1. INTRODUCTION
Changing climatic conditions and frequent recurrence of perturbed environmental conditions contribute major crop loss worldwide and severely affect crop productivity [1]. Climatic changing conditions belong to wide array of abiotic as well as biotic stress. Abiotic stresses such as salinity, drought, heat, and cold are among the most common stresses which severely affect plant growth and crop productivity adversely causing more than 50% of yield loss of major crops every year worldwide [2]. There is a serious concern and a big challenge for agriculture scientists to ensure food security for growing population globally. It is expected that human population will reach 10 billion by 2050 [3]. Although there is sufficient food production in the present time, the agricultural productivity may not match the increasing population in coming years and will put pressure on food security for human population. The world is facing stagnancy of the crop productivity or even decline in crop production, which could be due to several factors including climate change and reducing agriculture land, decreasing crop diversity [4]. Therefore, there is a pressing need to focus on agriculture sustainability through developing climate-resilient crops [5]. However, developing improved crop varieties for different agronomic traits, exhibiting resilience against abiotic and biotic stresses is a big challenge among the scientists and identifying the various omics approaches for underutilized crops for future food security under changing climate conditions [6]. In the present situation, to maintain crop production during adverse environmental conditions, climate change and global warming create the pressing need to adopt and implement sustainable solutions in terms of technology development, varietal improvement, agricultural management practices, and resource managements [7].
Largely, the crops are improved through four major breeding techniques, i.e., cross breeding and mutational breeding, transgenic breeding, and breeding by genome editing (GE) to create diversity [8]. Because the variations at genome level in crops are the primary basis of crop improvement, therefore breeding techniques use these variations for genetic improvement. In the last few decades, emergence of transgenic technology has offered a potential solution quick crop improvement, although it is very challenging and has several limitations [9]. The advances of technological assets and tools have made a significant effect on agriculture research, from methods of farming system to crop improvement approaches toward agriculture sustainability [10]. Over the last decade, the emergence of transgene free GE technology has provided a new dimension in precise genetic modifications of plant genome to achieve genetic diversification and desired trait improvement. Recently reviewed the diverse approaches of gene editing tools for increasing the crop trait for nitrogen used efficiency and other application of GE technology for sustainable agriculture and future food security [11]. Recent GE is defined as next generation plant breeding tools [8]. The development of genome edited crops with desired traits could be cheaper and more publicly acceptable in contrast to the development of genetically modified (GM) crops as identified based on the survey and expert opinions. This also revealed that genome-edited crops are superior for agronomic performance and food quality as compared to alternatives and conventional approaches of crop breeding, however, the relevance of other alternative approaches cannot be diminished over GE as they have relevance in present time also. Moreover, the estimated cost may be crop wise and case specific in some cases. However, in general, development of genome-edited crops is less costly as compared to developing GM crops [12-14]. Crop development through trait improvement is the major thrust to produce resilient crops to mitigate challenges of climate change and biotic and abiotic stresses on agriculture production [15]. Therefore, it is imperative to enhance the climate resilience of the agricultural crops and such improved crop could be a major benefit to agriculture system in the regions with frequent incidences of abiotic stress to mitigate the adverse effect. In the last decade, technology development helped scientific communities to decipher the underlying molecular mechanism of crop responses against abiotic stress and made a significant pool of information to work in focused direction [16-18]. The development of high throughput sequencing and functional genomic tools have made substantial progress in understanding the plant responses at the gene level including both functional genes and regulatory genes respond against abiotic stress [19]. However, there is a major drawback to understand the exact mechanism of abiotic stress tolerance due to the mutagenic control of physiological traits. This makes abiotic stress tolerance in plants a complex phenomenon which is governed by cascades of gene expression in response to abiotic stresses [20]. Moreover, transgenic and GE technologies are potentially better and provide a quick solution to develop abiotic stress-tolerant crops [21]. However, recent studies have suggested that the development of abiotic stress-resilient crops with multiple genetically engineered genes may be better option to achieve effective abiotic stress-resilient plants rather than single gene manipulation [22]. Furthermore, working on those regulatory genes which are involved in abiotic stress response could be an alternative better choice for the controlled expression of several genes associated with response against abiotic stress because transcription factors are key regulators for several abiotic stress-responsive genes [23]. There are several transcription factors belonging to different families, i.e., AP2/EREBP, MYB, WRKY, NAC, and bZIP have been found to be involved in different abiotic stress responses and are capable to improve stress resilience in crop plants [24].
