35], accepted the model when the amino acid in allowed region was around 82.7%.
3.4. Molecular Docking with Pesticide
Molecular docking has effectively been used over the past few years to perform the biodegradation process of environmental contaminants remediation. It is an extremely relevant and low-cost approach to understanding ligands reaction mechanisms with high accuracy of proteins or enzyme binding [37]. The purpose of docking is to produce and evaluate plausible intermolecular complex structures. Docking assay allows examining how protein molecules (enzymes) and ligands (pollutants) interact to form a stable docked complex. The compound strongly binds to the active site of the target molecule with minimum binding energy (ΔG) serves as a potential molecule. Before performing in vitro or on-site bioremediation, docking studies can predict the binding efficiencies of toxic pollutants with potent detoxifying enzymes present in microorganisms. Several researchers screened microbial proteins and enzymes for their ability to bind and form stable complexes with toxic pollutants using a similar approach [20,21,38,39]. The active site or binding site of particular enzymes needed to be explored for understanding its structural features and functions towards ligand binding. Before docking, the binding site of the hypothetical protein was predicted using BiteNet. The site with highest score (0.82) was chosen for the study. Molecular docking of ThPON1-like with 5 OP pesticides and 1 common degradation intermediate revealed their interaction with reasonable binding energy score [Table 3]. The binding free energy of the pesticides was in the order Anilofos (−5.8) > Profenofos (−5.6)> MCP (−5.1)> Ethion (−4.8)> Dimethylphosphate (−4.4)> DM (−4.1). Anilofos showed the best interaction and DM had the lowest interaction with ThPON1-like which is due to differences in their chemical structure, bond, and size [Figures 6 and 7].
Table 3: Molecular docking of ThPON1-like with selected pesticides and degradation intermediates.
| Pesticide | Bonds | PubChem ID | Structure | Binding energy (kcal/mol) |
|---|---|---|---|---|
| Monocrotophos | P-O, P=0 | 5371562 |  | −5.1 |
| Dimethoate | P-O, P-S, P=S | 3082 |  | −4.1 |
| Profenofos | P-O, P-S, P=S-Cl, -Br | 38779 | 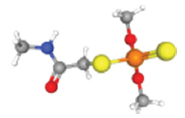 | −5.6 |
| Ethion | P-O, P-S, P=S | 3286 |  | −-4.8 |
| Anilofos | P-O, P-S, P=S | 91687 |  | −5.8 |
| Dimethyl phosphate | P-O, P=O | 13134 |  | −4.4 |
ThPON1-like: Trichoderma harzianum paraoxonase 1 like
3.5. Comparative Analysis of In silico and Plant-based Studies with Selected Pesticides
Some studies compared results from in silico and in vitro experiments. For instance, Aamir et al., 2018 [40], have identified the fungal SDR as novel targets for fungicides. They observed similar results between in silico analysis and in vitro assessment of the fungicide on the pathogen. In a separate study, Singh et al., 2021 [41], investigated how carbamate pesticide intermediates affected membrane architecture in Escherichia coli using both in vitro and in silico techniques. In the above-mentioned studies, the results obtained from in silico and in vitro studies were consistent. As part of the current study, the researchers compared results of in vitro tolerance assay, seed germination experiment, and in silico analysis using two selected pesticides, MCP and DM, to see if the results were comparable. T. harzianum isolate showed higher tolerance to MCP (lethal dose [LD50] > 1900) compared to DM (LD50 > 300). In silico studies also displayed the same pattern as binding with MCP (Binding energy −5.1 Kcal/mol) was more stable than DM (Binding energy −4.1 Kcal/mol). The lower the binding energy the better is the interaction between enzyme and pesticide ligand [37].
Seed germination assays with MCP and DM pesticides revealed that both pesticides are toxic to plants, but better seed germination was observed in T. harzianum treated seeds compared to respective pesticide controls. At 300 ppm concentration of MCP, the germination percentage of untreated seeds was 45% while T. harzianum treated seeds had 80% germination. When seeds were treated with 300 ppm DM, they had a 25% germination rate, whereas T. harzianum treated seeds showed a 40% germination rate [Figure 8]. Germination study results were similar to those from in silico studies. Under MCP and DM stress conditions, the T. harzianum treatment increased seed germination by 35% and 15%, respectively. The germination restoration activity of T. harzianum is better in the presence of MCP than DM, because T.harzianum contains an enzyme that binds more efficiently to MCP than DM (as evidenced by in silico studies).
4. CONCLUSION
Multicompetent plant-friendly fungus T. harzianum is a key component in sustainable agriculture, but its bioremediation potential is not well explored, especially in the case of OP pesticides. This study suggests that PON1 type of OP pesticide hydrolyzing enzyme (ThPON1-like) is present in T. harzianum. In addition, this study proves that results obtained from and in silico and in vitro studies are comparable. In silico bioremediation could reliably help to identify the potential microbial detoxifying enzymes for detoxification of pesticides before going for onsite bioremediation.
