1. INTRODUCTION
Actinomycetes are the most widely dispersed group of microorganisms in nature and widely distributed in soil, water, and affiliation with plants. Actinomycetes are Gram-positive microorganism allotted shape of branching hyphae with high G+C content material in their genes along with severe metabolic possibilities [1,2]. Actinomycetes preserve a distinguished purpose applications range in screening programs due to their diversity in the production of novel antibiotics of pharmaceutical interest. The rhizosphere is the narrow region influenced by plant roots and is a hot spot for numerous organisms, such as bacteria, fungi, and algae. Rhizosphere organisms that have been well studied for their really helpful effects on plant increase and health are the nitrogen-fixing bacteria, mycorrhizal fungi, plant growth-promoting rhizobacteria, bio-control microorganisms, mycoparasitic fungi, and protozoa [3].
Actinomycetes are current appreciably in the plant rhizosphere soil and produce a sturdy antimicrobial potential of pathogenic microorganisms, saprophytic in nature of plant amplify promoters and quite a number of agro-active compounds [4]. The best-known Actinomycetes in the soil are the genus Streptomyces, although others like Norcardia, Microbispora, Micromonospora, Actinoplanes, and Streptosporangium have additionally been isolated from soil [5]. Streptomyces is the biggest genus of Actinobacteria, out of 500 species; the Streptomyces species are aerobic, spore formers with DNA affluent and large branching substrate, aerial mycelia, and widely dispersed in soil [6]. They have been diagnosed as a prolific supply of bioactive microbial metabolites as they produce 75% of biotic active compounds. One of the first antibiotics used was streptomycin produced through the ability of Streptomyces griseus [7]. Nocardia species are a part of the soil microflora and play a feature in the turnover of organic matter; there is moreover proof that some strains produce secondary metabolites of attainable industrial purpose [8].
The genus Microbacterium was characterized largely by their marked heat resistance, presence in dairies, and production of small portions of L (+) lactic acid from glucose [9,10]. The Actinomycetes isolated from the soils accrued from Kolli Hills show antibacterial or extraordinary matters to do (anti-tumor, antifungal) or might additionally function as herbicides [11]. The microorganisms are having herbal count variety and many bioactive compounds, such as Avermectin, enzymes, such as amylase, lipase, and cellulases produced from Actinomycetes play a vital function in therapeutic medicines, foods, fermentation, textile, and paper industries. They are used as plant growth promoters, bio-control agents, bio-pesticides and additionally as a grant of agro-active compounds [12].
The current research was involved in assessment of efficacy of antibiotic sensitivity and antimicrobial activity of Actinomycetes isolated from Alternanthera sessilis (L.) R. Br. ex DC weed plant rhizosphere soils.
2. MATERIALS AND METHODS
2.1. Isolation and Screening of Pigment-Producing Actinomycetes from Rhizosphere Soil Samples
Total of 26 Actinomycetes strains were isolated as per standard microbiological methods. The soil samples were collected in sterile polythene bags from plant rhizosphere of A. sessilis (L.) R. Br. ex DC from the agricultural field of Gudugeri region, Dharwad, Karnataka, India. The samples were stored in sterile polythene bags, labeled, and stored at 4°C until use. Every sample was dried at room temperature before isolation. Actinomycetes were isolated by serial dilution technique; each soil sample (1 g) was diluted with 99 ml sterile water (1:100) and shaken for 1 hour. The supernatant was diluted into 10-4, 10-5, and 10-6 [13]. 0.1 ml of each dilution was spread on starch casein agar (SCA) medium and czapekdox agar (CZA) medium supplemented with Nalidixic acid and Cycloheximide to prevent the growth of fungi and bacteria and incubated for 6 to 10 days at 28°C. Each one of the plates was morphologically observed for the Actinomycetes colonies. The plates were incubated at 30°C for 3 days and observed periodically for the growth of Actinomycetes and the isolated Actinomycetes were purified and maintained in starch casein slants at 4°C for subsequent studies.