The emergence of transgenic technology has changed the entire scenario of breeding technology, but despite having immense potential for quick and effective trait improvement, it suffers several limitations (i.e., involvement of transgene, high cost, environmental concerns, divided social acceptance, stringent policies and regulations etc.) for its wide application and acceptance [25]. The increasing awareness about GE crops its acceptance among the consumers have raised though in smaller proportions. However, still, large proportions of consumer are in lurk and hesitate in acceptance [26]. However, policies and regulations now being framed which distinguished genome-edited crops from GM crops based on the absence of transgene, i.e., genome-edited crops that do not contain DNA from foreign origin. Such genome-edited crops now being not considered as genetically modified organisms (GMO) and regulated as conventional plants unless they contain foreign gene in several countries [27]. Recently, Pixley et al. [28] discussed about the genome-edited crops and the concern associated with risk, causing low acceptance in the light of regulatory legal and traded frame work for genome-edited crops. Ahmad et al. [29] focussed on the current status and perception about genome-edited crops by regulatory authorities, whether genome-edited crops (transgene free) and genetic modified (with transgene) crops are same or different. Focussed on the legal consideration of genome-edited crops as GMO or non-GMO in different countries. Importantly, there is a need to develop a universal, plausible scalable regulatory framework for clustered regularly interspaced short palindromic repeat (CRISPR)-edited plants including public awareness for acceptance. Moreover, several countries now trying to make consensus about non-transgenic genetic edited crops to be treated as conventional bred varieties and accordingly worldwide for genome-edited plant varieties and therefore emphasized on the need for exemption of genome-edited crops from GMO stringent regulations [30].
In the line of trait improvement, emergence of exogene or transgene-free GE technology has taken crop improvement drive to the new height (Guidelines of Ministry of Science and Technology, New Delhi (2022), Ministry of Environment, Forest and Climate Change, New Delhi (2022). In the last decade, advent of new approaches to breeding has opened a plethora of opportunities for genetic manipulations and introduction of improved traits in crop plants to develop crops with desired traits. Such techniques involved precision genome-modification platforms such as transcription activator-like effector nuclease (TALEN) and CRISPR/CRISPR-associated protein (Cas)-based methods [31]. Moreover, the development of CRISPR-associated tools in the recent time like base editors (BE) and prime editors (PE) have further broadened the application and utility of GE, which allow precise nucleotide substations and site-specific deletion and addition of nucleotides [32]. Such approaches are helpful in not only reducing breeding duration but also effective introduction of improved traits [33]. The use of sequence-specific nucleases (SSNs) in GE has revalorized the entire scenario of genome engineering. This can give major thrust in agriculture productivity through desired trait change. Beside increasing crop productivity and yield, GE is important tool for study of functional and reverse genetics [34]. GE is a technique, which involves the specific and precise cleavage of nucleotide sequences through precise double-strand breaks (DSBs) at or near the target site in the genome [35]. Moreover, such SSNs have become the magic molecules to perform precision cleavage in the target genome to get desired GE. There are four broad classes of SSNs, namely meganucleases, zinc-finger nucleases (ZFNs), TALEN, and Cas proteins. These SSNs exhibit immense potential in plant breeding through precision GE which includes, Gene knock-out, Gene knock-in, Targeted mutagenesis, etc. SSNs offer great advantages over conventional breeding approaches [8]. The development of mutational diversity through GE is successfully implemented in several crops to improve agronomic traits; however, the reports for development of abiotic stress using gene editing are very scanty until development of new GE tool called CRISPR/Cas, which open an immense opportunity in GE plants to improve abiotic stress tolerance in crops [36,37]. The present review article is focussed on the different tools and strategies for GE and their applications in developing better crop varieties for exhibiting resilience against abiotic stress to augment agriculture sustainability.
