1. INTRODUCTION
In ancient times, various types of enzymes are being used for making various food and dairy products, leather, textiles, biodiesel production, pharmaceuticals, and other valuable products for human beings [1]. A new technique (submerged culture, solid state fermentation, and immobilized cell culture) was then introduced to produce enzymes in the purified form at a commercial level from microbial sources [2]. In the world, the global enzyme market was developed a few million dollars since the 1960s. From those days, the global market has been continuously developed miraculously. Today, different types of 200 enzymes are used at the commercial level. The demand for the enzyme market is increased by study enzyme biochemistry and found new methods of fermentation process and product recovery methods. There are six classes of enzymes, of which lipases belong to the EC 3, hydrolases. Lipase hydrolyses fats and lipids (triglycerides) into monoacylglycerols and free fatty acids [3]. Some important bacterial species (Bacillus, Pseudomonas, Burlkholderia, Serratia, etc.) produced extracellular lipase by submerged fermentation and lipase production is increased by adding different lipid substrate [1,4]. The versatility of microbial lipase to choose promises application in the manufacture of paper, pharmaceutical products, cosmetics, detergents, textiles, dairy products, synthesis of fine chemicals, processing of fat and oil, and commercially [1,4,5].
2. MATERIALS AND METHODS
2.1. Soil Sample Collection
Samples of oil-contaminated were taken from different Industrial areas of Aravalli situated in Gujarat, India. Soil samples were collected in airtight sterile bags and stored in the laboratory at 4°C until further use.
2.2. Screening of Lipolytic Bacteria
For isolation of lipolytic bacteria, 1 g of soil samples was taken in pure 100 ml distilled water. Serial dilution was made from this suspension from 10-1 to 10-6. Take 1 ml suspension from each dilution and inoculated on tributyrin agar medium (Composition g%: Peptone-0.5, Yeast extract- 0.3, NaCl-0.1, Tributyrin-1.0, and Agar- 2.0) by spread plate method [6]. The plates were then incubated (37°C for 48 h). All plates were studied after 48 h of incubation for lipase production in an agar plate. Observed halo zone of clearance (lipid hydrolysis) surrounded bacterial colonies. All isolates were further screened as for maximum lipase producer were inoculated on tributyrin agar medium. The best lipase producer was selected based on the maximum zone of clearance for further studies.
2.3. Identification of Lipolytic Bacteria
The selected bacterial isolate (KKSC12) was identified morphologically by Gram staining and genetically by 16S rRNA sequencing. The molecular identification (16S rRNA sequencing) was performed at Biokart PVT Ltd. Bangalore.
2.4. Production of Lipase Enzyme
Lipase was produced from a selected bacterial isolate (KKSC12) by submerged fermentation. Production medium (Composition g/L; Peptone [2.0 g], NH4H2PO4 [1.0 g], NaCl [2.5 g], MgSO4.7H2O [0.4 g], CaCl2.2H2O [0.4 g], Olive oil [2%(v/v)]), Tween 80 (1–2) drop, and D/W (1000 ml); pH 7.0) [7]. The activated culture of the isolate was inoculated into the production medium (100 ml) contained in Erlenmeyer flasks (500 ml) after sterilized in the autoclave at 121°C for 15 min. The flask was placed on a metabolic shaker (150 rpm) at 37°C for 48–72 h. After incubation, the crude enzyme extract was obtained at 10,000 rpm (centrifugation) for 20 min, and for analysis of lipase enzyme quantitatively, the supernatant was used.
2.5. Spectrophotometric Assay of Lipase Activity
The lipase activity in the supernatant was examined by the spectrophotometric method using phenyl acetate using as a substrate [8].