2.2. Antagonistic Activity by Cross Streak Method
Preliminary screening of Actinomycetes isolates was tested by cross streak method against microbial pathogens, viz., Aspergillus fumigatus (MTCC 8877), Enterobacter aerogenes (ATCC 2822), Staphylococcus epidermidis (MTCC 435) and Shigella flexneri (MTCC 1457) used to be inoculated as a single streak at the center of the petri-dish consisting of media and incubated at 30 ± 2°C for 3–6 days. Later, the test pathogens were inoculated by means of streaking perpendicular to the streak of isolate, in addition, further incubation analyzed by observing the inhibition zone of activity [14].
2.3. Phenotypic Characterization of Potent Isolated Actinomycetes
Characterization of potent Actinomycete isolates was carried out by morphological, biochemical and physiological studies as described in the International Streptomyces Project (ISP). Morphological and physiological characterization were done by checking the growth in the range of temperature from 20°C to 40°C and at different pH level from pH 6.0 to 8.0. Biochemical tests like gelatine hydrolysis, starch hydrolysis, urea and catalase test along with acid production from different sugars and NaCl resistance tests were done [15].
2.4. Antibiotic Sensitivity Profiling
The antibiotic resistance of the isolates was tried utilizing five distinctive standard anti-microbial plates on Muller–Hinton agar medium. The isolates were inoculated in tryptone yeast extract broth (ISP 1) and incubated at 28°C at 150 rpm for 10 to 15 days. The developed cultures were exposed to centrifugation at 8,000 rpm for 3 minutes and the supernatant was spread with sterile L-shape spreader over the plates of Muller–Hinton agar media. The antibiotic discs, such as Chloramphenicol, Gentamycin, Penicillin, Streptomycin, and Tetracycline, were set on the plate and incubated at 30°C for 24 hours. Antibiotic affectability design was seen by estimating the width of inhibition zone. Actinomycetes isolates were either considered as sensitive (S), intermediate (I) or resistant (R) to the tried antibiotic drugs [16].
2.5. Molecular Characterization and Phylogenetic Analysis
The selected isolate RSA-14 was subjected for 16S rRNA molecular sequencing. The genomic DNA was isolated, extracted, and amplified using PCR and product was sequenced bi-directionally using the 16S primers. The 16S rRNA gene fragment was amplified using universal primers (forward primer; 5-AGAGTTTGATCTGGCTCAG-3 and reverse primer 5-TACGGTACCTTGTTACGACTT-3). The PCR product (1,500 bp) of the sample was identified by 1% agarose gel electrophoresis. Computerized sequencing was carried out concurring to the connected biosystems robotized by di-deoxy chain termination strategy utilizing cooperative energy logical administrations [17]. The arrangement was compared for closeness level with the reference strains of Actinomycetes from genomic database banks, utilizing the NCBI impact accessible at the http://www.ncbi.nlm.nih.gov/ impact web location. The structures were examined utilizing the CLUSTAL Wv.1.4 program and MEGA 7.0 software program [18].
3. RESULTS
3.1. Isolation and Screening of Pigment-Producing Actinomycetes from Rhizosphere Soil Samples and Antimicrobial Activity
In the present study, a total of 26 Actinomycetes are isolated from the rhizosphere soil collected from A. sessilis (L.) R. Br. ex DC a weed plant from some regions of Gudugeri, Karnataka, India. These strains were sub-cultured on SCA slants and preserved in cooled refrigerator for further study purposes. All the 26 strains were tested for a Gram-positive or Gram-negative test. The 26 strains that produced different pigments on the media were shown in (Fig. 1). The color varied among the isolates, such as RSA-1, 15 and 24 observed with white pigment, RSA-2, 12, and 23 with green pigment, RSA-14 and 21 orange pigment, while RSA-3, 10, and 22 with grey pigment, RSA-4, 7, 16, and 25 with yellow, RSA-5, 9, 17, 18, and 19 with brown, RSA-6 and 11 with red, RSA-8 purple pigment and RSA-13, 20, and 26 with black pigment. The observation was analyzed with the help of the RAL code and listed (Table 1). These isolates were further subjected to a cross streak technique for preliminary screening of antimicrobial undertaking for extraordinary pathogenic strains and results were shown in (Table 2). Out of 26 strains, RSA-14 strain showed the wide antimicrobial activity of pathogenic organisms as proven (Fig. 2) and hence subjected to further analysis.