2. MEGANUCLEASES
The meganucleases are obtained from transposable elements of microorganism and several variants of meganuclease are developed through protein engineering to recognize various sequence combinations. This is the class of site-specific endonucleases, which can recognize and cut the DNA sequence more than 12 bp long at specific site. Meganucleases are broadly classified into five families according to their sequence and motifs present in it namely, i.e., LAGLIDADG, His-Cys box, GIY-YIG, PD(D/E) XK, and HNH [38]. Although all families of meganucleases exhibit endonuclease activity, LAGLIDADG family of meganucleases is primarily used for GE purpose. Meganucleases exhibit less polymorphism due to occurrence of very few restriction sites in the genome, perhaps, due to long recognition sequences and target site. Meganucleases act by creating a DSB at specific recognition site in the genome followed by repair through non-homologous end joining repair (NHEJ) system of the host leading to deletion or micro-insertion at the target site [23,39]. I-SecI and I-CreI are the most widely used nuclease protein among all the members of the different families of meganucleases. The gene I-SecI encoding I-SecI protein is found in 21 s RNA encoding gene of mitochondrial DNA of Saccharomyces cerevisiae whereas the gene I-CreI encoding I-CreI protein is found in 23s RNA encoding gene of chloroplast of algae Chlamydomonas reinhardtii [23]. Mega-nucleases are not capable to target the endogenous genes originally. However, alteration in maganuclease binding capabilities could develop endogenous genes targeting meganucleases [40]. Although research is available for the use of meganucleases in GE, for example, development of site-specific mutagenesis in maize, it has limited use in GE because of the tedious process of developing engineered meganuclease for each new target site in the genome for editing purpose [41].
3. ZFNs
ZFNs are designed restriction enzymes, which are composed of fusion protein with two important domains: One is sequence-specific zinc finger DNA binding domain, which composed of 3–6 zinc finger repeats and the second is Flavobacterium okeanokoites I (FokI) cleavage domain comprises non-specific restriction enzyme [42,43]. Active form of ZFNs functions in dimer form, where two monomers of ZFN bind to 18 or 24 bp sequence with 5–7 nucleotide spacer. ZFNs functions through sequence specific protein-DNA binding, therefore for each new editing site in the genome, a new ZFN is needed to be created [8]. ZFNs can be effectively used for site specific deletions at target site in the genome to create concurrent DSB in the DNA. Targeted insertions/deletions of intervening DNA sequence can be obtained by ZFNs used to create concurrent DSBs followed by NHEJ or Homology Direct Repair (HDR) repair pathways [44]. Recently, the site-specific single transgene integration has been demonstrated in soybean (Glycine max) crop using nucleases followed by NHEJ and HDR. Bonawitz et al. [45] demonstrated the application of ZFNs in targeted integration of multiple transgenes into single loci of soybean (G. max) to generate fertile transgenic soybean plant using NHEJ-mediated targeted insertions of multigene donors at an endogenous genomic locus. In the recent years, GE has made a potential imprint in crop improvement for several agronomic traits which to address the challenges of climate change, biotic and abiotic stress tolerance, increasing productivity, yield, nutritional contents, etc., in several cereal crops, i.e., rice, maize, wheat, and barley, etc. [46]. ZFNs offers advantages over other tools for GE due to its efficiency, high specificity, and less non-target effects [47]. However, there is limited use of ZFNs because of certain limitations such as high cost, limited target sites in the genome, and construction of ZFN for each target site.
4. TALEN
In the last decade, gene editing potential of transcription activator-like effectors (TALE) was realized because of its capability to specifically bind to the promoter of targeted gene to activate transcription [48]. Toward the development of new improved tools for GE, TALEN were identified in 2011, they were developed from fusion of nuclease to TALE proteins [49]. Moreover, the presence of repeat sequences in TALEN enhances the efficacy. TALEs were first introduced into the plant cell by the bacterium Xanthomonas spp. to modulate the plant cell gene expression to meet the bacterial need TALE are type III effector proteins produced by bacterium Xanthomonas spp. [50,51]. Realizing the targeted DNA binding potential of TAL effectors, the engineered TAL effectors were developed fused with Fok1 nuclease activity domain, hence known as TALEN, these were the new kind of SSN [34,52]. TALEN later provided the immense opportunities and wider applications for GE in specific locus including in plants [53]. There are reports available indicating the application of TALEN gene editing technology in plants as an effective GE tool to generate improved traits in crops such as maize [54], soybean [55,56], tomato [57], wheat [58], and barley rice [59,60]. Although TALEN gene-editing tool has plenty of potential for creating GE for crop improvement, yet is suffers certain drawbacks and limitations for its effective applications in crops. This includes inconvenient large size of TALEN encoding cDNA, difficult construction of repetitive sequences, and delivery in plant cell [61]. However, the most convenient method for the development of TALEN molecule is a ligation-based “Golden Gate System” which is widely accepted and used [61,62].