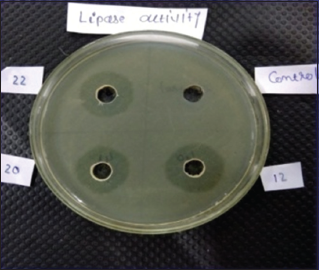 | Figure 1: Zone of clearance of KKSC12. [Click here to view] |
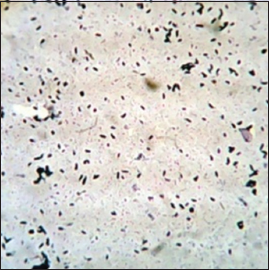 | Figure 2: Morphological Identification of KKSC12. [Click here to view] |
2.6. Lipase Enzyme Purification
(NH4)2SO4 precipitation and Diethylaminoethyl (DEAE) cellulose anion exchange chromatography were used for purification of extracellular lipase from bacterial isolate (KKSC12) [9]. The supernatant was precipitated at different ammonium sulfate saturation. Precipitation of protein was done by addition of ammonium sulfate at 4°C with stirring for 2 h to obtain fractions, that is, 40% and 60%. At the end of each fraction, precipitates were collected and dissolved in phosphate buffer pH 7.2. The collected ammonium sulfate precipitate was dissolved in 50mM Tris-HCl buffer, pH 8.0, and applied onto the DEAE-cellulose anion chromatography for further clarification.
2.7. Molecular Mass Determination of Lipase Enzyme
According to the procedure of Laemmli [10], the Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis method was used to the determined molecular mass of purified bacterial lipase. For gel electrophoresis, 4% stacking gel was used. Electrophoresis was performed at 200v for 2 h. After gel electrophoresis, the gel was removed from plates and stained with 0.025% Coomassie brilliant blue dye. Then molecular mass was determined.
3. RESULTS AND DISCUSSION
3.1. Screening of Lipase Producing Bacteria
The best bacterial isolate (KKSC12) was chosen from 46 bacterial based on the production of the maximum zone of clearance as detected by hydrolysis of lipid on tributyrin agar medium due to the extracellular lipase production. This bacterial isolate (KKSC12) was produced highest zone of clearance, 25 mm diameter on tributyrin agar medium [Figure 1] among the isolates used in the study. Olawaseya et al. [11] isolated Pseudomonas aeruginosa from soil and Bacillus sp. from water using tributyrin agar, rhodamine olive oil agar, and phenol red agar medium. Leelatulasi et al. [12] reported that cold-adapted Pseudomonas sp. LSK25 containing lipase activities was isolated on tributyrin, triolein, and rhodamine olive oil agar from Antarctica.
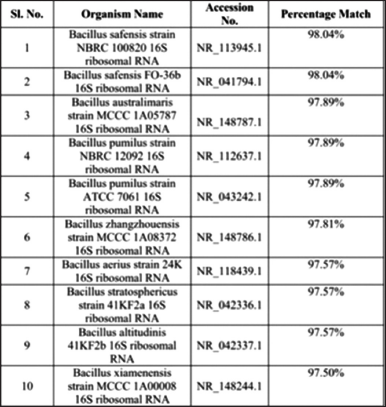 | Table 1: Blast analysis report of the bacterial isolate KKSC12 (Bacillus safensis). [Click here to view] |
 | Table 2: Purification steps of lipase from Bacillus safensis. [Click here to view] |
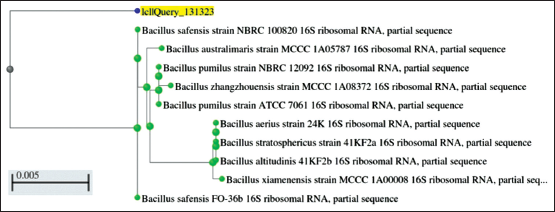 | Figure 3: Phylogeny of KKSC12 (Bacillus safensis). [Click here to view] |
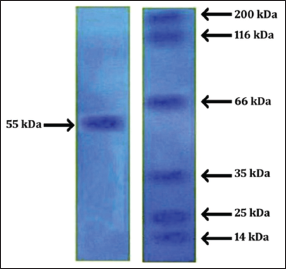 | Figure 4: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of Bacillus safensis [Click here to view] |
3.2. Identification of Lipolytic Bacteria
The selected potential isolate was morphologically identified by performing Gram staining. The isolated bacteria (KKSC12) were observed gram-positive, big rods [Figure 2]. Using the 16S rRNA gene sequencing method isolated bacterial KKSC12 was identified as Bacillus safensis [Figure 3] as 98.4% of sequence similarity was matched [Table 1].
