1. INTRODUCTION
Neem (Azadirachta indica), a tree in the mahogany family Meliaceae, in the genus Azadirachta, is native to India and Burma, growing in tropical and semi-tropical regions [1]. It has been used as a traditional medicine to cure multiple diseases [2]. Before history was documented, neem has also been widely used in agriculture field to improve soil health as well as crop productivity [3]. In several ways, neem has been considered to be one of the most valuable plants having a broad range of biological properties [1]. In agricultural sector, neem has been used as fertilizer, manure, pesticide and soil conditioners [3]. Neem pest control is very beneficial for proper crop and pest management. Neem cake and neem extract widely used in agriculture as organic pesticides and also helps in the colonization of microbes. Neem is also used as urea coating agent, which is natural, environmental friendly, non-toxic, reduces urea consumption, increases the yield of crops, and is convenient to apply. The use of the neem extract was reported to enhance the growth of soil and rhizosphere microorganisms. This could be because some protein or carbohydrate residues present in the extract which is beneficial to the microorganisms [4]. Carney and Matson (2005) [5] reported that variations in the abundance of soil microorganisms might occur as a result of soil carbon processes. The carbon percentage of the soil was most strongly associated with changes in the composition of the microbial community. Soil microorganisms play a crucial role in improving plant growth and nutrition. The variety of microorganisms present in a particular soil is usually an indicator of soil quality. Neem extract is involved in promoting growth of beneficial soil microorganism and, at the same time, is also capable of controlling pathogenic microorganisms [4]. Studies showed that neem plant parts contain many microbes which demonstrated plant growth promotion (PGP) and antimicrobial activity.
Extensive research, as previously conducted, has shown that endophytes produce secondary metabolites that play an essential role in plant health [6]. Endophytes are plant-colonizing microorganisms and have been found in almost all plants [7]. Neem tree is well known to have beneficial microflora and studies have also detected unique microbial spectrum in neem stem, roots, bark, and leaves [8-10]. They serve as defenses against pathogens, and some develop useful secondary anti-microbial substances [6]. It was reported previously that plant microbiome continuously exchanged genetic material with other species in an ecological niche which is beneficial for plant growth and microbes adaptation [11]. There is growing attention in the plant microbiome and their bioactive compounds due to their utility in agriculture, industry, and in healthcare [12]. As a result, the endophytic bacteria extracted from neem had significant applications in agriculture, biology, bio-energy, and pharmacy due to the availability of secondary metabolites such as epoxy/hydroxyazadiradione, nimbin, and salanin [13]. The neem tree offers a very favorable niche for microorganisms to flourish due to its profuse plant development [10]. Few microbes such as Serratia, Enterobacter, and Bacillus species were isolated from external exudates of papaya and snap bean plants [14]. Gums are water-soluble, complex carbohydrates (such as galactose, xylose, and rhamnose) that transform viscous gels and mucilages [15]. The stem of the neem tree contains a viscous brown fluid called neem gum, which has many functional carbohydrates and polysaccharides. In certain plants, they are secreted by cells contained in the bark and act as the plant’s natural sheath providing protection against external microbial attacks [16]. This study is done under the thought that diverse and more efficient microbes may be found in neem gum. This research aimed to isolate classify and describe bacterial isolates present in neem gum and explore their PGP activities.
2. MATERIALS AND METHODS
2.1. Sampling and Isolation of Bacteria
Neem gum was collected from Neem plants located at Maunath Bhajan (25.90oN, 83.49oE), Uttar Pradesh, India under sterile conditions and processed for bacterial isolation. Different media such as nutrient agar (NA), 1/10NA, King’s B, starch casein agar (SCA), tryptic soya agar, R2A, actinomycetes isolation agar, streptomyces isolation agar, SCA with antibiotics (20 μg/mL Cycloheximide, 50 μg/mL Rifampicine), 1/100 NA, international streptomyces project medium 4 (ISP4), and 10% neem leaf media were all procured from HiMedia (India), which were used in the current analysis to isolate the diverse bacteria present in the neem gum. Serial dilutions 10-3, 10-4, and 10-5 dilution were plated on the different media and incubated at 37°C for 3 days. Different bacterial morphotypes were purified and used for further analysis.
2.2. Amplified Ribosomal DNA Restriction Analysis (ARDRA) and Phylogeny
Genomic DNA was extracted from the bacterial isolates following the procedures described by Ausubel et al. (1992) [16]. The 16S rRNA gene was amplified using the primer pA (5′-AGA GTT TGA TCA TGG CTC AG-3′) and pH (5′-AAG GAG GTG ATC CAG CCG CA-3′) [17]. The obtained PCR products were visualized on 1.5% agarose gel using ethidium bromide and documented using a gel documentation unit. PCR products were purified using Wizard SV Gel/PCR product purification kit (Promega, USA). The purified 16S rRNA gene amplicons were digested with two restriction endonucleases – MspI and HaeIII (Promega, USA) in a 20 μL reaction volume following the manufacture’s protocol. After digestion of the purified amplicons, the samples were resolved on 2% agarose gel, and amplified rDNA restriction analysis (ARDRA) was performed [18]. Each sample’s restriction pattern was visualized and compared to a 100 bp DNA ladder (Thermo Fischer Scientific) and documented using a Bio-Rad imager. Binary data clustering and pattern analysis were performed using a numerical taxonomy analysis program (NTYSIS) package (version2.02e, Exeter Software, Setauket, NY). Jaccard’s coefficients were used to differentiate the isolates obtained from neem gum, and a UPGMA dendrogram was constructed using the binary data [19].
2.3. Phylogenetic Analysis
The purified amplicons of the selected OTUs were sequenced from Eurofins India Pvt. Ltd., Bengaluru. The partial 16S rRNA gene sequences were compared with sequences available in the NCBI database using the BLASTn tool. Isolates were identified to species level based on 16S rRNA gene sequence similarity of ≥97% with the sequences in GenBank. Sequence alignment and comparison used the multiple sequence alignment tool CLUSTAL W2 [20] with default parameters. The phylogenetic tree was constructed on aligned data sets using the neighbor-joining (NJ) method and MEGA 7 [21]. Bootstrap analysis was performed on 1000 random samples taken from multiple alignments.