Further toward improvisation more advanced system, i.e. platinum TALEN and platinum gate coordination were also developed to create mutations in eukaryotic system [63]. TALEN is important in improving the agronomic traits of interest. Clasen et al. [64] demonstrated that TALEN-mediated knockout of vacuolate invertase (Vlnv) gene on potato tuber leads to enhanced quality of potato during cold storage conditions by preventing the production and accumulation of reducible sugars. Developing the technology for introducing nucleic acid-free SSN in plant for GE has provided more possibilities for crop improvement. Therefore, the nucleic acid-free introduction of TALEN makes it possible for GE in plants [65]. The use of TALEN is preferred over ZFN in genome editing because of certain advantages, like TALENS are less toxic and their engineering is easy due to presence of TALEN repeats corresponding to repeat-variable di-residues and their easy DNA sequence recognition [34].
Although due to the ease in CRISPR tool, TALEN is still an attractive GE tool for targeted base editing in organelles (plastids, mitochondria) in plants [66]. However, it has certain limitations, like, designing TALENs that need reengineering of a new protein for each of the targets is very tedious as compared to CRISPR whereas designing and use of CRISPR are simple [47]. The first time the TALEN GE technology was used in technology cancer treatment in humans. There are very few genome-edited plants that could come to market, i.e., a soybean cultivar by calyx and canola by cibus, however, many more crops are underway to market in coming years. Only few genome-edited plants (a soybean cultivar by calyx and canola by cibus) have reached the market so far, but more crops are in the pipelines of various companies and may soon appear on the market.
5. CRISPR/CAS
Being a complex trait abiotic stress responses in the plant are governed by several genes and processes. Hence, rather than transgenic approaches, targeted GE in the single or multiple target sites could be the better solution for developing abiotic resilience in the crops [67]. The GE approach has opened plethora of opportunities in genetic modifications and crop improvement for desired traits within a small period of the time [68,69]. In the recent years, application of gRNA-directed Cas9 nuclease for GE has made a significant contribution to genetic modifications and crop improvement in several crops. In the last decade, the emerging utility of CRISPR/Cas9 system for targeted GE has made impeccable impact in plant genetic engineering making this approach a most potential tool toward developing improved crops with better tolerance against adverse environmental conditions (Wada et al. [21], Nascimento et al. [70]) and its applications in crop abiotic stress management [36]. Its utilization in GE was first demonstrated in 2013 by (Zaidi et al. [5], Shan et al. [71], Li et al. [72]). GE in plants using CRISPR involves certain steps which include (1) DSB at a target site in the genome through SSN and endonuclease CRISPR/(Cas9, Cas 12a or others) (2) repairing of DSB at target DNA site through NHEJ method or HDR method, and (3) followed by creation of small insertion or deletion of nucleotides at target DNA site by NHEJ pathway. This insertion or deletion of nucleotides creates the frame shift mutations in the DNA, whereas accurate DNA repair is achieved through HDR by inserting a donor strand of desired sequence [73-75].
The CRISPR/Cas9 approach has been used for crop improvement in different crops [Figure 1]. This approach has been used for crop improvement in several crops to improve the abiotic stress tolerance, i.e., development of salt-tolerant rice using CRISPR/Cas9 targeting the OsRR22 gene in rice [76], wheat [77], tomato [78], maize [79], and soybean [80], etc. Several studies also revealed the leading use of these techniques to improve the crops with agronomically and economically important traits using transgene-free system [8]. The reduced diversity of food crops due to extensive domestication and adverse impact of climate change has further aggravated the reduction in crop productivity and yield to challenge food security [81]. Adopting gene editing tools, the challenges of diversity and climate changes can be mitigated to make the agronomic traits more resilient. CRISPR-Cas gene editing promotes site-specific precise mutagenesis in the target loci of the genome to modify the desired agronomic traits in several crops and generate genetic diversity in staple food crops [82].