3.3. Production of Lipase
Extracellular lipase enzyme was produced by submerged fermentation. The production medium containing olive oil as a lipid substrate and fermentation was carried out at pH 7.0, at 37°C temperature, for 48–72 h in shaking condition at 150 rpm. The crude enzyme was assayed by the spectrophotometric method using phenyl acetate as a substrate from production medium. The selected B. safensis was produced the highest lipase activity such as 113.33U/ml after 48 hours. Pushpinder S. [4] reported using submerged fermentation to produce Bacillus methylotrophicus PS3 lipase using lipidic substrate (tributyrin) and the maximum activity was 320.0IU at 40°C and pH-7 with 1% tributyrin. Venkata et al. [13] reported maximum lipase produced from Stenotrophomonas maltophilia using tributyrin as a substrate. Naif et al. [14] shown that the highest lipase activity from Streptomyces sp. was found 253U/ml at pH-11.0 and 35°C temperature with olive oil. Amira et al. [15] also reported that extracellular lipase production from Aspergillus niger using tween 80 broth by submerged fermentation.
3.4. Partial Purification and SDS-PAGE Gel Electrophoresis
The ammonium sulfate fractionation method was used for partial purification of lipase enzyme using crude extract of B. safensis. Crude enzyme filtrate was subjected to different fractions of ammonium sulfate precipitation at 40% and 60% saturation. The fractions obtained at 40% and 60% followed by DEAE-cellulose chromatography showed that maximum activity, specific activity, and yield were detected [Table 2]. The molecular mass of the purified lipase from B. safensis was 55kDa using different marker proteins as confirmed by the appearance of a single band on SDS- PAGE [Figure 4]. Gulhan et al. [16] studied that the 17.8 fold obtained from Bacillus subtilis LP2 was partially purified by the Ammonium fractionation method. Iqbal and Rehman [17] reported that B. subtilis I-4 was purified by thin-layer chromatography, and molecular weight was determined to be 53kDa by SDS-PAGE gel electrophoresis. Loreni et al. [18] reported that lipase from Pseudomonas helmanticensis was purified resulting in 18.78 fold purification, 5.58% yield and high specific activity of 3368 U/mg. Anil et al. [19] reported that extracellular lipase from Psychrotrophic bacterium Chryseobacterium polytrichastri was purified by ammonium sulfate precipitation, size exclusion, and hydrophobic interaction chromatography method.
4. CONCLUSION
Lipase production by lipolytic bacteria is worth exploiting for detailed investigations and further applications at the industrial level. Lipase enzyme from bacteria is suggested for commercial production due to its constitutive nature and easy purification. Limited research reports are available on lipase production from bacteria and this would be one of them in this direction.
5. ACKNOWLEDGMENTS
A grant from the Department of Microbiology, Sir P. T. Science College, Modasa, Gujarat, India, supported the author’s research. The author would like to express her gratitude to the Microbiology Department for providing the laboratory facilities required to complete the research.
6. AUTHORS’ CONTRIBUTIONS
All authors made intellectual contribution to the completion of this research work. They also took part in an experimental design, analyzed the research data, and played a role in composing the describing result in an article.
7. FUNDING
Research Grant received from Department of Microbiology, Sir P. T. Science College, Modasa, Gujarat, India.
8. CONFLICTS OF INTEREST
The authors report no conflicts of interest.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subject.