2.4. Physiological and Biochemical Characterization
All identified isolates were physiologically characterized based on their ability to tolerate various salt and pH concentration. Isolates were spotted on nutrient agar supplemented with varying salt concentrations (2, 4, 6, 8, and 10% NaCl) and with a wide range of pH (5, 7, 9, and 10) plate and incubated at 37°C for 72 h. Isolates were also biochemically characterized based on Gram reaction, oxidase, IMViC test, nitrate reduction, Triple sugar utilization test, and H2S production per Bergey’s Manual of Determinative Bacteriology [22].
2.5. In Vitro Screening for Plant Growth Promoters
The plant growth-promoting attributes such as Indole acetic acid (IAA), HCN, siderophore production, and phosphate solubilization were determined as described earlier by Ahmad et al. [23]. Further, the potassium and zinc solubilization was performed as method of Gupta and Pandey [24]. Briefly, the bacterial cultures grown in nutrient broth with supplementation of L-Tryptophan for 48 h at 37°C were used to estimate the production of IAA. Similarly, chrome azurol S (CAS) agar plates were used to estimate the production of siderophore. HCN production was determined by growing the isolates on King’s B agar plate amended with 4.4 g glycine, and placing a Whatman filter paper no. 1 soaked in 2% sodium carbonate in 0.5% picric acid solution on the top of the plate. Phosphate (P) solubilization was performed on Pikovskaya’s agar media containing tri-calcium phosphate as insoluble source of phosphate was used. Similarly, potassium (K) solubilization was determined by growing the cultures on Aleksandrov medium containing potassium alumino silicate as source of K. Further, the bacterial strains were screened for Zn solubilization on modified Tris-minimal agar media supplemented with 0.1% ZnSo4.
2.6. Hydrolytic Enzyme Production
The bacterial isolates were characterized for hydrolytic enzyme production such as amylase, cellulase, lipase, and pectinase through an agar plate assay [25]. Briefly, amylase production was tested qualitatively on starch agar (peptone 0.5%, yeast extract 0.2%, soluble starch 2%, and NaCl 0.5%). Isolates were spotted on starch agar plates and incubated at 30°C for 48 h. After incubation, plates were flooded with iodine solution (10 g potassium iodide, 5 g iodine crystals, and 100 mL distilled water). Isolates showing a zone of clearance were recorded as positive for amylase production. A positive culture was further tested for quantitative amylase production at pH 7.0 following the methods described by Bernfeld et al. [26]. The cellulolytic activity of bacterial isolates was performed using CMC agar (0.2% NaNO3, 0.1% K2HPO4, 0.05% MgSO4, 0.05% KCl, 0.2% carboxymethylcellulose [CMC] sodium salt, 0.02% peptone, and 1.7% agar), inoculated plates were incubated for 3 days at 30°C. For visualization of the hydrolysis zone, the plates were flooded with Congo red solution (1 mg/mL) for 15 min. This solution was then drained off, and the plates were destained twice by 1 M NaCl solution for 15 min. The yellow zone around the bacterial growth indicated the presence of cellulolytic activity. A positive culture was, further, tested for quantitative cellulase production at pH 7.0 following the protocols described by Singh et al. [27]. The pectinase activity of bacterial isolates was performed in pectinase agar media [28] containing yeast extract-1gm, ammonium sulfate-2 g, Na2HPO4- 6 g, KH2PO4- 3 g, citric pectin-5 g, and Agar-20 g, pH- 4, after incubation for 3 days colony showed clear zone on flooding with 1% cetyltrimethylammonium bromide confirmed pectinase producer.
3. RESULTS AND DISCUSSION
3.1. Isolation and Identification Bacteria from Neem Gum
In plants, it has been discovered that many endophytic microbes are present and need further taxonomical classification [29]. Studies also used classical approaches to count on the total number of microbial organisms. It is broadened the range of possibilities for microbes as well as new functions, particularly in agriculture, health, and the environment [30]. In the present study, a total of 130 different bacterial morphotypes were obtained from neem gum [Table 1]. A maximum number (13.84%) of isolates were obtained from nutrient agar and R2A media followed by AIA (13.07%) and 1/10NA (12.30%). The least number (0.76%) of the bacterial count was noticed in SCA with antibiotics. The dendrogram generated using the banding patterns obtained with MspI and HaeIII showed 50 operational taxonomic units (OTUs) at 100% similarity level. Sequencing of 16S rRNA (partial) amplicons of these OTUs was performed, and their taxonomic status was revealed by BLAST search. The most common bacterial phylum’s recovered from neem gum were Firmicutes (Bacillus sp. [11], Paenibacillus sp. [3], and Planococcus [1]); Actinobacteria (Rhodococcus [2], Brevibacterium, Streptomyces, Mycobacterium, Micromonas, Curtobacterium, Kytococcus, Brachybacterium [2], Isoptericola, Rothia, Kocuria); and Proteobacteria (Pseudomonas [10], Stenotrophomonas, Acinetobacter, Pantoea [2], Enterobacter [7], and Trabulsiella).
Table 1: Media used for the isolation of different bacteria isolated from Neem Gum.
| Media | No. of isolates | Percentage |
|---|---|---|
| Nutrient agar | 18 | 13.84 |
| 1/10 NA | 16 | 12.30 |
| KING’S B | 10 | 7.69 |
| SCA | 13 | 10.0 |
| TSA | 2 | 1.53 |
| R2A | 18 | 13.84 |
| AIA | 17 | 13.07 |
| Streptomyces isolation agar | 8 | 6.15 |
| SCA with antibiotic | 1 | 0.76 |
| 1/100 NA | 10 | 7.69 |
| ISP4 | 7 | 5.38 |
| Neem leaf media | 10 | 7.69 |
| Total | 130 | - |
The phylogenetic tree constructed with the partial sequence of 16S rRNA generated three clusters, of which each cluster represented the different phylums, that is, Firmicutes, Actinobacteria, and Proteobacteria [Figure 1]. The result obtained suggests the presence of diverse bacteria in neem gum. Some researchers doubted whether phenotypic methods would accurately classify isolates in the presence of the complex community [31]. As a piece of evidence for the molecular investigation, we have employed in our study 16S rRNA gene amplification and sequencing for selected isolates. Several microbiological and physiological identification methods were also used for the chosen bacterial isolates of neem gum to ascertain their diversity. The composition of neem gum is complex and might have enabled the growth of certain microbes. To date, we believe our study is novel, as no studies on neem gum microbial diversity were reported.