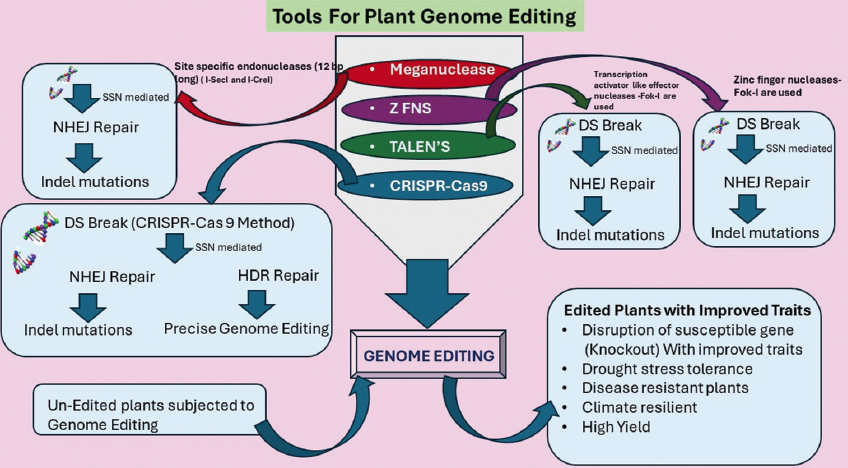 | Figure 1: General description of some important genome editing tools in plants for trait improvement. ZFNs: Zinc finger nucleases, TALENs: Transcription activator-like effector nucleases, SSN: Site-specific nucleases, DS break: Double-strand break, DNA repair pathways: NHEJ: Non-homologous end joining, HDR: Homology directed repair. [Click here to view] |
There are studies available, which revealed the role of certain genes as negative regulators in plant’s responses under abiotic stress including drought and salt stress, etc., for instance, Zhang et al. [76] developed salt-tolerant rice cultivar using an engineered Cas9-OsRR22-gRNA expressing vector, to target OsRR22 gene and developed knocked down mutant using CRISPR/Cas9 gene editing. The OsRR22 gene encodes a type-B response regulator involved in cytokinin signaling, therefore, knockdown of OsRR22 leads to improved tolerance in rice cultivars under salt stress. The T2 homozygous mutant line exhibited enhanced salt tolerance as compared to the wild type at the seedling stage.
Zhang et al. [76] developed the improved mutant rice variety exhibiting better yield, grain size, and enhanced cold stress resilience using CRISPR-Cas9 gene editing technology in three important genes, i.e., osPIN5b, GS3, and OsMYB30 controlling important agronomic traits. Recently, the precise gene editing using CRISPR/Cas gene editing tool in drought and salt stress (OsDST) gene in rice cultivar helped to develop the elite improved rice cultivar exhibiting better tolerance for drought and salt stress. The OsDST gene encodes zinc finger transcription factor. Therefore, CRISPR/Cas was used to generate mutant of dst (drought and salt tolerance) gene, which leads to the development of drought as salt stress tolerance in indica rice Cv MTU1010 [83].
Roca Paixão et al. [84] demonstrated the improvement of drought stress tolerance in the A. thaliana using CRISPRa dCas9HAT as tool to improve drought stress tolerance through the positive regulation of the abscisic acid (ABA)-responsive element binding protein 1/ABRE binding factor (AREB1/ABF2). This is a vital positive regulator of the drought stress response. AREB1 activity exhibited enhanced drought stress tolerance in other crops also, i.e., rice and soybean, while AREB1 loss of function causes drought stress sensitivity. In Arabidopsis AREB1, AREB2, and AREB3 mutants exhibited reduced drought stress tolerance indicating their role in osmotic stress tolerance [85]. Similarly, in soybean over-expression, AtAREB1 is associated with improved drought stress tolerance [86] and in rice [87]. In the past few years, CRISPR/Cas9 GE technology has gained momentum for trait improvement in developing improve desired crops. Development of drought stress tolerant wheat variety generated through GE in important drought stress-responsive gene, i.e., TaDREB2 gene which is responsible for encoding a dehydration responsive element binding protein-2 in the protoplast of wheat using CRISPR/Cas9 techniques [88]. Similarly, CRISPR/Cas9 mutagenesis in wheat ethylene-responsive factor 3 (TaERF3) also creates abiotic stress tolerance. The study suggested that CRISPR/Cas9 techniques can be effectively used for GE in TaDREB2 and TaERF3 genes in wheat protoplast. The study revealed the potential effectiveness of gene editing in wheat and opens possibilities of trait improvement in polyploid crops [89].