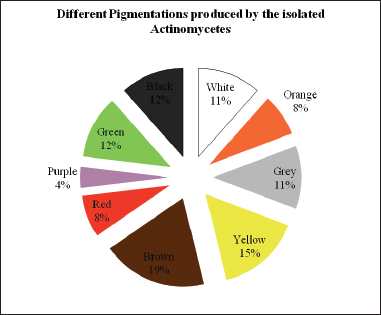 | Figure 1: Percentage distribution of pigmentations produced by the isolated Actinomycetes [Click here to view] |
3.2. Characterization and Identification of Actinomycetes
The primary importance was to identify cultural characteristics and further subjected to morphology, physiology and biochemical characteristics of Actinomycetes strain RSA-14. The color of aerial mycelium and substrate mycelium was observed with white and orange, respectively. The pH range of 6.0–8.0 was chosen for the growth of the organism and pigment production, in which the increased growth observed at pH 7.0. The isolated strain RSA-14 showed good growth and pigment production at 30°C temperature. For gram staining, the active isolate stood Gram-positive and was non-motile. The biochemical tests were carried out and the isolate RSA-14 confirmed advantageous effects for NaCl, starch, casein, urea and gelatin, carbohydrates and nitrate reduction tests and results were shown in Table 3.
 | Table 1: Isolates of pigment producing Actinomycetes. [Click here to view] |
3.3. Antibiotic Sensitivity Profiling
Total of 26 Actinomycetes were isolated and were tested for their ability to grow in the presence of antibiotics, such as Chloramphenicol, Gentamycin, Penicillin, Streptomycin and Tetracycline. The strain RSA-14 was examined for its capacity to grow in the presence of antibiotics and results were recorded (Table 4).
3.4. Molecular Identification and Phylogenetic Analysis
The genomic DNA was isolated from the strain RSA-14 and analyzed by 16S rRNA gene sequencing with the help of PCR amplification. The isolated DNA was confirmed with 1% agarose gel. The 16S rRNA gene sequence of strain RSA-14 nucleotide sequence obtained was 1,409 bp in length which was later, subjected to the search of close relatives through Nucleotide BLAST (BLASTN) in NCBI database (Accession No MK905528). The 16S rRNA gene sequencing and analysis of the phylogenetic tree showed 100% sequence similarity with Streptomyces cinereoruber strain P.B.373 (Fig. 3).