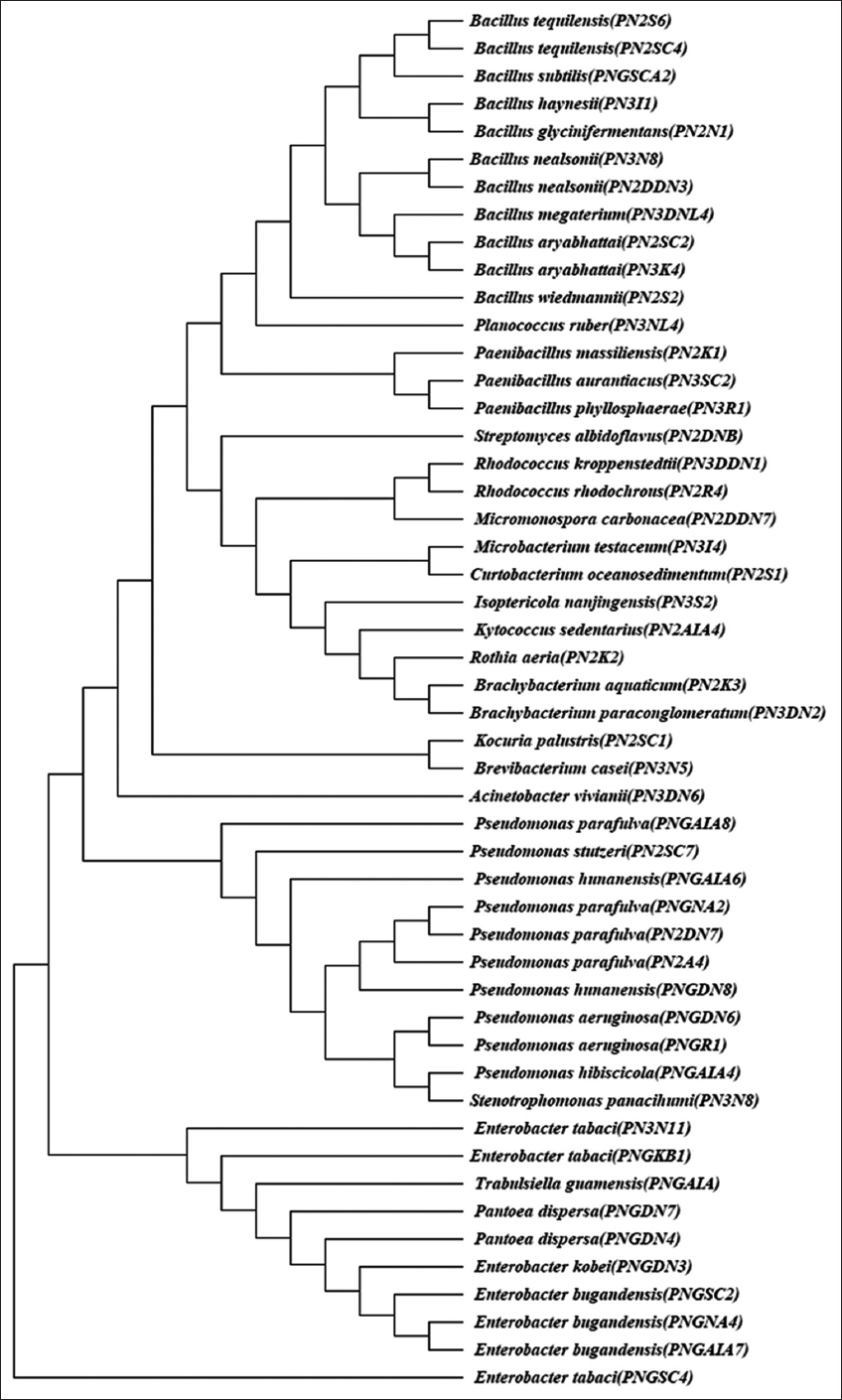 | Figure 1: Phylogenetic dendrogram based on comparison of 16S rRNA sequences of bacterial isolates isolated from neem gum. The phylogenetic tree was constructed on aligned datasets using the neighbor joining method in the program MEGA 7. Bootstrap analysis was performed on 1000 random samples taken from multiple alignments. [Click here to view] |
To provide a more comprehensive view of the overall bacterial diversity associated with the neem gum, the bacterial growth experiments were carried out with a wide range of salt concentrations [Figure 2a]. The obtained results were represented using a Venn diagram for bacterial isolates growth features at different salt and pH concentration [Figure 2]. Based on the molecular, physiological, and biochemical features, it was observed that the neem gum has a diverse microbial population. Overall, the Bacillus and Pseudomonas species were found to be predominant among the 50 isolates identified. These isolates belonged to different species of Bacillus, Paenibacillus, Brevibacterium, Rhodococcus, Kytococcus, Streptomyces, Pseudomonas, Enterobacter, Trabulsiella, Acinetobacter, Brachybacterium, Pantoea, Steonotrophomonas, Rothiaseria, Kocuria, Isoptericola, Curtobacterium, Micromonas, Microbacterium, and Planococcus. Earlier, various species of Bacillus and Pseudomonas were reported from leaves and roots of neem [32,33]. Similarly, a large number of actinomycetes belonging to Streptomyces, Streptospoarangium Saccharomonospora, Microbispora, and Nocardia were reported from the roots of neem [34,35].