Bouzroud et al. [90] revealed that downregulation of SlARF4 enhances salinity and osmotic resilience in tomato; moreover, the knocking out of SlARF4 function results into improved root length and density further helping drought stress tolerance. Furthermore, auxin response factors-4 (ARF4) mutant generated by CRISPR/cas9 technology (arf4-cr) exhibits similar responses as ARF4 antisense. The study also revealed the auxin involvement in stress tolerance in tomato and the involvement of ARF4 in this process. Recently, Abdallah et al. [91] demonstrated that CRISPR/Cas-mediated mutagenesis can be applied in wheat for the development of drought stress tolerance. In the recent study, Zhu et al. [92] revealed the role of ZmMPKL1 in the regulation of seedling responses to drought stress in the maize. The maize ZmMPKL1 knockout mutant created through CRISPR/Cas9 exhibited reduced drought tolerance under drought conditions which suggested that ZmMPKL1 positively regulates seedling drought sensitivity in maize.
Zhang et al. [76] developed the improved mutant rice variety exhibiting better yield, grain size, and enhanced cold stress resilience using CRISPR-Cas9 gene editing technology in three important genes, i.e., osPIN5b, GS3, and OsMYB30 controlling important agronomic traits. Recently, the precise gene editing using CRISPR/Cas gene editing tool in drought and salt stress (OsDST) gene in rice cultivar helped to develop the elite improved rice cultivar exhibiting better tolerance for drought and salt stress. The OsDST gene encodes zinc finger transcription factor. Therefore, CRISPR/Cas was used to generate mutant of dst (drought and salt tolerance) gene, which leads to the development of drought as salt stress tolerance in indica rice Cv MTU1010 [83]. Recently, Chennakesavulu et al. [75] extensively reviewed the CRISPR/Cas GE technology in the development of desired cultivars with improved tolerance for various abiotic stress and also focused on some of the important advances and updates in the plant GE such as BE, PE, epigenome editing, tissue-specific, and inducible GE for trait improvement. Shii et al. (2017), developed ARGOS8 variants of maize using CRISPR/Cas9 technology for allelic variations. The genomic editing in ARGOS8 locus leads development of maize cultivar with enhanced grain yield even under drought stress conditions. The ARGOS8 is negative regulator of ethylene response in plant [93]. Singh et al. [94] explored the potential of GE in chick pea for trait improvement to combat the threat of climate changes. There are studies available, which revealed the role of certain genes as negative regulator in plant’s responses under abiotic stress including drought and salt stress, etc., for instance, Zhang et al. [76] developed salt-tolerant rice cultivar with OsRR22 knocked-down mutant using CRISPR/Cas9 gene editing tool. RR22 gene which encodes a type-B response regulator involved in cytokinin signaling, therefore, knockdown of OsRR22 leads to improved tolerance in rice cultivar under salt stress.
The study on tomato revealed specific and targeted gene editing in SlLBD40 gene [Table 1]. The knockout mutant of tomato exhibited enhanced drought tolerance by improving crop water holding capacity. As SlLBD40 knockout lines wilted slowly as compared to the SlLBD40 overexpressing lines, which wilted very quick under drought stress. The study also revealed the role of SlLBD40 as negative regulator in drought tolerance, therefore knocking out of the SlLBD40 leads to enhanced drought tolerance [95]. Wang et al. [96] revealed the role of SIMAPK3 in the induction of drought stress tolerance in tomato, and development of slmapk3 mutant (loss of function) using CRISPR/Cas tool leads to reduce drought stress tolerance in tomato, suggesting the involvement of SIMAPK3 in regulation of drought tolerance in tomato. In another study, CRISPR/Cas9 mediates gene mutagenesis in SlNPR1 gene. The mutant of SINPR1 exhibited reduced drought tolerance in tomato which was corroborated by reduced level of antioxidant enzymes, enhanced malondialdehyde and hydrogen peroxide level, and increased stomatal aperture. The study suggested the involvement SlNPR1 in regulating tomato plant drought response [97]. Debbarma et al. [98] discussed the about improvement of abiotic stress tolerance in plant by gene editing of ethylene responsible factor (ERF, a transcriptional factor) of the AP2/ERF superfamily using CRISPR/Cas9. The regulatory role of phytohormone ABA in plant responses against abiotic stress responses is well studied. The important AITRs act as a family of novel transcription factors, which exhibit an important role in regulating abiotic stress responses in the plant. The study revealed that mutants generated through targeted gene editing of AITRs using CRISPR/Cas9 tool lead to enhanced abiotic stress tolerance and reduced ABA sensitivities in Arabidopsis plants [99]. Similarly, CRISPR/Cas9 mutagenesis (small insertion/deletion mutations into two target genes) in an Acquired Osmotolerance (ACQOS; AT5G46520) and ACQOS/VICTR established the association with osmotic tolerance and salt resistance. Further, the plants that possess wild-type ACQOS alleles were sensitive to salt stress after acclimation as compared to those harboring knockout ACQOS and showed better salinity tolerance. Considerable tolerance to salt stress, suggesting that (ACQOS; AT5G46520) suppresses osmotic tolerance [100]. Zheng et al. [101] identified the role of histone acetyltransferase TaHAG1 that acts as an important regulatory factor for enhancing salinity stress tolerance in hexaploid wheat (Triticum aestivum).
