1. INTRODUCTION
Improvement in next-generation sequencing (NGS) technologies enabled researchers to decipher the complex communities associated with each microbiome. Metagenomic DNA from different microbiome helps to identify host-associated microbial diversity, and this is essential in vertebrate models like zebrafish. Isolation of quality metagenomic DNA from diverse sources is challenging to microbiologists. Gut microbes have vital functions in improving immunity, disease resistance, and nutrient absorption in vertebrates [1]. Gut microbiome of fish has major roles in nutrient metabolism, synthesis of vitamins, amino acids, enzymes, digestion, and absorption [2]. Furthermore, the microorganisms also involve in the development of the gut, regulation of endocrine system, as well as immune system modulation [3]. The effect of immunostimulants on beneficial gut microbiota is less studied but promising area of future research in fish [4]. For gut microbial diversity analysis, conventional culture-based approaches, as well as molecular techniques, have been used [5]; these are less popular, being prolonged, labor-intensive, inferior in accuracy, and unable to determine microbial profile in whole [6]. Recently, metagenomics based molecular techniques were widely used to identify the rare and uncultivated bacterial community associated with different microbiomes [7], in an array of biological materials such as soil, fecal matter, and specific organs of animals [8]. Pyrosequencing technologies helped to investigate the gut-associated microbiota in humans [8] and other animals such as mice [9] cats, dogs, and fish [6,10]. The unraveling of intricate host-microbe symbioses enables the application of a potential healthy microbiome to benefit fish culture, as well as to gain deeper insight into the role of microbiomes in vertebrate health.
One reason for studying gut microbiome of fishes is the idea that modification of these communities could enhance host health. Considering the increased application of NGS to study gut microbiome of species other than human and mouse, there is need for comparative assessment of DNA extraction from other species used as model organism, like zebrafish, in gut microbiome studies with 16S rRNA amplicon sequencing as the downstream application.
The present study aimed at standardizing a protocol for the extraction of good quality, high-molecular-weight, and metagenomic DNA from zebrafish gut. The study was focused on zebrafish gut microbiome metagenomic DNA due to its significance in downstream processing like 16S rRNA sequencing and analysis that can unfold bacterial diversity.
2. MATERIALS AND METHODS
2.1. Zebrafish Husbandry
AB wild-type zebrafish strains with an average weight of 1.5 ± 0.12 g were purchased from local aquarium shop. Fish were acclimatized in a recirculating tank under laboratory conditions at 28.5°C with a 14-h light/10-h dark cycle for 2 weeks and fed commercial fish diet to achieve total daily feed input of 3% of body weight per day.
2.2. Zebrafish Dissection
Zebrafish were starved 24 h before dissection so that DNA isolation is unhindered by food material. Euthanization by immersion in 500 mg/L of 3-aminobenzoic acid ethyl ester (MS-222) buffered to pH 7.4 (Sigma-Aldrich, USA) was followed by rinsing in RO water to remove traces of MS-222 and placed on paper towels to remove excess water. Fish surface was disinfected with 100% ethanol and then the skin and underlying muscle from above the operculum region posteriorly along the side of the fish and down the anal fin was cut and removed; the whole intestine was separated from the rest of the organs and put in sterile 2.0 mL microfuge tubes and kept on ice.
2.3. DNA Extraction
Metagenomic DNA was isolated by three methods such as QIAamp DNA Stool Mini Kit, modified method of QIAamp DNA Stool Mini Kit, and the manual method [11] with minor modification in the purification steps.
2.3.1. Method 1: Kit method
Microbes adhering the intestinal walls were isolated. For this pooled whole intestine from five euthanized fish in the same tank was cut into small pieces and homogenized in 10-fold dilution of cold sterile ×1 phosphate-buffered saline (PBS) solution using autoclaved mortars and pestles, and centrifuged (Hermle, Germany) at 20,000 g for 20 min. The pellet with the microbial cells was used for metagenomic DNA isolation using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA). DNA concentration was determined at 260 nm using NanoDrop™ 1000 spectrophotometer.