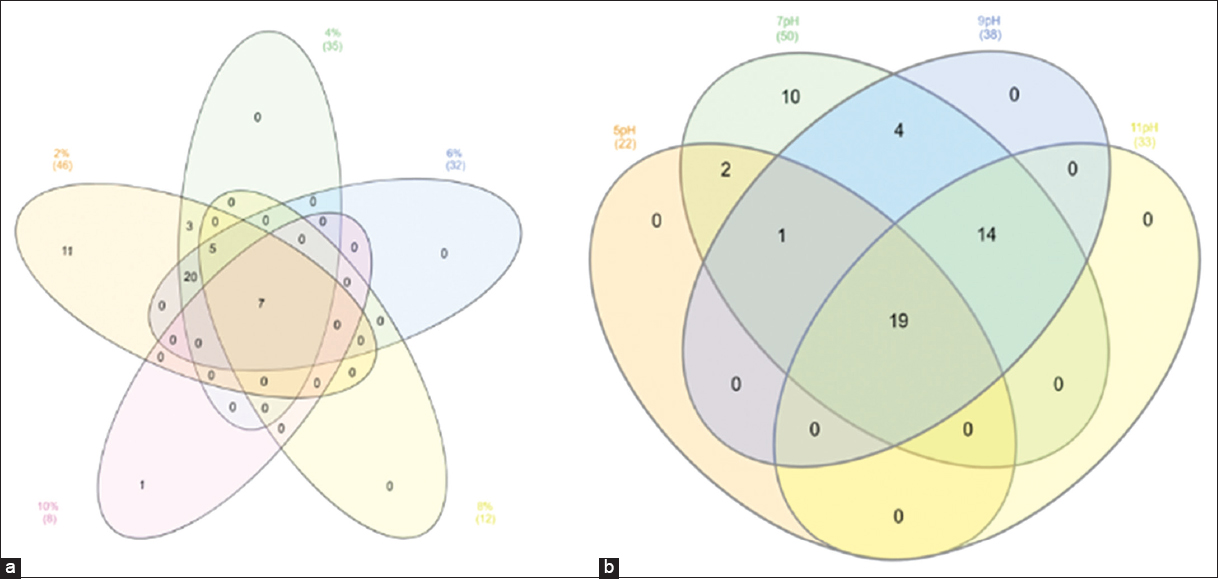 | Figure 2: Venn plot showing physiological characterization of selected isolates at different salt (a) and pH (b) concentrations. [Click here to view] |
It was noted that seven isolates were able to utilize 0.5 to 10% salt concentration. Among the 50 isolates, it was observed that 19 bacterial cultures could grow at pH 5 to 10, while ten isolates could grow only until pH 7 [Figure 2b]. Physiological characterization permitted us to classify promising microbes and concentrated on the relationships between particular bacterial behaviors and strain features. Few isolates were able to survive under high salt concentrations and a wide range of pH, suggesting their ability to grow under osmotic stress. The biochemical diversity was also noticed among the 50 isolates per Bergey’s Manual [Figure 3]. A total of 22 Gram-negative and 28 Gram-positive isolates were isolated from neem gum. The biochemical features such as citrate utilization, acid and gas production, IMViC assay, lactose, glucose, and sucrose fermentation experiments were performed for the selected isolates. Most of the isolates were able to ferment glucose and produced catalase. Only two of the Pseudomonas parafulva isolates, PN2A5 and PN2DN7, produced gas, while all other isolates showed no gas production. Such traits might have helped the microorganisms to colonize and survive under high osmotic potential of neem gums due to presence of sugars and polysaccharides. Endophyte enables the plants to provide many benefits to the surrounding environment. Most endophytic microbes can retrieve various nutrients and protection from the invading parasites and fungal organisms [36].
 | Figure 3: Biochemical characterization pattern of the selected isolates. The black box represents positive results. [Click here to view] |
3.2. PGP Activity
Bacterial isolates obtained from neem gum exhibited that various PGP activities are presented in Table 2. IAA production was found to be positive for 44 isolates. IAA production ranged of 2–730 μg/mL. Up to 36.3% of bacteria could produce IAA between the ranges 50–100 μg/mL [Figure 4b]. Highest (730.7 μg/mL) IAA production was noticed for Trabulsiella guamensis (PNGAIA), followed by Enterobacter bugandensis (PNGNA4) (564.4 μg/mL) with [Figure 4a]. Most of the Enterobacter spp. (such as E. tabaci, and E, bugandensis) were able to produce IAA in a range of 431.3–158.5 μg/mL. All these isolates belonged to the Proteobacteria. The IAA production was highest in isolates of T. guamensis, followed by Enterobacter, Pseudomonas hunanensis, Kytococcus sedentarius, Microbacterium testaceus, and Bacillus aryabhattai. Enzymatic activity is present in many endophytic bacteria, including pectinase, cellulase, amylase, and many more [25,37]. Around 40% of the bacteria found in this study had enzymes that made at least one of the four hydrolytic products. Similarly, high hydrolytic enzyme especially pectinase production potential might have helped the microbial population to break down the complex polysaccharides of the gum and use them as carbon source.
Table 2: Functional characteristic of bacterial isolates isolated from Neem gum on the basis of Plant growth promotion (PGP) traits.
| Organism | Isolate | Solubilization | Biocontrol activity | Phytohromone | |||
|---|---|---|---|---|---|---|---|
| Phosphate | Potassium | Zinc | Siderophore | HCN | IAA | ||
| PNGNA2 | Pseudomonas | - | - | - | + | + | + |
| PNGNA4 | Enterobacter | - | + | + | + | - | + |
| PNGDN3 | Enterobacter | - | + | - | + | - | + |
| PNGDN4 | Pantoeadispersa | + | - | + | + | - | + |
| PNGDN6 | Pseudomonas aeruginosa | - | - | + | - | + | + |
| PNGDN7 | Pantoeadispersa | - | - | - | + | + | + |
| PNGDN8 | Pseudomonas hunanensis | - | - | - | + | - | + |
| PNGKB1 | Enterobacter tabaci | + | + | + | + | - | + |
| PNGSC2 | Enterobacter bugandensis | - | - | - | - | - | + |
| PNGSC4 | Enterobacter tabaci | - | - | + | - | + | + |
| PNGR1 | Pseudomonas aeruginosa | - | - | + | - | + | + |
| PNGAIA4 | Pseudomonas | - | - | - | - | - | + |
| PNGAIA6 | Pseudomonas hunanensis | - | - | - | + | - | + |
| PNGAIA7 | Enterobacter bugandensis | + | + | + | + | - | + |
| PNGAIA8 | Pseudomonas parafulva | - | - | - | + | + | + |
| PNGAIA | Trabulsiella guamensis | + | + | + | + | - | + |
| PN2N1 | Bacillus wiedmannii | - | - | - | - | - | + |
| PN2S1 | Rhodococcuskroppenstedtii | - | - | - | - | - | + |