Table 1: Gene editing through CRISPR/Cas tool in different crops for abiotic stress resilience.
| S. No. | Crop | Gene-editing tool | Abiotic stress | Targeted gene/genes | Type of responses | Role of genes | References |
|---|---|---|---|---|---|---|---|
| 1. | Wheat (Triticum aestivum) | CRISPR/Cas9 | Drought | TaSal1 Upregulated | Enhanced Tolerance | Regulate stomatal closure, increased stomata width, and increase in the size of the bulliform cells. | Abdallah et al. [91] |
| 2. | Wheat (Triticum aestivum) | CRISPR/Cas9 | Salt tolerance | TaHAG1 | Salt tolerance | Enhance salt tolerance through histone modification | Zheng et al. [101] |
| 3. | Rice (Oryza sativa) | CRISPR/Cas9 | Drought | OsSYT-Downregulated | Enhanced Tolerance | Role in regulating expression of various genes involved in ABA-associated stress signaling | Shanmugam et al. [106] |
| 4. | Arabidopsis thaliana | CRISPR/Cas9 | Drought and salinity | AITR3 and AITR4 mutant (knockouts) | Enhanced tolerance | Role in regulating ABA response. Leading to reduced sensitivity to ABA and enhanced tolerance to drought and salt. | Chen et al. [99] |
| 5. | Arabidopsis thaliana | CRISPR/Cas9 | Salt stress | ACQOS/VICTR mutant (knockouts) | Enhanced salt tolerance | Regulation of osmotic tolerance and salinity stress tolerance. | Kim et al. [100] |
| 6. | Chick Pea (Cicer arietinum ) | CRISPR/Cas9 | Drought Tolerance | At4CL, AtRVE7 Knockout mutagenesis | Reduce drought tolerance | Role in regulation lignin biosynthesis metabolism and plant circadian rhythm. | Badhan et al. [102] |
| 7. | Indica mega rice cv. MTU1010 | CRISPR/Cas9 | Drought and Salt stress | OsDST gene encodes zinc finger transcription factor | Enhanced Drought and salinity tolerance | Involved in the regulation of stomatal developmental genes, therefore mutation leads to enhanced drought and salt tolerance | Santosh Kumar et al. [83] |
| 8. | Tomato (Solanum lycopersicum, L.) | CRISPR/Cas9 | Salinity and osmotic | SlARF4 and ARF4 | Down regulation and loss of function leading to improve salt and osmotic tolerance | Auxin response transcription factor role in genes associated with stress. Down regulation and loss of function lead to improve salt and osmotic tolerance | Bouzroud et al. [90] |
| 9. | Tomato (Solanum lycopersicum, L.) | CRISPR/Cas9 | Drought | SlLBD40 gene (knock down) | Knock out/loss of function, enhanced drought tolerance | It is involved in JA signaling and negative regulator of drought stress tolerance. | Liu et al. [95] |
| 10. | Maize (Zea mays) | CRISPR/Cas9 | Drought stress | MAPK-like protein 1 ZmMPKL1 knockout mutagenesis | Enhanced Drought tolerance in seedlings | Positively regulates drought sensitivity by modulating the ABA biosynthetic and catabolic genes, and ABA homeostasis. | Zhu et al. [92] |
| 11. | Brassica napus | CRISPR/Cas9 | Drought stress | bnaa6.rga-D BnaA6.RGA, gain of function mutant | Induced Drought tolerance | Involved in as negative regulator of gibberellins (GA) signalling and induced drought tolerance. | Wu et al. [103] |
| 12. | Arabidopsis thaliana | CRISPR/Cas9 | Drought stress | AREB1 | Enhanced drought tolerance | It is a key positive regulator of the drought stress response through histone modulation. | Roca Paixão et al. [84] |
| 13. | Rice (Oryza sativa) | CRISPR/Cas9 | Cold stress | osPIN5b, GS3, and OsMYB30 | Enhanced grain size, yield, and cold stress tolerance | They are involved in panicle length, grain size, and cold tolerance, respectively. Their knockdown mutants lead to enhanced traits including cold stress tolerance. | Zeng et al. [107] |
