1. INTRODUCTION
Public eating places like restaurants and canteens, fast foods, and popular foods on street have become today the major elements of urban development. They allow more than 80% of the cities’ populations to satisfy their nutritional needs [1]. There is no law or legislation on the universities’ public restaurants. Despite their social-economic importance, the hygienic affairs in the common eating places systems are still a major worry for the consumers. In fact, taking into account the huge quantity of daily cooked foodstuffs, low instruction, and training of manpower, the hygiene rules are most of the time neglected in our African countries particularly, where the lower level of training is recorded [2].
If the universities’ public eating places are kept up and continue to grow, this means an increase in needs. However, the multiplication of serious food-infected diseases has been reported in recent years in several African countries, related to the proliferation of restaurants according to the WHO in 2009. Actually, more than 250 diverse diseases coming from food poisoning are described and the bacteria are reported to be the cause of 2/3 of these epidemics [3]. Among the predominant bacteria, enterobacteria are involved in these diseases. Enterobacteria are the normal hosts found in human and animal digestive tubes [4].
Klebsiella pneumoniae is an important enterobacteria. It is an opportunistic pathogen causing several diseases and is acquiring resistance to several antibiotics [5,6]. This organism is responsible for about one-third of all Gram-negative infections such as urinary tract infections, cystitis, pneumonia, surgical wound infections, endocarditis, and septicemia [7–9]. It also causes necrotizing pneumonia, pyogenic liver abscesses, and endogenous endophthalmitis [10,11]. Klebsiella pneumoniae can be found in the respiratory and intestinal tracts of humans and animals. It is frequently isolated from foods, including fresh legumes, powder foods for babies, meats, fish, and street foods [12–14]. Infections with K. pneumoniae can be transmitted through contaminated food or water and can be associated with community-acquired infections or between persons and animals involved in hospital-acquired infections.
The emergence of antimicrobial resistance is causing serious health problems nowadays worldwide. In Côte d’Ivoire, bacteria resistance to antibiotics and the emergence of bacteria producing the ELSB are becoming a serious public health problem [15–17]. Some previous works had reportewd the presence and expansion of human enterobacteria strains producing ELSB [18,19]. Klebsiella pneumoniae has developed resistance to several antibiotics through the production of enzymes such as extended-spectrum β-lactamase (ESBLs) and carbapenemase [20,21]. The production of ESBLs is encoded by several genes such as blaSHV, blaTEM, and blaCTM [22,23].
The aim of this study was to determine the prevalence, identify the antimicrobial resistance genes, and determine the resistance profile of K. pneumoniae isolated from cooked and sold food samples.
2. MATERIALS AND METHODS
2.1. Sample Collection
This study was conducted from June to September 2017 and then from November 2017 to February 2018. It was carried out on 160 of cooked and sold food samples of rice and cassava powder called “attiéké,” fish soups, fried fish, and raw fresh vegetables collected from the public and private restaurants of the two main public universities, Nangui Abrogoua University (UNA) and Félix HouphouëtBoigny University (UFHB). The choice of the private collective restaurants was based on their attendance by the students and the choice of meals consumed by them. The samples were taken in sterile bowls and sent to the laboratory for analysis.
2.2. Isolation and Identification of K. pneumoniae
Ten (10) g of each sample was suspended in aseptic conditions in 90 ml of buffered peptone water previously autoclaved for 15 minutes at 121°C. The sample suspensions were homogenized for 30 seconds. From the suspension, a serial dilution was conducted up to 10−5. From each fold dilution, 0.1 ml was streaked directly onto VRBG agar plates and incubated for 24 hours at 37°C. Red violet colonies of 0.5 mm in size were transferred to nutrient agar plates. All the suspected K. pneumoniae isolates were checked for their Gram-staining reaction, indole production, and motility. The selected colonies were further identified by biochemical tests using API 20E kits (BioMérieux, France).