| PN2S2 | Bacillus tequilensis | - | - | - | - | - | + |
| PN2S6 | Microbacteriumtestaceum | - | - | + | - | - | + |
| PN2R4 | Streptomyces albidoflavus | + | - | - | - | - | - |
| PN2SC1 | Kocuria palustris | - | - | - | - | - | + |
| PN2SC2 | Bacillus aryabhattai | - | - | + | - | - | + |
| PN2SC4 | Bacillus nealsonii | - | - | - | + | - | + |
| PN2SC7 | Pseudomonas stutzeri | - | - | - | - | - | + |
| PNGSCA2 | Bacillus nealsonii | - | - | - | - | - | + |
| PN2K1 | Brachybacterium aquaticum | - | - | - | - | - | - |
| PN2DDN3 | Paenibacillus aurantiacus | - | - | - | - | - | - |
| PN2DDN7 | Paenibacillus phyllosphaerae | - | - | - | - | - | + |
| PN2DNB | Micromonospora carbonacea | - | - | + | + | + | - |
| PN2A5 | Pseudomonas parafulva | - | - | + | - | - | + |
| PN2DN7 | Pseudomonas parafulva | - | - | - | + | + | + |
| PN2AIA4 | Bacillus megaterium | - | - | + | + | + | + |
| PN2K2 | Planococcus ruber | - | - | - | - | - | + |
| PN2K3 | Bacillus glycinifermentans | - | - | - | + | + | + |
| PN3I4 | Bacillus glycinifermentans | - | - | + | - | + | + |
| PN3K4 | Rothiaaeria | - | - | - | - | - | + |
| PN3N8 | Stenotrophomonaspanacihumi | - | - | - | - | - | + |
| PN3S2 | Bacillus aryabhattai | - | - | - | + | + | + |
| PN3DN6 | Acinetobacter vivianii | - | - | - | - | - | + |
| PN3N3 | Rhodococcusrhodochrous | - | - | - | - | - | + |
| PN3N5 | Bacillus subtilis | - | - | - | - | - | + |
| PN3DNL4 | Kytococcus sedentarius | - | - | + | + | - | + |
| PN3I1 | Curtobacterium oceanosedimentum | - | - | - | + | - | + |
| PN3SC2 | Bacillus tequilensis | - | - | + | - | - | + |
| PN3N11 | Enterobacter tabaci | + | + | + | + | - | + |
| PN3R1 | Isoptericola nanjingensis | - | - | - | + | - | + |
| PN3DDN1 | Paenibacillus massiliensis | - | - | - | - | - | + |
| PN3NL4 | Brevibacterium casei | - | - | - | - | - | - |
| PN3DN2 | Brachybacterium paraconglomeratum | - | - | + | - | - | - |
The isolates which showed positive response for more than three functional properties were highlighted in grey color. - Indicates negative response, + indicates positive response of isolates for PGP activity. IAA: Indole acetic acid
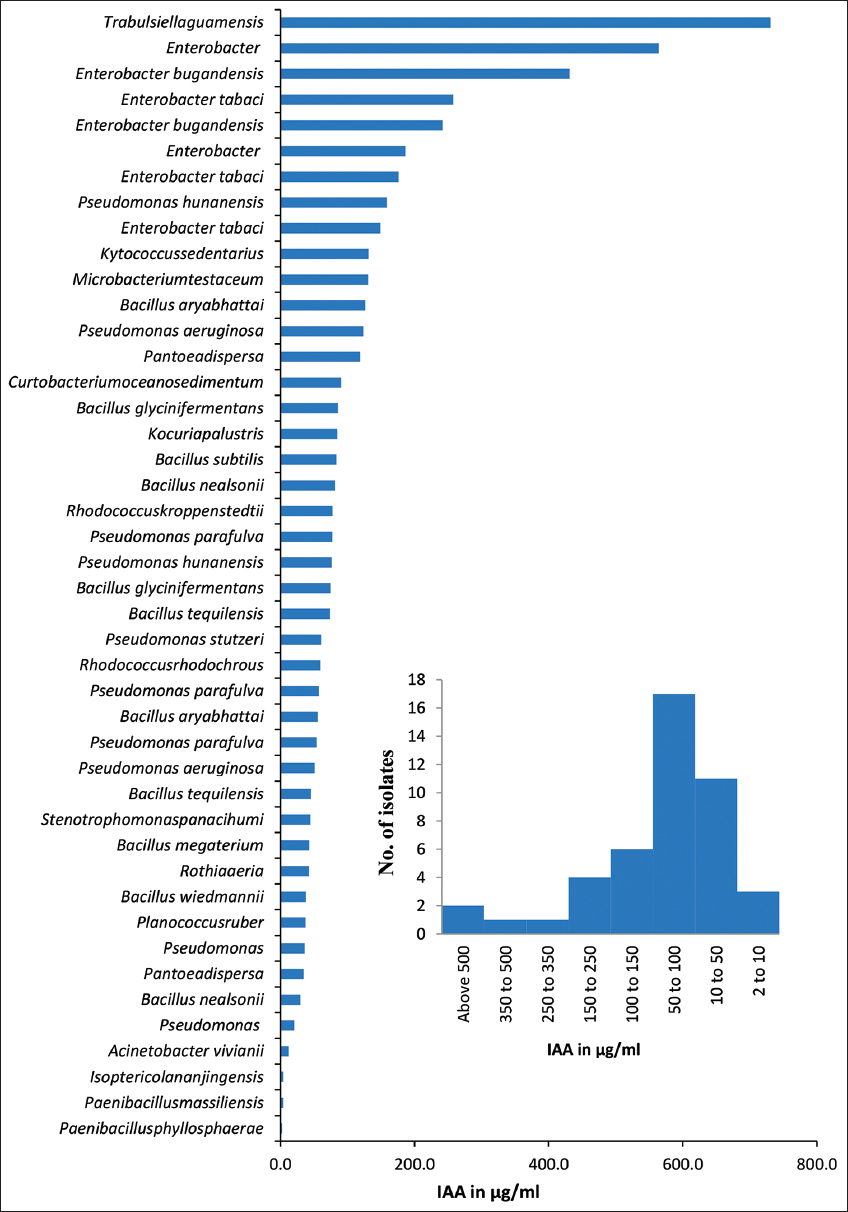 | Figure 4: Graph represents quantification of Indole acetic acid produced by among diverse bacterial isolates isolated form neem gum. [Click here to view] |
Out of 50 bacterial isolates, 21 isolates showed siderophore production, and 12 isolates were found positive for HCN production. Most of the Pseudomonas (PNGNA2, PNGDN6, PNGR1, PNGAIA8, and PN2DN7) and Bacillus (PN2AIA4, PN3K3, PN3I4, and PN3S2) species were predominant HCN producers. In case of siderophore producers, Enterobacter (PNGNA4, PNGDN3, PNGKB1, PNGAIA7, and PN3N11), Pseudomonas (PNGNA2, PNGDN8, PNGAIA6, PNGAIA8, and PN2DN7), Bacillus (PN2SC4, PN2K3, PN2AIA, and PN3S2), and Pantoea dispersa (PNGDN4 and PNGDN7) were found to be abundant. Among all, six different isolates (Pantoea dispersa PNGDN, T. guamensis PNGAIA, Streptomyces albidoflavus PN2R4 and three isolates of Enterobacter sp. PNGKB1, PNGAIA7, and PN3N11) were able to solubilize phosphate. In potassium solubilization, most of the Enterobacter spp. (PNGNA4, PNGDN3, PNGKB1, PNGAIA7, and PNGN11) and T. gumensis (PNGAIA) have shown a positive reaction. Similarly, 18 bacterial isolates are found to be Zn solubilizers. Distribution of various plant growth promoting attributes among the isolates is presented using Venn. Diagram [Figure 5]. Potential PGP and disease resistance properties, when present, are usually considered to be correlated with endophytic bacteria [38]. Some have suggested that a variety of hypotheses may explain the PGP of plant-associated endophytes. Several of which enhance the availability of nutrients for plant growth, such as nitrogen and phosphorus, or metals, or help to supply the plant with biosynthetic metabolites that regulate development, production, and plant response to protection [39]. It was observed that about 98%, 42%, and 24% of isolated bacteria were able to produce IAA, siderophore, and HCN. Many plants which possess such bacteria with biocontrol activity would compete with phytopathogens and protect the plant from infections [38]. These bacterial endophytes were distinguished based on their PGP activity, IAA production, and solubilization. In the present study, T. guamensis PNGAIA was tested positive more most of the parameters, except for lipase, pectinase, and HCN production.