2.3.2. Method 2: Modified kit method
Intestine from five euthanized fish was cut and ground in 600 μL ice-cold PBS using autoclaved mortar and pestle and collected in 2.0 mL microfuge tubes. Added lysozyme at 50 mg/mL and incubated at 37°C in a water bath for 30 min. DNA was extracted using the QIAamp Stool Mini Kit (Qiagen, Valencia, CA, USA), eluted in 200 μL of AE buffer, followed by addition of 1/10 volume of 3 M sodium acetate (pH5.2), 0.6–0.7 × volume of room temperature isopropanol and centrifuged at 20,000 g for 30 min at 4°C to reduce coprecipitation of salts. The supernatant was discarded and DNA pellet was rinsed with 70% ethanol and centrifuged at 14,000 g for 15 min at 4°C. Pellet was air dried after discarding the supernatant and dissolved in AE buffer. DNA concentration was determined at 260 nm using a NanoDrop™ 1000 spectrophotometer.
2.3.3 Method 3: Using manual method
Intestine isolated from five euthanized fish was cut into small pieces, collected in 700 μL of lysis buffer 500 mM NaCl, 50 mM Tris-HCl, 50 mM EDTA, and 4% SDS (pH 8) in 2.0 mL microfuge tubes, vortexed for 3 min and incubated at 70°C for 20 min in water bath, with gentle shaking every 5 min, then centrifuged at 4°C for 5 min at 16,000× g. Transferred the supernatant to a fresh 2 mL microfuge tube and the previous steps were repeated, to pool the supernatant. Added 260 μL of 10 mM ammonium acetate to each tube, mixed well, and incubated on ice for 5 min then centrifuged at 4°C for 10 min at 16,000× g. Supernatant was transferred to two 1.5 mL microfuge tubes, added one volume of isopropanol, mixed well, incubated on ice for 30 min, and centrifuged at 4°C for 15 min at 16,000× g. The supernatant was removed and pellet washed with 70% ethanol, dried, and dissolved in 100 μL of Tris-EDTA (TE) buffer and the aliquots pooled. Added 2 μL of DNase-free RNase (10 mg/mL) and incubated at 37°C for 15 min. Added 15 μL of proteinase K and 200 μL of buffer AL (from the QIAamp DNA Stool Mini Kit), mixed well, and incubated at 70°C for 10 min. Added 200 μL of ethanol and mixed well. Transferred to a QIAamp column and centrifuged at 16,000× g for 1 min. Discarded the flow through, added 500 μL of buffer AW1 (Qiagen), and centrifuged for 1 min at room temperature. Discarded the flow through, added 500 μL of buffer AW2 (Qiagen), and centrifuged for 1 min at room temperature. Added 100 μL of buffer AE (Qiagen) and incubated at room temperature for 2 min. The DNA concentration was determined at 260 nm using a NanoDrop™ 1000 spectrophotometer.
2.4. Quantitative and Qualitative Assessment of Metagenomic DNA
The isolated metagenomic DNA was analyzed by agarose gel electrophoresis using λ/Eco RI + Hind III double digest ladder as marker. Purity and concentration of DNA were estimated by Nanodrop™ 1000 spectrophotometer. Yield of extracted DNA was determined by measuring absorbance at 260 nm. The purity of corresponding DNA samples was determined by A260/A280 (DNA/protein) and A260/A230 (DNA/humic acid) ratios to determine protein and humic acid contamination, respectively [12].
2.5 Gel Electrophoresis
The extracted DNA was analyzed by agarose gel electrophoresis run in 0.8% gel containing 10 mg/mL ethidium bromide in 1X TAE buffer at 60 V, and the image was captured using gel documentation system (Syngene, USA).