2.3. Molecular Characterization of Beta-Lactamase Genes
The presence of acquired extended-spectrum beta-lactamase genes (blaTEM, blaSHV, and blaCTX-M15) was investigated by multiplex polymerase chain reaction (PCR) assays. Representative colonies of K. pneumoniae were cultured on nutritive broth at 37°C for 24 hours. Then, the cell pellet was centrifuged at 13,000 rpmfor 10 minutes. The supernatant was discarded and 400 μl of lysed buffer was added to the cell pellet and then mixed for 1 minute. Three hundred microliters of chloroform was added and then mixed again for 1 minute. Twenty μl of sodium acetate and 400 μl of ethanol were added to the supernatant and incubated at −20°C for 1 hour. The mixture was centrifuged at 12,000 rpm for 10 minutes. The supernatant is discarded, and 200 μl of isopropanol was added to the cell pellet and then centrifuged at 12,000 rpm for 10 minutes. The next step was the elimination of alcohol by drying the pellet at room temperature or for 30 minutes. Finally, 50 μl of TE buffer was added to the pellet and incubated at −20°C until use. The total DNA in the supernatant was thereafter precipitated with 70% ethanol and used as the template for PCR.
All the oligonucleotide primers have been synthesized by Sangon Biotech (Table 1). The mixture (25 μl) used for PCR included 12.5 μl of Dream Taq TM Green PCR Master Mix (Fermentas, Waltham, MA), 2 μl of primers, and 3 μl of the template DNA. PCR amplification was carried out in a thermocycler, Bio-Rad PTC-200 (Bio-Rad, Hercules, CA), according to the following cycles: initial denaturation at 95°C for 5 minutes, followed by 40 cycles at 95°C for 30 seconds; for SHV gene, 40 cycles of hybridization for 30 seconds at 53°C; for TEM gene, 40 cycles of hybridization at 50°C for 30 seconds; for CTX-M-15 gene, 40 cycles of hybridization at 49°C for 30 seconds; and the initial elongation at 72°C for 1 minute and one final elongation cycle at 72°C for 5 minutes in a thermocycler. Klebsiella pneumoniae T300, obtained from Pasteur Institute of Abidjan, was used as a positive control. The amplified products were analyzed by electrophoresis on 1.5% agarose gel composed by Gold View (0.005% v/v) (SBS Genentech, Beijing, Chine) in Tris-buffered saline (TBS) buffer 1× (Tris-HCl 40 mM, 1,18 ml acetate acid, ethylenediaminetetraacetic acid (EDTA) 2 mM pH 8,0), and the bands were visualized using a system of capture, Image Quant 350 (GE Healthcare, Waukesha, WI).
 | Table 1: Primers used for PCR amplification. [Click here to view] |
2.4. Antibiotic Susceptibility Tests
The susceptibility test of K. pneumoniae to antibiotics was determined on Mueller Hinton agar by diffusion method on disk Kirby–Bauer according to Clinical and Laboratory Standards Institute instructions. Commercially available antimicrobial discs that were used included amoxicillin (30 μg), amoxicillin + clavulanic acid (30 μg), ticarcillin + clavulanic acid (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ertapenem (30 μg), gentamicin (15 μg), tetracycline (10 μg), and ciprofloxacin (30 μg). The measurement of the inhibition zone diameter around the antibiotic discs was carried out and compared with the Tables of international measurements.
 | Table 2: Identification of enterobacteria. [Click here to view] |
3. RESULTS
3.1. Identification of K. pneumoniae
A total of 24 strains of K. pneumoniae out of 118 enterobacteria were isolated from cooked rice, “attiéké,” fried fish, fish soup, and raw fresh Vegetables (Table 2).
3.2. Contamination Rate of Food Samples with K. pneumoniae Isolates
The contamination rates of K. pneumoniae in cooked and sold food at the universities’ private and public restaurants are presented in Table 3. In total, 160 cooked food samples were tested in this study, and K. pneumoniae was isolated from 24 samples. Overall, 15% of the food samples were positive for K. pneumoniae. Klebsiella pneumoniae was isolated from 8.3% of rice samples, 20.8% of fish soup samples, 4.2% of “attiéké” samples, 37.5% of raw fresh vegetable samples, and 29.2% of fried fish samples.