| 14. | Rice (Oryza sativa) | CRISPR/Cas9 | Salt stress | OsRR22Mutagenesis (knockout) | Enhanced salinity stress tolerance | Involved in salinity stress tolerance. | Zhang et al. [76] |
| 15. | Tomato (Solanum lycopersicum, L.) | CRISPR/Cas9 | Drought | SINPR1 mutagenesis | Reduced drought tolerance | It is involved in positive regulation of tomato plant drought response through modulation of stomatal aperture, MDA level, H2O2 level, etc., up-regulated expression of drought-related key genes, including SlGST, SlDHN, and SlDREB. | Li et al. [97] |
| 16. | Soybean (Glycine max) | CRISPR/Cas9 | Drought | GmMYB118 mutagenesis (loss of function) | Reduced tolerance to drought and salt stress (Knock out function) | Involved in positive regulation drought and salinity stress. through modulation of, MDA level, proline level, ROS, etc. | Du et al. [104] |
| 17. | Tomato (Solanum lycopersicum, L.) | CRISPR/Cas9 | Drought | SlMAPK3 gene mutagenesis | Reduced drought tolerance | Its role is involved in drought response through protecting cell membranes from oxidative damage and modulating the transcription of stress-related genes. | Wang et al. [96] |
| 18. | Maize (Zea mays) | CRISPR/Cas9 | Drought stress | ARGOS8-negative regulator of ethylene response | Enhanced grain yield under drought stress | It is a negative regulator of ethylene responses. A overexpression leads to reduced ethylene sensitivity and improved grain yield under drought stress conditions. | Shi et al. [93] |
| 19. | Rice (Oryza sativa) | CRISPR/Cas9 | Salinity stress | RAV2 Abiotic stress-responsive transcription factor | Enhanced salinity tolerance | Abiotic stress-responsive transcription factor. | Takagi et al. [108] |
Recently, a study reported the CRISPR/Cas 9 gene editing in two important genes, namely 4-coumarate ligase (4CL) and Reveille 7 (RVE7), which are associated with drought tolerance in chickpeas, the knockdown mutagenesis of these genes in the chickpea protoplast through CRISPR/Cas9 system opens the opportunities for targeted mutagenesis in chickpea and deciphering of the roles of genes under stress [102]. In Brassica napus, there are four BnaRGA genes that code for DELLA proteins, negative regulators of GA signaling. Among them, expression of BnaA6.RGA was greatly induced by drought and ABA. The CRISPR/Cas9-mediated mutagenesis led to an enhanced functional mutant of bnaa6.rga-D, and the knockdown function of quadruple mutant, bnarga. This leads to the developed enhanced drought tolerance in bnaa6.rga-D mutant while bnarga mutant exhibited reduced drought tolerance and was less sensitive to ABA treatment [103]. The role of GmMYB118-overexpressing (OE) soybean plants exhibited better drought and salt tolerance. However, plants subjected to CRISPR/Cas9-mediated mutagenesis in GmMYB118 gene led to reduced drought and salt stress tolerance suggesting the involvement of the targeted gene in drought and salt stress tolerance enhancement [104].
The GE tools as discussed, i.e., Meganuclease, ZFNs, TAL effector nucleases (TALENs), and CRISPR/Cas9 can create suitable changes in the genes with precision to obtain desired trait. But except CRISPR/Cas9, other initial GE tools such as meganuclease, ZFN, and TALENs are labor intensive and tedious because of their prerequisites that involve protein engineering, hence limiting their application. However, CRISPR/Cas9 is the one that can be used with ease in GE using different gRNAs making it most sought-after technology for GE [94]. Therefore, the recent time CRISPR/Cas-based GE system is most potent for precise GE and manipulation of gene expression in plants, leading to wide options for crop [105].