2.6. Polymerase Chain Reaction (PCR) Ability of DNA as Criteria for Purity
Furthermore, the quality of metagenomic DNA isolated from zebrafish was assessed for PCR amplification using 16S rRNA gene-specific universal primers 5’AGAGTTTGATCCTGGCTCAG 3’ and 5’ ACGGCTACCTTGTTACGACTT 3’ [13] to assess its suitability for PCR with the following parameters: 94°C(05:00)+ (94°C(00:30)+56°C(00:30)+72°C(02:00)) × 34 cycles + 72°C(10:00). PCR was performed in a thermal cycler (Bio-Rad, USA). The amplicons were electrophoresed on 1.5% agarose gel and visualized using gel documentation system (Syngene, USA).
2.7. Statistical Analysis
All experiments were repeated thrice and GraphPad Prism Version 6.01 was used for statistical analysis (values are mean ± standard deviation of three independent experiments).
3. RESULTS AND DISCUSSION
A variety of commercial kits and modified protocols is reported for bacterial metagenomic DNA extraction. However, only few methods yield optimum amount of DNA and most of these methods are time consuming, costly, and use toxic chemicals giving unsatisfactory results in downstream applications like PCR. Most of the studies done so far have proved that kit-based methods yield more DNA than manual methods [14]. Stool kits are commonly used in diversity studies because of its ability to detect higher diversity than tissue kit. Initial sample weight taken in tissue kit is much less than that in the stool kit.
In the present study, we modified the kit protocol to get good quality community DNA from fewer number of zebrafish. Only five fish of approximate weight 1.5 ± 0.12 g were used in the given three methods, and the samples weighed approximately 150 mg compared to the minimum sample of 220 mg needed in the stool kit. Kit protocols normally use enzymatic methods, particularly for the lysis of Gram-positive bacteria which have a thick peptidoglycan layer. In the modified kit method, we gave an additional lysozyme treatment along with an extended incubation in lysis buffer which helped in the lysis of more bacterial cells, therefore, giving a higher yield of community DNA. Further, for concentrating the eluted DNA in AE buffer, precipitation was done using 3M sodium acetate and isopropanol at room temperature. Manual method was modified slightly in the final steps using spin column from QIAamp DNA Stool Mini Kit to check for improved DNA yield. Treatment with DNase-free RNase and Proteinase K was done before transfer to QIAamp spin column. DNA binds specifically to the QIAamp silica gel membrane while contaminants pass through. These modifications helped in improving the purity of DNA but not the yield. An overview of the methods used in the study is shown [Table 1].
Studies on gut microbiome are significantly dependent on the DNA extraction protocols [15,16]. It was shown that commercial DNA extraction kits which are optimized for the removal of inhibitors perform better with feces and intestinal mucus samples and also that the method of DNA extraction influences the downstream applications in sequencing studies. The effect of the method chosen for DNA extraction and its impact on microbiota is often neglected and has not been studied in fish [14]. Three methods of metagenomic DNA isolation from zebrafish gut were compared in terms of DNA yield, purity, and suitability for PCR. The highest yield was obtained by modified kit method giving 288 ng/mg of gut sample, while the lowest was by kit method (28.48 ng/mg). The DNA was visualized in 0.8% agarose gel [Figure 1]. Visible band was not observed for method I due to low yield of DNA. In gut microbiome studies, gut content is used to analyze non-adherent (allochthonous) communities, and rinsed intestinal tissue is used to analyze adherent (autochthonous) bacterial communities [17]. In all the three methods, samples were processed so that both adherent and non-adherent communities were isolated during the procedure. Several studies have been reported for the isolation of metagenomic DNA from zebrafish [11,18-20]. However, the yield and purity of DNA isolated remained a major difficulty in downstream processing of metagenomics. Results from our study showed that the modified kit method gave better yield of DNA compared to the other two. The higher yield was attributed to the lysozyme treatment before using kit.