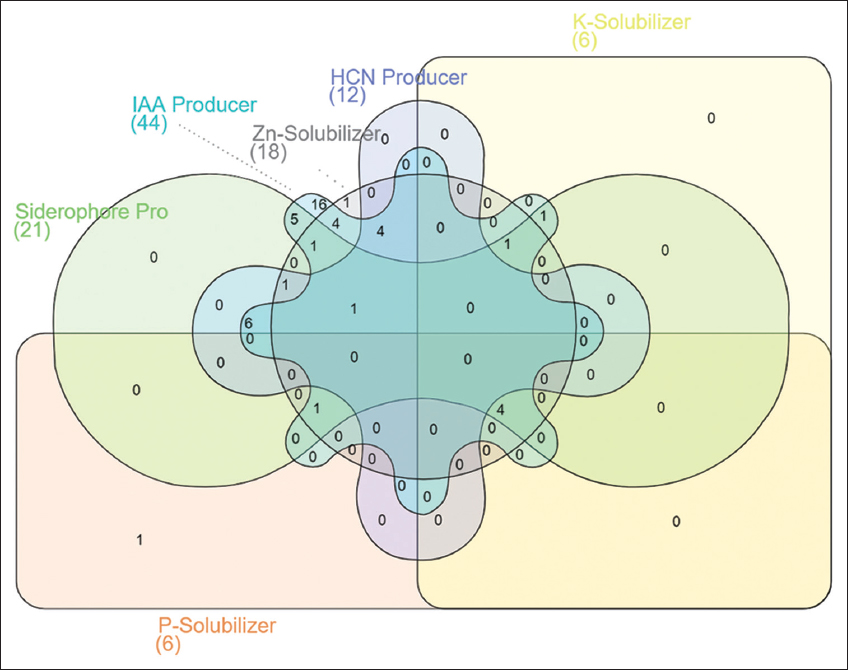 | Figure 5: Venn diagram showing analysis of plant growth promotion activity of the bacteria isolates obtained from neem gum. [Click here to view] |
3.3. Hydrolytic Enzyme Production
Bacterial isolates obtained from neem gum were tested for their hydrolytic enzymes such as amylase, cellulase, pectinase, and lipase. Plate assays revealed that, out of 50 isolates, 13, 12, 14, and 31 isolates were positive for amylase, cellulase, lipase, and pectinase, respectively, at pH 7 [Table 3]. The overall data for the production of hydrolytic enzymes is presented using Venn Diagram [Figure 6]. Among all, pectinase-producing organisms were found to be abundant in neem gum. Among the 14 lipase-producing organisms, isolates of Pseudomonas, Bacillus, Rhodococcus, Enterobacter tabaci, Brachybacterium aquaticum, Rothia aeria, Acinetobacter viviani, Kytococcus sedentarius, and Curtobacterium oceeanosedimentum were predominant. It was observed that Pseudomonas spp., Bacillus spp. E. tabaci, T. guamensis, Microbacterium testaceum, Streptomyces albidoflavus, Paenibacillus phyllosphaerae, and Brevibacterium casei were found to produce amylase. Highest (36.4 IU/mL) amount of amylase was produced by B. arybhattai PN3S2, and followed by Paenibacillus phyllosphaera PN2DDN7, Pseudomonas PNGNA2, and E. tabaci PNGSC4 with 22.17, 13.5, and 11.8 IU/mL, respectively. The microorganisms such as Enterobacter sp., Pantoea dispersa, T. guamensis, Bacillus sp., Micromonospora carbonacea, R. aeria, and K. sedentarius were able to produce cellulase. The isolate B. arybhattai PN3S2 was also found to produce highest (1.5 IU/mL) amounts of cellulase.
Table 3: Functional characteristic of bacterial isolates isolated from Neem gum on the basis of hydrolytic enzyme production.