At UNA, 13 (54.2%) K. pneumoniae strains were isolated from public and private restaurants. Klebsiella pneumoniae was mostly isolated from all the food samples: rice samples [2 (8.3%)], fish soup samples [4 (16.7%)], “attiéké” [1 (4.2%)], raw fresh vegetables [3 (12.5%)], and fried fish [1 (4.2%)] from private restaurants. Klebsiella pneumoniae was isolated only from fried fish samples [2 (8.3%)] from the public restaurant. At UFHB, 11 (45.8%) K. pneumoniae strains were isolated from public and private restaurants. Klebsiella pneumoniae was mostly isolated from three food samples fish soup samples [1 (4.2%)], raw fresh vegetables [2 (8.3%)], and fried fish [1 (4.2%)] from private restaurants. Klebsiella pneumoniae was isolated from two food samples, raw fresh vegetables [4 (16.7%)] and fried fish samples [3 (12.5%)] from public restaurants (Table 4).
 | Table 3: Contamination rates of K. pneumoniae isolates from all the food samples. [Click here to view] |
 | Table 4: Contamination rates of K. pneumoniae isolates per sold food from universities restaurants. [Click here to view] |
 | Table 5: Contamination rates of K. pneumoniae isolates per sold food from private and public restaurants. [Click here to view] |
The contamination rates of K. pneumonia isolates per meal served in private and public restaurants were reported in Table 5. In the public restaurants, K. pneumoniae was isolated only from 2 (8.3%) and 7 (29.2%) of “attiéké”–vegetables–fried fish samples from UNA and UFHB, respectively. K. pneumonia was isolated from 6 (25%) of rice–fish soup samples and 5 (20.8%) of “attiéké”–vegetables–fried fish samples served in the private restaurants of UNA. Klebsiella pneumoniae was isolated from 1 (4.2%) of rice–fish–soup samples and 3 (12.5%) of “attiéké”–legume–fried fish samples served in the private restaurants of UFHB.
3.3. Antimicrobial Resistance Determinants of the K. pneumoniae Isolates
According to the results of antimicrobial susceptibility testing, 24 strains were tested for ESBL genes and 3 β-lactamase genes were amplified. As shown in Figure 1, 13 out of 24 strains isolated carried only the blaSHV gene. BlaTEM and blaCTX-M-15 genes were not detected in these isolates.
3.4. Antibiotic Resistance Profile of K. pneumoniae
Antimicrobial susceptibility testing was conducted for the 13K. pneumoniae isolates producing the blaSHV gene, and detailed information on the resistance rates to all of the tested antimicrobials is listed in Table 6. The susceptibility profile of K. pneumoniae strains revealed high resistances to beta-lactams: amoxicillin (92.3%), amoxicillin + clavulanic acid (76.9%), ticarcillin + clavulanic acid (76.9%), ceftriaxone (92.2%), cefotaxime (92.2%), and ceftazidime (92.2%). The resistance rates observed for the other antibiotics were 61.5% to aminosids and 92.2% to quinolones. None of the strains were resistant to tetracycline (84.7%) and ertapenem (67.9%).
 | Table 6: Antimicrobial resistance rate of (n = 13) K. pneumoniae ESBL isolated. [Click here to view] |
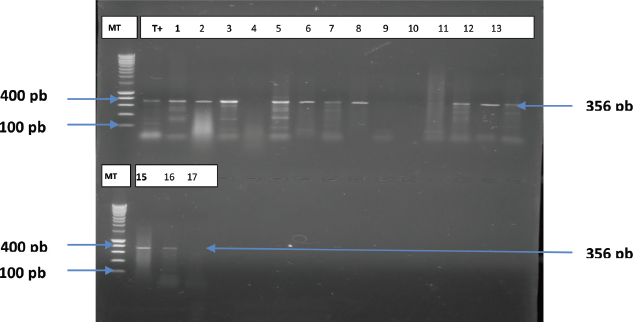 | Figure 1: Electrophoretic profile indicating the presence of the gene in K. pneumoniae isolates MT: marker height (100–1,000 bp), lines 1–17: isolates tested, and T+: positive control. [Click here to view] |
4. DISCUSSION
This study revealed a high prevalence of enterobacteria with a dominance of K. pneumoniae in cooked and sold foods in private and public restaurants of the two universities. Haryani et al. [25] also found a high prevalence of K. pneumoniae in street foods. According to Mélanie [26], the high prevalence of K. pneumoniae was due to the lack of hygiene in the cooking of foods and to the contamination of raw products (hand preparation, use of poor water quality, and insufficient cleaning of fish and vegetable). Barro et al. [1] reported that, in collective food, the hygiene rules are most of the time neglected in accordance with the enormous quantity of foods daily cooked and the low level of education of restaurant staff. According to Tereza Gelbí?ová et al. [27], Klebsiella spp is not generally considered as a major foodborne pathogen but a hygiene indicator with regard to food production. Our study revealed the presence of K. pneumoniae in cooked food from private and public restaurants. According to the Public Health Agency of Canada [28] and Sri Harminda et al. [29], this may result from its thermotolerance. These authors found that complete inactivation of the bacteria may not always occur at 60°C.