The A260/280 ratio is used to assess the purity of DNA and RNA [12,21] and a ratio of ~1.8 is generally accepted as “pure” for DNA, while that of ~2.0 is generally accepted as “pure” for RNA. If the ratio is appreciably lower in either case, it indicates the presence of protein, phenol, or other contaminants that absorb strongly at or near 280 nm. Comparative analysis revealed that A260/280 ratios for kit, modified kit, and manual method were 2.52, 1.93, and 1.82, respectively. In manual method, the higher purity was due to modification in purification steps included in the protocol, but the yield of DNA was less than that from modified kit method. The A260/230 ratio is generally used as a secondary measure for accessing nucleic acid purity. The 260/230 values are often higher than respective 260/280 values, which are commonly in the range of 2.0–2.2. Both kit and manual method gave higher ratios for A260/230 indicating the presence of contaminants which absorbed at 230 nm. Studies have stated that modification of protocols using commercial kits or manual methods which utilize commercial kit only for purification of DNA, greatly affects the quantity and quality of DNA isolated [22-24]. The yield and purity of DNA isolated are summarized [Table 2].
Quality of the isolated metagenomic DNAs was further analyzed for downstream application mainly as template for PCR amplification using 16S rRNA primer which showed expected size amplicons of 1.5 kb size [Figure: 2]. The major contribution in natural diversity studies of microorganisms is supported by methods that are based on PCR amplification and sequencing of 16S rRNA gene sequences. The analysis of 16S rRNA isolated from different microbiome has led to the identification of several new phylogenetic lineages of high taxonomy level of bacteria and archaea [25]. 16S rRNA gene sequences are the most widely used housekeeping genetic marker for reasons such as (1) its occurrence in almost all bacteria in which they exist as operon, (2) the function of 16S rRNA gene has not changed over time, and (3) the gene size is large (1500 bp) which makes it suitable for bioinformatics studies [26]. In the study, DNA isolated by all the three methods could be PCR amplified, with DNA isolated by method I giving a faint band due to low yield of DNA. In general, a high-quality DNA free from contaminants is a prerequisite for PCR amplification. PCR amplification from template DNA obtained from method I gave a faint band compared to the other two methods which may be due to coextraction of RNA during DNA extraction. This may be due to the absence of RNAse treatment in method I. Overall, from the current study, it was evident that modification in protocols when using commercial kits can result in better results, and this modified method can be used for the isolation of metagenomic DNA from zebrafish gut.
 | Table 1: An overview of the methods used in the study. [Click here to view] |
 | Table 2: Comparison of DNA purity and yield obtained by three different methods [Click here to view] |
 | Figure 1: Agarose gel electrophoresis of metagenomic DNA from three methods. Lane 1. λ/Eco RI + Hind III double digest ladder, Lane 2. DNA isolated by kit method, Lane 3. DNA isolated by modified kit method, Lane 4. DNA isolated by manual method [Click here to view] |
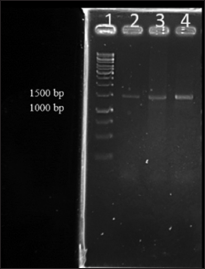 | Figure 2: Polymerase chain reaction (PCR) amplification of 16S rRNA using genomic DNA isolated from three methods. Lane 1. Gene ruler 1 kb DNA ladder, Lane 2. 16S PCR products of DNA isolated by kit method, Lane 3. 16S PCR products of DNA isolated by modified kit method, Lane 4. 16S PCR products of DNA isolated by manual method [Click here to view] |
4. CONCLUSION
The modified kit method can be used for the isolation of metagenomic DNA from gut of zebrafish which is the model organism. To the best of our knowledge, this is the first study reported from India as a part of metagenomics studies in gut microbiome of zebrafish. This study was done as part of diversity studies in gut microbiome of zebrafish as a result of probiotic administration. Future studies should examine the relation between the amount of sample, yield of DNA, and gut microbial diversity in downstream analysis.
5. ACKNOWLEDGMENTS
The first author acknowledges University Grants Commission (UGC) of the Government of India for providing financial support in the form of UGC NET-JRF. This study was also supported by Cochin University of Science and Technology.