10. DATA AVAILABILITY
Data is available online as well as mentioned in references in bibliographic notes.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Prem CE, Ranjan S, Pankaj KA. Microbial lipases and their industrial applications: A comprehensive review. Microb Cell Fact 2020;19:169. CrossRef
2. Sindhu R, Binod P, Sabeela BV, Amitha A, Anil KM, Aravind M, et al. Application of microbial enzymes in food industry. Food Technol Biotechnol 2018;56:16-30.
3. Yasaman P, Sandip S. Biochemistry, Lipase. Treasure Isaland. FL: Star Pearls Publishing; 2021.
4. Pushpinder S, Shweta H. Purification and characterization of lipase by Bacillus methylotrophics PS3 under submerged fermentation and its application in detergent industry. J Genet Eng Biotechnol 2017;15:369-77. CrossRef
5. Anitha M, Leena GB. Extracellular lipase production, purification and characterization using Bacillus subtilis in submerged fermentation. Biomed J Sci Tech Res 2018;5:7641-5.
6. Jair CP, Blanca E, Rivera C, Norma RB, Luisa IM, Guadalupe V, et al. Improved method for qualitative screening of lipolytic bacterial strains. MethodsX5 2018;5:68-74. CrossRef
7. Veerapagu M, Shankara NA. Ponmurugan K, Jeya KR. Screening, selection, identification and optimization of bacterial lipase from oil spilled soil. Asian J Pharm Clin Res 2013;6:62-7.
8. Abd-Elhakeem MA, Elsayed A, Alkhulaqi TA. New colorimetric method for lipase activity assay in microbial media. Am J Anal Chem 2013;4:442-4. CrossRef
9. Shivika S, Shamsher SK. Purification and Biochemical characterization of a solvent tolerant and highly thermostable lipase of Bacillus licheniformis strain SCD11501. Proc Natl Acad Sci India B Biol Sci 2017;87:411-9. CrossRef
10. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680-5. CrossRef
11. Olawaseya LL, Abiodan EA, Japhel AO, Efemena MO. Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci Afr 2020;8:297. CrossRef
12. Leelatulasi S, Raja NZ, Abu BS, Fairolniza S, Peter C, David P. Isolation, characterization, and lipase production of a cold-adapted bacterial strain Pseudomonas sp. LSK25 Isolated from Signy Island, Antarctica. Molecules 2019;24:715. CrossRef
13. Venkata NY, Jamal A, Sudheer B, Shravani RS, Prakasham RS. Production and characterization of lipase from novel bacterium Stenotrophomonas maltophilia RSP-09. Curr Trends Biotechnol Pharm 2015;9:89-101.
14. Naif A, Galal AE, Abdul-Kareem MG, Mariadhas VA. Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi J Biol Sci 2020;27:474-9. CrossRef
15. Allabdalall AH, Alanazi NA, Aldakeel SA, Abdil AS, Borgio JF. Molecular, physiological, and biochemical characterization of extracellular lipase production by Aspergillus niger using submerged fermentation. Biochem Biophys Mol Biol 2020;8:e9425. CrossRef
16. Gulhan Y, Unzile GG, Elif G, Fatih A. Screening, partial purification and characterization of the hyper-thermophilic lipase produced by a new isolate of Bacillus subtilis LP2. Biocatal Biotransformation 2020;38:367-75. CrossRef
17. Iqbal SA, Rehman A. Characterization of lipase from Bacillus subtilis I-4 and its potential use in oil contaminated wastewater. Braz Arch Biol Technol 2015;58:789-97. CrossRef
18. Loreni CP, Rounak C, Megha K, Krishnen TG, Sahoo D, Binod P, et al. Production and characterization of lipase for application in detergent industry from a novel Pseudomonas helmanticensis HS6. Bioresour Technol 2020;30:123352. CrossRef
19. Anil K, Shrijana M, Neeraj K, Vishal A, Sanjay K, Rakshak K. A broad temperature active lipase purified from a psychrotrophic bacterium of Sikkim Himalaya with potential application in detergent formulation. Front Bioeng Biotechnol 2020;8:642. CrossRef