A high prevalence of K. pneumoniae was observed in the samples of “attiéké” with vegetables (tomato, pepper, and onion) and fried fish in the public and private restaurants of the two universities. These results are in agreement with the results from the survey of Falomir et al. [30] on ready-to-eat food samples, which showed that K. pneumoniae was detected in mixed vegetables rice dishes (25%, 13 out of 52). Anoman [31] also reported the presence of pathogens into “garba” (a kind of “attiéké”) with raw fresh vegetables and fried tuna fish sold in Côte d’Ivoire. Belomaria et al. [32] also found that fresh vegetables are more involved in toxic food poisoning. Consumption of raw or slightly cooked vegetables can increase the risk of foodborne disease. According to some researchers [33,34], poor hygienic practices and improper handling and cooking are considered major factors for contamination of foods. The presence of K. pneumoniae in the cooked and sold foods by the university restaurants indicated potential fecal contamination, possible cross-contamination between food handlers, food preparation, surfaces, and the food itself [25]. The attiéké meal is also served with fried tuna or horse mackerel fish. These types of fish can be contaminated with harmful enteric bacteria. According to Duflos [35], histamine-producing bacteria such as enterobacteria are commonly found in tuna fish. Insufficient cooking and improper handling and storage after cooking by the kitchen staff can contaminate the fish product. Also, K. pneumoniae may be spread from person to person or from person to item during food preparation and serving.
In our study, 13 strains of K. pneumoniae isolates harbored the blaSHV gene only. Perilli et al. [36] and Dayan et al. [37] also found that the blaSHV gene was the most dominant ESBL genotype in their studies carried out in Italy and Spain, respectively. However, ESBL blaCTX-M is the most prevalent gene found in a high proportion of samples from different parts of the world [38,39]. But in our study, blaCTM was not detected in the isolates. Finding K. pneumoniae isolates that carry antimicrobial resistance genes could potentially be an indirect public health hazard, regardless of their pathogenicity, because the available genetic pool of resistance is increased [40]. Interestingly, 13 out of 24 strains of K. pneumoniae isolates positive for the blaSHV gene were resistant to beta-lactams (amoxicillin, amoxicillin + clavulanic acid, ticarcillin + clavulanic acid, ceftriaxone, and cefotaxime). These data corroborate the finding of Gadou [41] who reported that 96.7% of K. pneumoniae strains were resistant to ceftriaxone and 95.6% to cefotaxime. Mathlouthi et al. [42] also reported that 84% of K pneumoniae strains were resistant to ceftriaxone and 97% to cefotaxime. This study reports that K. pneumoniae from cooked food samples can be resistant to multiple antibiotics. The major reasons for K. pneumoniae resistance to antibiotics including beta-lactam antibiotics are the production of a broad-spectrum beta-lactamase (ESBL), the changing of the permeability barrier or the target site represented by penicillin-binding protein, or alteration of outer membrane protein [43]. Additionally, K. pneumoniae have many efflux pumps that expel the antibiotic to the outside [44,45]. In this study, K. pneumoniae was also found to be highly resistant to quinolones at the rate of 92.2% and to aminosids at 61.5%. The resistance of K pneumoniae to aminosids and quinolones has also been reported by Gadou [41], at the respective resistance rates of 90% and 86.7%. Kouassi-M’bengué et al. [46] also found that the resistance rates were 85.7% for quinolones and 66.7% for aminosids. These results were in disagreement with those of Zhang et al. [47] who found that more than 90% of 61 K. pneumoniae isolates from retail foods in China were susceptible to quinolone antibiotics, including ciprofloxacin. The resistance of K. pneumoniae to most of the tested antibiotics mostly the beta-lactams may be the consequence of the abusive consumption of antibiotics in Côte d’Ivoire. Beta-lactams have always been the drugs of choice for the treatment of Klebsiella infections in healthcare institutions around the world, because aminoglycoside-modifying enzymes, macrolides esterases, and efflux systems render many other classes of drugs ineffective [48].