| Organism | Isolate | Enzymatic activity | |||
|---|---|---|---|---|---|
| Lipase | Pectinase | Amylase | Cellulase | ||
| PNGNA2 | Pseudomonas | - | + | + | - |
| PNGNA4 | Enterobacter | - | + | - | + |
| PNGDN3 | Enterobacter | - | - | - | - |
| PNGDN4 | Pantoeadispersa | - | - | - | + |
| PNGDN6 | Pseudomonas aeruginosa | - | + | - | - |
| PNGDN7 | Pantoeadispersa | - | + | - | - |
| PNGDN8 | Pseudomonas hunanensis | - | + | - | - |
| PNGKB1 | Enterobacter tabaci | - | - | - | + |
| PNGSC2 | Enterobacter bugandensis | - | + | - | - |
| PNGSC4 | Enterobacter tabaci | + | + | + | - |
| PNGR1 | Pseudomonas aeruginosa | + | + | + | - |
| PNGAIA4 | Pseudomonas | + | + | - | - |
| PNGAIA6 | Pseudomonas hunanensis | - | + | - | - |
| PNGAIA7 | Enterobacter bugandensis | - | - | - | + |
| PNGAIA8 | Pseudomonas parafulva | - | + | - | - |
| PNGAIA | Trabulsiella guamensis | - | - | + | + |
| PN2N1 | Bacillus wiedmannii | - | + | + | - |
| PN2S1 | Rhodococcus kroppenstedtii | + | + | - | - |
| PN2S2 | Bacillus tequilensis | - | + | + | + |
| PN2S6 | Microbacterium testaceum | - | - | + | - |
| PN2R4 | Streptomyces albidoflavus | - | - | + | - |
| PN2SC1 | Kocuriapalustris | - | - | - | - |
| PN2SC2 | Bacillus aryabhattai | - | - | - | - |
| PN2SC4 | Bacillus nealsonii | - | + | - | - |
| PN2SC7 | Pseudomonas stutzeri | - | - | - | - |
| PNGSCA2 | Bacillus nealsonii | - | + | - | - |
| PN2K1 | Brachybacterium aquaticum | + | + | - | - |
| PN2DDN3 | Paenibacillus aurantiacus | - | - | - | - |
| PN2DDN7 | Paenibacillus phyllosphaerae | - | + | + | + |
| PN2DNB | Micromonospora carbonacea | - | + | - | + |
| PN2A5 | Pseudomonas parafulva | + | + | - | - |
| PN2DN7 | Pseudomonas parafulva | - | + | - | - |
| PN2AIA4 | Bacillus megaterium | + | - | - | - |
| PN2K2 | Planococcusruber | - | + | - | - |
| PN2K3 | Bacillus glycinifermentans | - | - | - | |
| PN3I4 | Bacillus glycinifermentans | + | + | + | + |
| PN3K4 | Rothiaaeria | + | + | - | + |
| PN3N8 | Stenotrophomonas panacihumi | - | + | - | - |
| PN3S2 | Bacillus aryabhattai | - | - | + | + |
| PN3DN6 | Acinetobacter vivianii | + | + | - | - |
| PN3N3 | Rhodococcusrhodochrous | + | + | - | - |
| PN3N5 | Bacillus subtilis | - | - | - | - |
| PN3DNL4 | Kytococcussedentarius | + | - | - | + |
| PN3I1 | Curtobacteriumoceanosedimentum | + | - | - | - |
| PN3SC2 | Bacillus tequilensis | + | - | + | - |
| PN3N11 | Enterobacter tabaci | - | + | - | - |
| PN3R1 | Isoptericola nanjingensis | - | + | - | - |
| PN3DDN1 | Paenibacillus massiliensis | - | - | - | - |
| PN3NL4 | Brevibacterium casei | - | + | + | - |
| PN3DN2 | Brachybacterium paraconglomeratum | - | - | - | - |
The isolates which showed positive response for more than three functional properties were highlighted in grey color. – indicates Negative response and+indicates Positive response of isolates for enzyme production
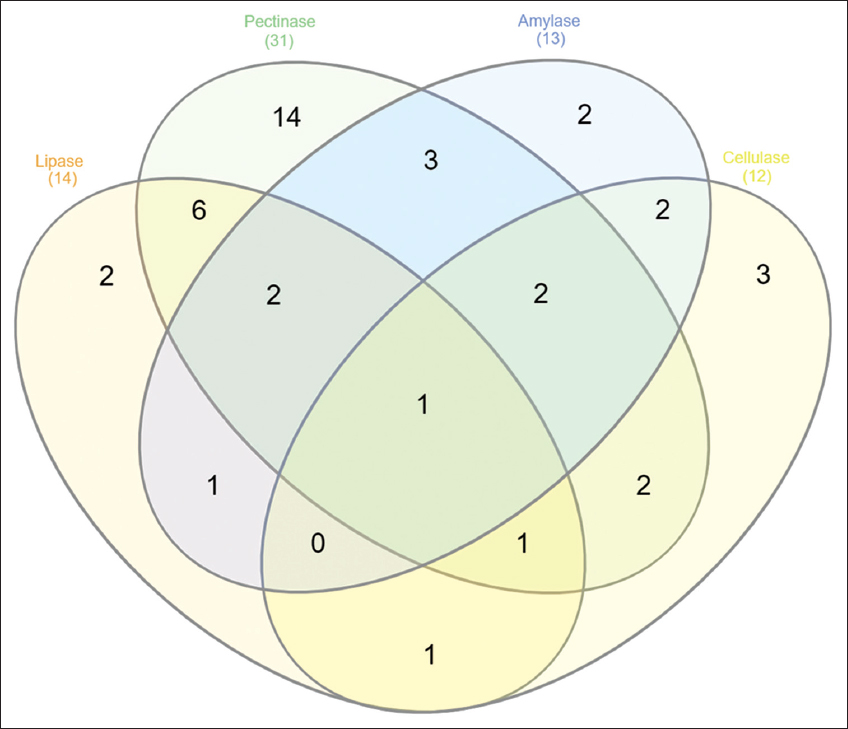 | Figure 6: Venn diagram showing analysis of hydrolytic enzyme production activity of bacterial isolates isolated from Neem gum. [Click here to view] |
The isolate B. glycinifermentans PN2DNB is the only bacteria that showed a positive response to all the enzymatic activity. Most of the Bacillus and Pseudomonas sp. were able to produce pectinase and amylase activity. Literature suggested that T. guamensis (an Enterobacteriaceae member) is a natural microbe residing in the termites gut with cellulolytic, ligninolytic, and nitrogen-fixing ability [40]. E. tabaci PNGKB1 and E. bugandensis PNGAIA7 showed no enzymatic activity but possessed solubilization and HCN and siderophore production activity. The presence of such PGP, enzymatic activity producing bacteria, increased the plant growth in four native Saudi plants [41]. The results demonstrate that the selected bacterial isolates have main PGP and enzymatic activity would substantially increase plant growth. These results have inspired us to study A. indica endophytic actinomycetes (neem gum). Simultaneously, there are several records about the neem’s endophytic fungal assembly [9]. Several studies have referred to endogenous actinomycetes’ function in phytopathogens defense and its impact on plant growth and physiology [42].