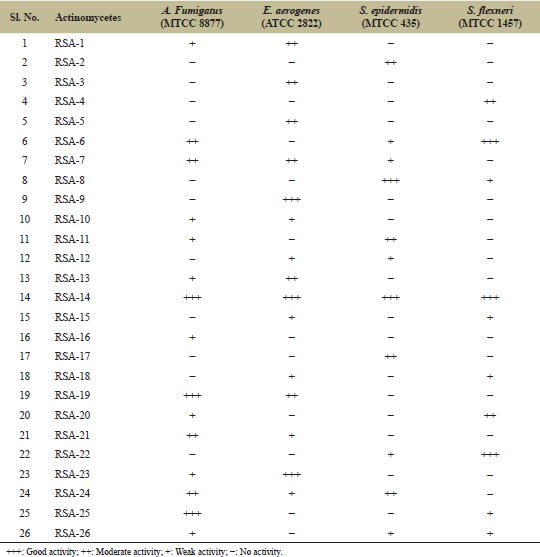 | Table 2: Activity of isolated strains against different test pathogens. [Click here to view] |
 | Figure 2: Zone of growth inhibition by cross streak-plating technique from isolated strain RSA-14. A. fumigatus (MTCC 8877), E. aerogenes (ATCC 2822), S. epidermidis (MTCC 435), and S. flexneri (MTCC 1457). [Click here to view] |
 | Table 3: Morphological, physiological and biochemical characteristics of the strain RSA-14. [Click here to view] |
 | Table 4: Antibiotics susceptibility pattern of isolated Actinomycetes from rhizospheric soil. [Click here to view] |
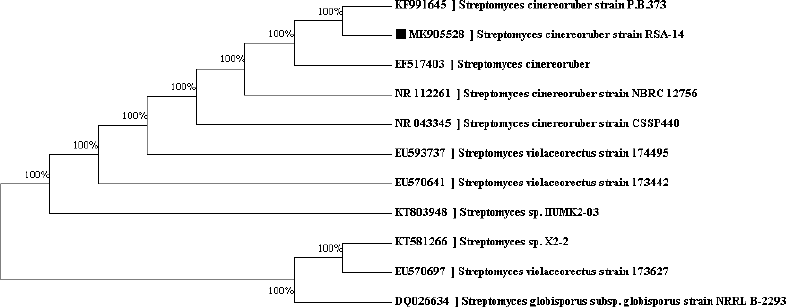 | Figure 3: Dendrogram indicating the phylogenetic relation of the S. cinereoruber strain RSA-14. [Click here to view] |
4. DISCUSSION
The screening of antibiotic producing Actinomycetes was isolated from soil. The thousands of Actinomycetes were isolated and screened in different research laboratories since 1937 [19]. These overviews brought about the separation of a few new genera and species just as revelation of new antibiotics [20]. In our experiments, different types of soils were taken from the Gudugeri region, Kundagol, Dharwad. According to Sarayza and Hemashenpagam [21], this might be because of their incubation period and diverse particular media incorporated with antibiotics. The morphological characterization of the Actinomycetes isolates was studied by using SCA and CZA. The morphological, cultural characteristics of 26 isolates showed different colors. Comparable discoveries were observed in prior reports [22,23], where chalky white, yellow, orange, red, violet, green, and gray colored series of Actinomycetes were found dominant in different soil samples. The aerial and structural mycelium color considered as characteristic features of Actinomycetes isolates. Kumar and Kanniban [24] used different media for isolation of Actinomycetes, and among the different media the SCA was proved to be effective for isolation. All Actinomycetes screened for antimicrobial activity against A. fumigatus, E. aerogenes, S. epidermidis, and S. flexneri. Preliminary screening of antimicrobial activity was done with all Actinomycetes isolates, only RSA-14 isolate revealed good inhibition zone. Essentially noticed the scope of antimicrobial activity among Streptomyces species has been recently seen in other soil isolates. Antibiotics susceptibility pattern were tabulated (Table 4). The strain RSA-14 showed excellent activity against Chloramphenicol, Penicillin and Tetracycline antibiotics. Gentamycin showed moderate activity, whereas the isolated strain RSA-14 showed resistance to Streptomycin. The morphological, physiological, and biochemical characteristics of RSA-14 classified it into Streptomyces sp. The pH which favored the ideal development of the strain RSA-14 was found at pH 7.0. There was no positive effect on growth at the pH below 7.0, but showed good growth at pH 8.0 [25]. The effect of temperature on the strain RSA-14 showed relatively good growth at 30°C. The strain RSA-14 showed a positive test for starch, casein, gelatine, and urea. The carbon sources most utilized by the strain RSA-14 were raffinose and galactose. The genomic DNA was isolated from the strain RSA-14 and analyzed by 16S rRNA gene sequencing with the help of PCR amplification [26]. The complete sequence obtained was 1,409 bp in length. The arrangement was submitted to NCBI with the accession number MK905528, which was later subjected to the search of close relatives through Nucleotide BLAST (BLASTN) in the National Centre for Biotechnology Information (NCBI) [27]. The analysis of the 16S rRNA gene sequence of the strain RSA-14 revealed a 100% sequence similarity with S. cinereoruber. Furthermore, a phylogenetic tree was built through neighbor getting strategy together with acquired quality groupings; the analysis of the phylogenetic tree revealed the strain RSA-14 fell within the cluster of the genus Streptomyces sp. and formed a branch with S. cinereoruber strain P.B.373.