5. CONCLUSION
This study shows that K. pneumoniae recovered from cooked and sold foods by private and public restaurants of two universities of Côte d’Ivoire harbored the blaSHV gene and were resistant to beta-lactams. Food containing antimicrobial-resistant K. pneumoniae has the potential to become a public health risk to consumers. The study also shows the importance of good hygiene and handling practices during food preparation to reduce the risk of consumers being contaminated by antimicrobial-resistant K. pneumoniae. Authorities of universities should undertake sanitation control and prevention strategies in order for restaurants to improve the food quality and hygiene of the restaurant environment and staff. Restaurant staff should be trained in good hygiene practices.A study of the pathogenicity of different species of K. pneumoniae isolated from foods should be undertaken to better estimate the risk associated with their consumption.
6. ACKNOWLEDGMENTS
The authors thank the directors of the various restaurants for their collaboration.
7. CONFLICTS OF INTEREST
None declared.
8. FUNDING
No funding was availed for this study.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Barro N, Ouattara CA, Nikiema PA, Ouattara AS, Traore AS. Microbial quality assessment of some street food widely consumed in Ouagadougou, Burkina Faso. Santé 2002;12:369–74.
2. Alassane A. Contribution to the study of hygiene in collective catering at the Dakar University Center (COUD). Veterinary Medicine Thesis, Dakar, Senegal, 150 p, 1988, No. 26.
3. Bhatia A, Zahoor S. Staphylococcus aureus enterotoxins. J Clin Diagnostic Res 2007;3:188–97.
4. Bakhoum IMNS. Evaluation of the microbiological quality of some street foods in the city of Ouagadougou in Burkina Faso. Health Noteb 2004;12(4):369–74.
5. Bi X, Xu WY. An investigation of food poisoning caused by Klebsiella pneumoniae. China Pract Med 2013;8:275–6; doi:10.3969/j.issn.1673-7555.2013.26.224
6. Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, et al. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol 2017;7(24):1–9; doi:10.3389/fcimb.2017.00024
7. Ryan KJ, Ray CG. Sherris medical microbiology. 4th edition, McGraw Hill, New York, NY, 2004.
8. Lopes ACDS, Rodrigues JF, Morais MAD. Molecular typing of Klebsiella pneumoniae isolates from public hospitals in Recife, Brazil. Microbiol Res 2005;160:37–46.
9. Amraie H, Shakib P, Rouhi S, Bakhshandeh N, Zamanzad B. Prevalence assessment of magA gene and antimicrobial susceptibility of Klebsiella pneumoniae isolated from clinical specimens in Shahrekord, Iran. Iran J Microbiol 2014;6(5):311–6.
10. Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, et al. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J Clin Microbiol 2008;46:2879–83.
11. Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, et al. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis J 2012;18(8):1322–5.
12. Overdevest IT, Heck M, Van der Zwaluw K, Huijsdens X, Van Santen M, Rijnsburger M, et al. Extended-spectrum β-lactamase producing Klebsiella spp. in chicken meat and humans: a comparison of typing methods. Clin Microbiol Infect 2014;20:251–5; doi:10.1111/1469-0691.12277
13. Kim HS, Chon JW, Kim YJ, Kim DH, Kim MS, Seo KH. Prevalence and characterization of extended-spectrum-β-lactamase-producing Escherichia coli, and Klebsiella pneumoniae, in ready-to-eat vegetables. Int J Food Microbiol 2015;207:83–6; doi:10.1016/j.ijfoodmicro.2015.04.049
14. Davies YM, Cunha MPV, Oliviera MGX, Oliviera MCV, Philadelpho N, Romero DC, et al. Virulence and antimicrobial resistance of Klebsiella pneumonia isolated from passerine and psittaccine birds. Avian Pathol 2016;45:194–201; doi:10.1080/03079457.2016.1142066
15. Dadie AT, Guessennd N, Tiekoura B, Faye-Kette H, Dosso M. Resistance to betalactamins of Escherichia coli originate from foods and human isolated in Abidjan. J Pharm Biol Sci 2003;4:62–9.