5. ACKNOWLEDGMENT
We would like to thank the Department of Biotechnology (DBT), the Government of India for providing funds and the Jaypee institute of information technology for providing basic facilities and infrastructure required for the execution of the project.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
Research was supported by the Department of Biotechnology, Government of India, Project grant number (BT / PR21299 /BCE /8 /1402/2016).
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
There are no animal or human subjects involved in this study.
10. DATA AVAILABILITY
The gene and proteins sequences are available online in the NCBI database (Accession numbers mentioned in the article).
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kumar M, Yadav AN, Saxena R, Paul D, Tomar RS. Biodiversity of pesticides degrading microbial communities and their environmental impact. Biocatal Agric Biotechnol 2021;31:101883. [CrossRef]
2. Aktar W, Sengupta D, Chowdhury A. Impact of pesticides use in agriculture:Their benefits and hazards. Interdiscip Toxicol 2009;2:1-12. [CrossRef]
3. Kumar S, Kaushik G, Dar MA, Nimesh S, Lopez-Chuken UJ, Villarreal-Chiu JF. Microbial degradation of organophosphate pesticides:A review. Pedosphere 2018;28:1-18. [CrossRef]
4. Sharma A, Kumar V, Shahzad B, Tanveer M, Sidhu GP, Handa N, et al. Worldwide pesticide usage and its impacts on ecosystem. SN Appl Sci 2019;1:1446. [CrossRef]
5. Bansal OP. Fate of pesticides in the environment. J Indian Chem Soc 2011;88:1525-32.
6. Kaushal J, Khatri M, Arya SK. A treatise on organophosphate pesticide pollution:Current strategies and advancements in their environmental degradation and elimination. Ecotoxicol Environ Saf 2021;207:11483. [CrossRef]
7. Kumar S, Kaushik G, Villarreal CJ. Scenario of organophosphate pollution and toxicity in India:A review. Environ Sci Pollut Res 2016;23:9480-91. [CrossRef]
8. Sarlak Z, Darani KK, Rouhi M, Garavand F, Mohammadi R, Sobhiyeh MR. Bioremediation of organophosphorus pesticides in contaminated foodstuffs using probiotics. Food Control 2021;126:108006. [CrossRef]
9. Ortiz HM, Sánchez SE, Dantán GE, Castrejón GM. Pesticide biodegradation:Mechanisms, genetics and strategies to enhance the process. Biodegrad Life Sci 2013?;10:251-87.
10. Raffa CM, Chiampo F. Bioremediation of agricultural soils polluted with pesticides:A Review. Bioengineering 2021;8:92. [CrossRef]
11. Zhao S, Xu W, Zhang W. Overview of a bioremediation tool:Organophosphorus hydrolase and its significant application in the food, environmental, and therapy fields. ApplMicrobiol Biotechnol 2021;105:8241-53. [CrossRef]
12. Thakur M, Medintz IL, Walper SA. Enzymatic bioremediation of organophosphate compounds –Progress and remaining challenges. Front Bioeng Biotechnol2019;7:289. [CrossRef]
13. Hassaan MA, Nemr AE. Pesticides pollution:Classifications, human health impact, extraction and treatment techniques. Egypt J Aquat Res 2020;46:207-20. [CrossRef]
14. Patel N, Chaudhary VK, Patel A, Singh A, Srivastav AL, Rai D. Application of microbes in bioremediation of pesticides. In:Inamuddin AM, Prasad R, editors. Application of Microbes in Environmental and Microbial Biotechnology. Singapore:Springer;2022. [CrossRef]
15. Mulla SI. Organophosphate pesticides:Impact on environment, toxicity, and their degradation. In:Saxena G, Bharagava R, editors. Bioremediation of Industrial Waste for Environmental Safety. Singapore:Springer;2020. 555-71. [CrossRef]
16. Chawla N, Bhardwaj J, Singh L. Bioremediation of organophosphate pesticides:Current status and future prospective. Plant Arch 2020;20:3405-12.
17. Mahajan R, Verma S, Chandel S, Chatterjee S. Organophosphate pesticide:Usage, environmental exposure, health effects, and microbial bioremediation. In:Das S, Dash HR, editors. Microbial Biodegradation and Bioremediation. Ch. 24. Amsterdam, Netherlands:Elsevier;2022. 473-90. [CrossRef]
18. Liu Z, Liu Y, Zeng G, Shao B, Chen M, Li Z, et al. Application of molecular docking for the degradation of organic pollutants in the environmental remediation:A review. Chemosphere 2018;203:139-50. [CrossRef]
19. Singh K, Muhammad B, Iqbal HM, Raj A. Trends in predictive biodegradation for sustainable mitigation of environmental pollutants:Recent progress and future outlook. Sci Total Environ 2021;770:144561. [CrossRef]
20. Kumar KD, Vyshnava SS, Shanthi BS, Bontha RR. In-silico analysis of the interaction of quinalphos and 2-hydroxyquinoxaline with organophosphate hydrolase and oxygenases. Biointerface Res Appl Chem 2022;12:608-17. [CrossRef]
21. Ritika A, Ritika G, Nikita J, Bableen K, Arunima M, Minakshi B, et al. In-silico prediction, characterization and molecular docking studies on glutathione-s-transferase as a molecular sieve for toxic agrochemicals explored in survey of North Indian farmers. Heliyon 2021;7:e07875. [CrossRef]
22. Coronado-Posada N, Mercado-Camargo J, Olivero-Verbel J. In-silico analysis to identify molecular targets for chemicals of concern:The case study of flocoumafen, an anticoagulant pesticide. EnvironToxicol Chem 2021;40:2034-43. [CrossRef]
23. Reyes-Espinosa F, Méndez-Álvarez D, Pérez-Rodríguez MA, Herrera-Mayorga V, Juárez-Saldivar A, Cruz-Hernández MA, et al. In-silico study of the resistance to organophosphorus pesticides associated with point mutations in acetylcholinesterase of lepidoptera:B. mandarina, B. mori, C. auricilius, C. suppressalis, C. pomonella, H. armígera, P. xylostella, S. frugiperda, and S. litura. Int J Mol Sci2019;20:2404.