5. CONCLUSION
It can be concluded that the soil of Gudugeri region may harbor a rich source of biologically potential Actinomycetes which has the capabilities of producing secondary metabolites with antimicrobial functions. Antibiotics produced from Actinomycetes of rhizosphere soils of A. sessilis (L.) R. Br. ex DC can be used to prevent plant bacterial and fungal diseases. Further, the characterization of Actinomycetes and their products for utilization in plant biotechnology, disease management, environmental biotechnology and some other applications yet to be done, the potential number of metabolites from Actinomycetes maybe discover in future
ACKNOWLEDGMENT
The authors gratefully acknowledge the instrumentation facilities for this work provided by USIC, Karnatak University, Dharwad through UGC-SAP-DSA-1, Department of Botany, Karnatak University, Dharwad, Karnataka, India.
CONFLICT OF INTEREST
Authors declare that they do not have any conflicts of interest.
FINANCIAL SUPPORT
None.
REFERENCES
1. Lo CW, Lai NS, Cheah HY, Wong NKI, Ho CC. Actinomycetes isolated from soil samples from the crocker range sabah. Asean Rev Biodivers Environ Conserv 2002;9:1–7.
2. Korn-Wendisch F, Kutzner HJ. The family Streptomycetaceae. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH (eds.). The prokaryotes. Springer-Verlag Inc, New York, NY, pp 921–95, 1992.
3. Rodrigo M, Paolina G, Jos M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 2013;37:634–63. CrossRef
4. Franco-Correa M, Quintana A, Duque C, Suarez C, Rodriguez MX, Barea JM. Evaluation of Actinomycete strains for key traits related with plant growth promotion and mycorrhiza helping activities. Appl Soil Ecol 2010;45:209–17. CrossRef
5. Isik K, Gencbay T, Ozdamir-Kocek F, Cil E. Molecular identification of different Actinomycetes isolated from east black sea region plateau soil by 16S rDNA gene sequencing. Afr J Microbiol Res 2014;8(9):878–87. CrossRef
6. Schatz A, Bugie E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Clin Orthop Relat Res 2005;437:3–6. CrossRef
7. Poornima R, Ponmurugan P. Studies on secondary metabolite production by Streptomyces species. Paper present in National seminar on “Emerging Trends in Industrial Biotechnology, Bioexplore’ 06.” Vivekananda College of Engineering, Tiruchengode, India, pp17–18, 2006.
8. Kinoshita N, Homma Y, Igarashi M, Ikeno S, Hori M, Hamada M. Nocardia vinacea sp. nov. Actinomycetologica 2001;15:1–5. CrossRef
9. Brennan NM, Brown R, Goodfellow M, Ward A, Beresford TP, Vancanneyt N, et al. Microbacterium gubbeenense sp. nov., from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 2001;51:1969–76. CrossRef
10. Young CC, Busse HJ, Langer S, Chu JN, Schumann P, Arun AB, et al. Microbacterium agarici sp.nov. Microbacterium humi sp. nov. and Microbacterium pseudoresistens sp. nov., isolated from the base of the mushroom Agaricus blazei. Int J Syst Evol Microbiol 2010;60:854–60. CrossRef
11. Thangapandian V, Ponmurugan P, Ponmurugan K. Actinomycetes diversity in the rhizosphere soils of different medicinal plants in Kolly hills-Tamilnadu, India, for secondary metabolite production. Asian J Plant Sci 2007;6(1):66–70. CrossRef
12. Salim FM, Sharmili SA, Anbumalarmathi J, Uma MK. Isolation, molecular characterization and identification of antibiotic producing Actinomycetes from soil samples. J App Pharm Sci 2017;7(9):69–75.