16. Akoua-Koffi C, Guessennd N, Gbonon V, Faye-Ketté H, Dosso M. Methicillin resistance of Staphylococcus in Abidjan 1998-2001: a new problem. Med Mal Infect 2004;34:132–6.
17. Gbonon VC, Guessennd KN, Kouassi M’bengue A, Kacou N’douba A, N’guessan KR, Faye-Kette H, et al. Bacteriological controls of the operating room environment in a developing country : the case of the Treichville University Hospital in Abidjan in the year 2000. Rev Bio Afr 2007;4:7–11.
18. Guessennd N, Kacou-N’Douba A, Gbonon V, Yapi D, Ekaza E, Dosso M, et al. Prevalence and resistance profile of extended-spectrum β-lactamase-producing enterobacteriaceae (ESBL) in Abidjan from 2005 to 2006. J Pharm Biol Sci 2008;9:63–70.
19. Yao HA, Yapi AD, Guessennd KN, Oga S, Ouattara M, Kacou-N’Douba A, et al. Phénotypes de résistance des entérobactéries productrices de βlactamases à spectre élargi à Abidjan de 2003 à 2004 et approches thérapeutiques. Rev Bio Afr 2010;8:39–45.
20. Babini GS, Livermore DM. Antimicrobial resistance amongst Klebsiella spp. collected from intensive care units in Southern and Western Europe in 1997-1998. J Antimicrob Chemother 2000;45(2):183–9.
21. Munita JM1, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spect J 2016;4(2):4–2; doi:10.1128/microbiolspec.VMBF-0016-2015
22. Yumei G, Haijian Z, Liyun Q, Zhizhao P, Tian Q, Hongyu R, et al. Frequency, antimicrobial resistance and genetic diversity of Klebsiella pneumoniae in food samples. PLoS One 2016;11(4):e0153561; doi:10.1371/journal.pone.0153561
23. Vasaikar S, Obi L, Morobe I, Bisi-Johnson M. Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in Mthatha, Eastern Cape Province, South Africa. Int J Microbiol 2017;2017:7. Article ID 8486742; doi:10.1155/2017/8486742
24. Neuberger A, Oren I, Sprecher H. Clinical impact of a pcr assay for rapid identification of Klebsiella pneumoniae in blood cultures. J Clin Microbiol 2008;46:377–9; doi:10.1128/JCM.00568-07
25. Haryani Y. Noorzaleha AS, Fatimah AB, Noorjahan BA, Patrick GB, Shamsinar AT, et al. Incidence of Klebsiella pneumonia in street foods sold in Malaysia and their characterization by antibiotic resistance, plasmid profiling, and RAPD–PCR analysis. Food Control 2007;17(7):847–53; doi:10.1016/j.foodcont.2006.04.009
26. Mélanie C. Bacteriological analysis of food in rural areas in Laos. Medical Thesis, National University of Laos, Vientiane, Laos, pp 44–5, 2006.
27. Tereza G, Kristýna K, Lucie PH, Nicol S, Renáta K. Dissemination and characteristics of Klebsiella spp. at the processed cheese plant. Czech J Food Sci 2021;39(2):113–21; doi: 10.17221/232/2020-CJFS
28. Public Health Agency of Canada. Klebsiella spp. pathogen safety data sheets. Public Health Agency of Canada, Ottawa, CA, 2011. Available via http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/Klebsiella-eng.php (Accessed 30 October 2017).
29. Hartantyo SHP, Chau ML, Koh TH, Yap M, Yi T, Cao DYH, et al. Foodborne Klebsiella pneumoniae: virulence potential, antibiotic resistance, and risks to food safety. J Food Prot 2020;83(7):1096–103; doi:10.4315/JFP-19-520
30. Falomir MP, Rico H, Gozalbo D. Enterobacter and Klebsiella species isolated from fresh vegetables marketed in Valencia (Spain) and their clinically relevant resistances to chemotherapeutic agents. Foodborne Pathog Dis 2013;10:1002–7; doi:10.1089/fpd.2013.1552 PMID: 23980710
31. Anoman AT. Microbiological and chemical dangers associated with garba, a traditional dish produced and sold in the Ivory Coast. Doctoral Thesis, Nangui Abrogoua University, Abidjan, Côte d’Ivoire, 92 p, 2019.