24. Sachdev S, Singh RP. Trichoderma:A multifaceted fungus for sustainable agriculture. In:Bauddh K, Kumar S, Singh R, Korstad J, editors. Ecological and Practical Applications for Sustainable Agriculture. Singapore:Springer;2020. 261-304. [CrossRef]
25. Sun J, Yuan X, Li Y, Wang X, Chen J. The pathway of 2,2-dichlorovinyl dimethyl phosphate (DDVP) degradation by Trichoderma atroviride strain T23 and characterization of a paraoxonase-like enzyme.Appl Microbiol Biot 2019;103:8947-62. [CrossRef]
26. Fiser A, Sali A. MODELLER:Generation and refinement of homology-based protein structure models. In:Carter CW, Sweet RM, editors. Methods in Enzymology. Vol. 374. San Diego:Academic Press;2003. 463-93. [CrossRef]
27. National Center for Biotechnology Information. PubChem Compound Summary for CID 3082, Dimethoate. Available from:https://pubchem.ncbi.nlm.nih.gov/compound/Dimethoate [Last accessed on 2022 Jan 12].
28. Trott O, Olson AJ. AutoDock Vina:Improving the speed and accuracy of docking a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31:455-61. [CrossRef]
29. Government of India. The Insecticides Act, 1968 (Act No. 46 of 1968). New Delhi;1968;Available from:http://www.krishi.bih.nic.in/Acts-Rules/Insecticides_Act_1968.pdf [Last accessed on 2021 Dec 05].
30. Production of Key Pesticides during 2014-15 to 2018-19, Official Site of Directorate of Plant Protection, Quarantine and Storage, Ministry of Agriculture and Farmer Welfare, Government of India. Available from:http://ppqs.gov.in/PMD.htm#variousPest [Last accessed on 2021 Jan 21].
31. Singh BK, Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 2006;30:428-71. [CrossRef]
32. Poirier L, Brun L, Jacquet P. Enzymatic degradation of organophosphorus insecticides decreases toxicity in planarians and enhances survival. Sci Rep 2017;7:15194. [CrossRef]
33. Matula, M, Kucera T, Soukup O, Pejchal J. Enzymatic degradation of organophosphorus pesticides and nerve agents by EC:3.1. 8.2. Catalysts 2020;10:1365. [CrossRef]
34. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer EL, et al. Pfam:The protein families database in 2021. Nucleic Acids Res2021;49:D412-9. [CrossRef]
35. Dutta A, Katarkar A, Chaudhuri K. In-silico structural and functional characterization of a V. cholerae O395 hypothetical protein containing a PDZ1 and an uncommon protease domain. PLoS One 2013;8:2. [CrossRef]
36. Wang Y, Zhang H, Zhong H, Xue Z. Protein domain identification methods and online resources. Comput Struct Biotechnol J 2021;19:1145-53. [CrossRef]
37. Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, et al. Insights into protein–ligand interactions:Mechanisms, models, and methods. Int J Mol Sci 2016;17:144. [CrossRef]
38. Kamil K, Kamil M, Daniel J, Eugenie N, Ondrej S, Jan K, et al. Oxime K074 –in vitro and in silico reactivation of acetylcholinesterase inhibited by nerve agents and pesticides. Toxin Rev 2018;39:157-66.
39. Kumar V, Singh S, Singh R, Upadhyay N, Singh J, Pant P, et al. Spectral, structural and energetic study of acephate, glyphosate, monocrotophos and phorate:An experimental and computational approach. J Taibah Univ Sci 2018;12:69-78. [CrossRef]
40. Aamir M, Singh VK, Dubey MK, Meena M, Kashyap SP, Katari SK, et al. In-silico prediction, characterization, molecular docking, and dynamic studies on fungal SDRS as novel targets for searching potential fungicides against fusarium wilt in tomato. Front Pharmacol 2018;9:1038. [CrossRef]
41. Singh P, Tripathi MK, Yasir M, Ranjan A, Shrivastava R. Effects of carbamate pesticides intermediates on Escherichia coli membrane architecture:An in vitro and in-silico approach. Environ Anal Health Toxicol 2021;36:3. [CrossRef]