13. Dandan H, Lanying W, Yanping L. Isolation, identification, and the growth promoting effects of two antagonistic Actinomycete strains from the rhizosphere of Mikania micrantha Kunth. Microbiol Res 2018;208:1–11. CrossRef
14. Reddy NG, Ramakrishna DPN, Rajagopal SV. Morphological, physiological and biochemical studies of marine Streptomyces rochei (MTCC 10109) showing antagonistic activity against human pathogenic microorganisms. Asian J Biol Sci 2011;4:1–14. CrossRef
15. Goodfellow M, Williams ST. Ecology of Actinomycetes. Annu Rev Microbiol 1983;37:189–216. CrossRef
16. Saadoun I, Muhana A. Optimal production conditions, extraction, partial purification and characterization of inhibitory compounds produced by Streptomyces Ds-104 isolate against multi-drug resistant Candida albicans. Cur Trends Biotechnol Pharm 2008;2:402–20.
17. Saravana KP, Preetam RJP, Duraipandiyan V, Ignacimuthu S. Antibacterial activity of some Actinomycetes from Tamil Nadu, India. Asian Pac J Trop Biomed 2012;2(12):936–43. CrossRef
18. Sreenivasa N, Bidhayak C, Pallavi SS, Meghashyama PB, Dattatraya A, Dhanyakumara SB, Muthuraj R, Halaswamy H, Shashiraj KN, Chaitra M. Isolation, characterization, and functional groups analysis of Pseudoxanthomonas indica RSA-23 from rhizosphere soil. J Appl Pharm Sci 2019;9(11):101–6. CrossRef
19. Kandasamy S, Muthusamy G, Thangaswamy S, Senthilkumar B. Screening and identification of antibiotic producing Actinomycetes and their antagonistic activity against common pathogens. World Res J Antimicrobial Agents 2012;1(1):7–10.
20. Hoodfellow M, Williams ST, Mordarski M. Actinomycetes in biotechnology. Academic Press, New York, NY, pp 78–82, 1988.
21. Sarayza S, Hemashenpagam N. Antagonistic activity and antibiotic sensitivity of lactic acid bacteria from fermented dairy product. Adv Appl Sci Res. 2011;2(4):528–24.
22. Pridham TG, Tenser HD, Waksman, Hinrici. Streptomyces. In: Bergey’s manual of determinative bacteriology. 8th edition, pp 339–748,1974.
23. Rizk M, Rahman TA, Metwally H. Screening of antagonistic activity in different Streptomyces species against some pathogenic microorganisms. J Biol Sci 2007;7:1418–23. CrossRef
24. Kumar S, Kannabiran K. Antifungal activity of Streptomyces VITSVK5 sp against drug resistant Aspergillus clinical isolates from pulmonary tuberculosis patients. J Mycol Med. 2010;20:101–7. CrossRef
25. Li D, Pang H, Sun L, Fan J, Li Y, Zhang J. Pseudoxanthomonas wuyuanensis sp. nov., isolated from saline-alkali soil. Int J Syst Evol Microbiol 2014;64:799–804. CrossRef
26. Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 1989;8:151–6. CrossRef
27. Sreenivasa N, Chethan JD, GirishBabu K, Ravichandra H, Pallavi SS, Bidhayak C. Molecular characterization and antimicrobial efficacy of Streptomyces gancidicus strain SN-3 from soil samples. J Microbiol Res 2018;8(4):97–102.