7. REFERENCES
1. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001;291:881-4.
2. Nayak SK. Role of gastrointestinal microbiota in fish. Aquacult Res 2010;41:1553-73.
3. Merrifield DL, Ringo E, editors. Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics. Chichester, UK: John Wiley & Sons; 2014.
4. Ringø E, Olsen RE, Vecino JG, Wadsworth S, Song SK. Use of immunostimulants and nucleotides in aquaculture: A review. J Mar Sci Res Dev 2012;1:104.
5. Silva FC, Nicoli JR, Zambonino-Infante JL, Kaushik S, Gatesoupe FJ. Influence of the diet on the microbial diversity of faecal and gastrointestinal contents in gilthead sea bream (Sparus aurata) and intestinal contents in goldfish (Carassius auratus). FEMS Microbiol Ecol 2011;78:285-96.
6. Carda-Diéguez M, Mira A, Fouz B. Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiol Ecol 2014;87:451-9.
7. Ravin NV, Mardanov AV, Skryabin KG. Metagenomics as a tool for the investigation of uncultured microorganisms. Russian J Gen 2015;51:431-9.
8. Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 2008;3:e2836.
9. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 2010;4:232.
10. Garcia-Mazcorro JF, Lanerie DJ, Dowd SE, Paddock CG, Grützner N, Steiner JM, et al. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol Ecol 2011;78:542-54.
11. Hart ML, Meyer A, Johnson PJ, Ericsson AC. Comparative evaluation of DNA extraction methods from feces of multiple host species for downstream next-generation sequencing. PLoS One 2015;10:e0143334.
12. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
13. Shivaji S, Bhanu NV, Aggarwal RK. Identification of Yersinia pestis as the causative organism of plague in India as determined by 16S rDNA sequencing and RAPD-based genomic fingerprinting. FEMS Microbiol Lett 2000;189:247-52.
14. Larsen AM, Mohammed HH, Arias CR. Comparison of DNA extraction protocols for the analysis of gut microbiota in fishes. FEMS Microbiol Lett 2014;362:pii: Fnu031.
15. Lantz PG, Matsson M, Wadström T, Rådström P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods 1997;28:159-67.
16. Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015;3:26.
17. Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015;3:26.
18. Ringø E, Zhou Z, Vecino JG, Wadsworth S, Romero J, Krogdahl Å, et al. Effect of dietary components on the gut microbiota of aquatic animals. A never ending story? Aquacult Nutr 2016;22:219-82.
19. Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, et al. Evidence for a core gut microbiota in the zebrafish. ISME J 2011;5:1595.
20. Borrelli L, Aceto S, Agnisola C, De Paolo S, Dipineto L, Stilling RM, et al. Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Sci Rep 2016;6:30046.
21. Udayangani RM, Dananjaya SH, Nikapitiya C, Heo GJ, Lee J, De Zoysa M. Metagenomics analysis of gut microbiota and immune modulation in zebrafish (Danio rerio) fed chitosan silver nanocomposites. Fish Shellfish Immunol 2017;66:173-84.
22. Gallagher SR, Desjardins PR. Quantitation of DNA and RNA with absorption and fluorescence spectroscopy. Curr Protoc Mol Biol 2006;76:A-3D.
23. Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, et al. DNA extraction from soils: Old bias for new microbial diversity analysis methods. Appl Environ Microbiol 2001;67:2354-9.
24. Lakay FM, Botha A, Prior BA. Comparative analysis of environmental DNA extraction and purification methods from different humic acidrich soils. J Appl Microbiol 2007;102:265-73.
25. Gutiérrez-Lucas LR, Montor-Antonio JJ, Cortés-López NG, del Moral S. Strategies for the extraction, purification and amplification of metagenomic DNA from soil growing sugarcane. Adv Biol Chem 2014;4:281.
26. Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J 2008;2:3.
27. Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn 2001;6:313-21.