32. Belomaria M, Ahiami AO, Aboussaleh Y, Elbouhalii B, Cherrah Y, Soulaymani A. Environnemental origin of collective foods intoxicationin Maroc. Study cas of Ghrab Chrarda Bnihssen region. Antropo 2007;14:83–8.
33. Ponniah J, Tunung R, Margaret SP, Son R, Farinazleen MG, Cheah YK, et al. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control 2010;21:774–8.
34. Tunung R, Ghazali FM, Noranizan M, Haresh KK, Lesley MB, Nakaguchi Y, et al. Rapid detection and enumeration of pathogenic Vibrio parahaemolyticus in raw vegetables from retail outlets. Int Food Res J 2011;18:67–78.
35. Duflos G. Histamin risks in fishing products. Bull Acad Vét Fr 2009;162(3):241–5.
36. Perilli M, Dell’Amico E, Segatore B, Rosaria de Massis M, Bianchi C, Luzzaro F, et al. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of enterobacteriaceae from an Italian nationwide survey. J Clin Microbiol 2002;40(2):611–4; doi:10.1128/JCM.40.2.611-614.2002
37. Dayan N, Dabbah H, Weissman I, Aga I, Even L, Glikman D. Urinary tract infections caused by community-acquired extended-spectrum β-lactamase-producing and nonproducing bacteria: a comparative study. J Pediatr 2013;163(5):1417–21.
38. Storberg V. ESBL-producing enterobacteriaceae in Africa a non-systematic literature review of research published 2008-2012. Infect Ecol Epidemiol 2014;4:1–6; doi:10.3402/iee.v4.20342
39. Barguigua A, Zerouali K, Katfy K, El Otmani F, Timinouni M, Elmdaghri N. Occurrence of OXA-48 and NDM- carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca, Morocco. Infect Genet Evol 2015;31:142–8.
40. Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistance bacteria. Infect Drug Resist 2015;8:49–61.
41. Gadou V. Moleculaire epidemiologyof enterobactersproducing de βlactamases with large spectralresistants to aminosids andto fluoroquinolones in Abidjan district, côte d’Ivoire. Thesis of University Félix Houphouet Boigny, Abidjan, Côte d’Ivoire, 111 p, 2019.
42. Mathlouthi N, Al-Bayssari C, El Salabi A, Bakour S, Ben Gwierif S, Zorgani AA, et al. Carbapenemases and extended-spectrum β-lactamases producing enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries 2016;10:718–27.
43. Aghamohammad S, Badmasti F, Solgi H, Aminzadeh Z, Khodabandelo Z, Shahcheraghi F. First report of extended-spectrum Betalactamase-producing Klebsiella pneumoniae among fecal carriage in Iran: high diversity of clonal relatedness and virulence factor profiles. Microbial Drug Resist 2018;26(3):261–9; doi:10.1089/mdr.2018.0181. Epub 2018 Oct 2.
44. Kouassi-M’bengue A, Folquet-Amorissani M, Nassirou F, Guessennd-Kouadio N, Kacou N’Douba A, Houenou Y, et al. Neonatal urinary tract infections in Abidjan: problematic of bacterial resistance. Mali Med 2008;23(1):34–7.
45. Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol 2018;9:289; doi:10.3389/fmicb.2018.00289. PMID: 29545778; PMCID: PMC5839057.
46. Kouassi-M’bengue A, Folquet-Amorissani M, Nassirou F, Guessennd-Kouadio N, Kacou N’Douba A, Houenou Y, Dosso M. Neonatal urinary tract infections in Abidjan: problematic of bacterial resistance. Mali Medical. 2008;23(1):34–7.
47. Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and Genotypic Characterization of Klebsiella pneumoniae Isolated From Retail Foods in China. Frontiers in Microbiology. 2018;1;9:289. doi: 10.3389/fmicb.2018.00289. PMID: 29545778; PMCID: PMC5839057.
48. Davies YM, Cunha MPV, Oliviera MGX, Oliviera MCV, Philadelpho N, Romero DC, et al. Virulence and antimicrobial resistance of Klebsiella pneumonia isolated from passerine and psittaccine birds. Avian Pathol 2016;45(2):194–201; doi:10.1080/03079457.2016.1142066