4. CONCLUSION
The neem gum was inhabited by genetically and functionally diverse specialized microbial assemblage. The bacterial population of neem gum was dominated by different species of Bacillus, Streptomyces, Pseudomonas, Enterobacter, Trabulsiella, and Acinetobacter. Many of the isolates were able to produce IAA, solubilize insoluble phosphate, potassium, and zinc. Some of the isolates were also able to produce siderophore and HCN. Their ability to tolerate high osmotic potential offered adaptive advantage to survive under osmotically stressed environment of neem gum. Furthermore, they could produce wide range of hydrolytic enzymes which ensured continuous supply of carbon through hydrolysis of complex polysaccharides of gum.
5. ACKNOWLEDGMENT
The authors thanks to Director, ICAR-NBAIM, Maunath Bhanjan (Uttar Pradesh) for providing necessary support for conducting the research work and acknowledge AMITY Institute of Biotechnology, Amity University, Noida (Uttar Pradesh), India.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
The authors gratefully acknowledge the financial assistance under project Application of Microorganisms in Agriculture and Allied Sectors (AMAAS) from Indian Council of Agricultural Research (ICAR)-NBAIM, India.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kumar VS, Navaratnam V. Neem (
2. Biswas K, Chattopadhyay I, Banerjee RK, Bandyopadhyay U. Biological activities and medicinal properties of neem (
3. Roshan A, Verma NK. A brief study on neem (
4. Sarawaneeyaruk S, Krajangsang S, Pringsulaka O. The effects of neem extract and azadirachtin on soil microorganisms. J Soil Sci Plant Nutr 2015;15:1071-83. [CrossRef]
5. Carney KM, Matson PA. Plant communities, soil microorganisms, and soil carbon cycling:Does altering the world belowground matter to ecosystem functioning?Ecosystems 2005;8:928-40. [CrossRef]
6. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K,
7. Fadiji AE, Babalola OO. Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front Bioeng Biotechnol 2020;8:467. [CrossRef]
8. Rajagopal R, Suryanarayanan TS. Isolation of endophytic fungi from leaves of neem (
9. Mahesh B, Tejesvi MV, Nalini MS, Prakash HS, Kini R, Subbiah V,
10. Verma VC, Gond SK, Kumar A, Mishra A, Kharwar RN, Gange AC. Endophytic actinomycetes from
11. Tiwari P, Bae H. Horizontal gene transfer and endophytes:An implication for the acquisition of novel traits. Plants 2020;9:305. [CrossRef]
12. Hardoim PR, van Overbeek LS, Berg G, PirttiläAM, Compant S, Campisano A,
13. Agasimundin VB, Rangiah K, Sheetal A, Gowda M. Neem Microbiome. Neem Genome;2019. 111-23. [CrossRef]
14. Irabor A, Mmbaga M. Evaluation of selected bacterial endophytes for biocontrol potential against
15. Kurt A, Cengiz A, Kahyaoglu T. The effect of gum tragacanth on the rheological properties of salep based ice cream mix. Carbohydr Polym 2016;143:116-23. [CrossRef]
16. Martínez M, Beltrán O, Rincón F, de Pinto GL, Igartuburu JM. New structural features of
17. Gomes LH, Duarte KM, Andrino FG, Tavares FC. A simple method for DNA isolation from
18. Chen J, Banks D, Jarret RL, Chang CJ, Smith BJ. Use of 16S rDNA sequences as signature characters to identify
19. Rai P, Sharma A, Saxena P, Soni AP, Chakdar H, Kashyap PL,
20. Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 1979;76:5269-73. [CrossRef]
21. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673-80. [CrossRef]
22. Kumar S, Stecher G, Tamura K. MEGA7:Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [CrossRef]
23. Holt JG, Krieg NR, Sneath PH, Staley JT. Bergy's Manual of Determinative Bacteriology. 9th ed. Baltimore, MD, United States:Williams and Wilkins Publication;1994.
24. Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 2008;163:173-81. [CrossRef]
25. Gupta S, Pandey S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (
26. Eid AM, Salim SS, Hassan SE, Ismail MA, Fouda A. Role of endophytes in plant health and abiotic stress management. In:Microbiome in Plant Health and Disease. Berlin:Springer;2019. 119-44. [CrossRef]
27. Bernfeld P. Amylases, a and b. In:Methods in Enzymology. Cambridge, Massachusetts:Academic Press;1955. 149-58. [CrossRef]
28. Singh A, Tiwari R, Sharma A, Adak A, Singh S, Prasanna R,
29. Namasivayam E, Ravindar. D J, K M, jiji A, Kumar M. Production of extracellular pectinase by
30. Cao L, Qiu Z, You J, Tan H, Zhou S. Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol Lett 2005;247:147-52. [CrossRef]
31. Dos Santos HR, Argolo CS, Argôlo-Filho RC, Loguercio LL. A 16S rDNA PCR-based theoretical to actual delta approach on culturable mock communities revealed severe losses of diversity information. BMC Microbiol 2019;19:74. [CrossRef]
32. Donelli G, Vuotto C, Mastromarino P. Phenotyping and genotyping are both essential to identify and classify a probiotic microorganism. Microb Ecol Health Dis 2013;24:20105. [CrossRef]
33. Singh MJ, Padmavathy S. Hydrocarbon biodegradation by endophytic bacteria from neem leaves LS. Int J Life Sci 2015;4:33-6. [CrossRef]
34. Verma VC, Gond SK, Kumar A, Mishra A, Kharwar RN, Gange AC. Endophytic actinomycetes from
35. Kaur M. Endophytic actinomycetes from
36. Dutta D, Puzari KC, Gogoi R, Dutta P. Endophytes:Exploitation as a tool in plant protection. Braz Arch Biol Technol 2014;57:621-9. [CrossRef]
37. ALKahtani MD, Fouda A, Attia KA, Al-Otaibi F, Eid AM, Ewais EE,
38. Ek-Ramos MJ, Gomez-Flores R, Orozco-Flores AA, Rodríguez-Padilla C, González-Ochoa G, Tamez-Guerra P. Bioactive products from plant-endophytic gram-positive bacteria. Front Microbiol 2019;10:463. [CrossRef]
39. Shahzad R, Waqas M, Khan AL, Asaf S, Khan MA, Kang SM,
40. Suman SK, Dhawaria M, Tripathi D, Raturi V, Adhikari DK, Kanaujia PK. Investigation of lignin biodegradation by
41. Eida AA, Ziegler M, Lafi FF, Michell CT, Voolstra CR, Hirt H,
42. Kolomiets EI, Zdor NA, Romanovskaya TV, Lobanok AG. Certain aspects of the phytoprotective activity of