Aeromonas caviae is a surging and opportunistic bacterium that causes gastrointestinal infections and septicemia. Due to the overuse of antibiotics, A. caviae has evolved to be antimicrobial resistant. This emergence has become a significant concern, as the effectiveness of antibiotics is threatened, demanding the need for alternative therapeutic approaches. The research employs immunoinformatics-based computational techniques to develop a multi-epitope-based vaccine for treating infections triggered by A. caviae. The whole proteome of A. caviae (strain: GSH8M-1) was retrieved and analyzed for anti-microbial resistance and was sequestered. Cytoplasmic membrane and periplasmic membrane proteins were compiled for B-cell and T-cell epitope prediction. Among those, six promiscuous epitopes (EIKPKDYPK, PYKFAPDGF, TTLGDDAKR, LPARAARTM, SLLPARAAR, FELDDKASL) were then connected using GPGPG linkers. Human β-defensin adjuvant was connected by an EAAK linker. Docking of the vaccine construct was performed with Human toll-like receptor 2 (TLR-2), TLR-3, Major Histocompatibility Class I (MHC-I), and MHC-II, and TLR2 with the lowest docking score was selected. Finally, the presumed vaccine was subjected to codon optimization, In silico cloning and immune simulations. This evaluation ensures the feasibility of the vaccine to elicit the immune response against antimicrobial resistance A. caviae infections, laying the preparation for future experimental investigations and clinical applications.
Yadava I, Baiju SK, Swaminathan P. Engineering a multi-epitope-based vaccine candidate against anti-microbial-resistant Aeromonas caviae using an integrated immunoinformatics approach. J Appl Biol Biotech 2025;13(6):160–170. http://doi.org/10.7324/JABB.2025.v13.i6.19
Antimicrobial resistance has become one of the top 10 global public health threats, according to the World Health Organization (WHO) [1]. The misuse and overuse of antimicrobials over the years has resulted in the development of many drug-resistant pathogens. Global Antimicrobial Resistance and Use Surveillance System report 2022 [2] stated antimicrobial resistance is estimated to kill approximately 700,000 people annually. The emergence of multi-drug-resistant pathogens has led to a dangerous level such that it has increased mortality for even common infections. Antimicrobial resistance affects everyone, irrespective of income and region. Recent research studies have predominantly observed the rise of antimicrobial-resistant Aeromonas species.
Aeromonas caviae is a Gram-negative bacterium that can cause gastrointestinal infections and extra-intestinal infections, including septicemia, traumatic wound infections, and lower respiratory tract infections [3]. In recent years, pathogenic A. caviae has shown resistance to penicillin, carbenicillin, ampicillin, and ticarcillin [4]. Since the multidrug-resistant bacteria show resistance against conventional antibiotics, treating an individual with this opportunistic A. caviae has become more challenging [5]. Hence, effective and long-lasting novel therapy is needed to encounter A. caviae infections. An immunoinformatics-based approach and reverse vaccinology techniques are considered to mitigate the emerging resistance in these bacteria. The process of discovering antigenic proteins (epitopes) from a genome is called reverse vaccinology. These reverse vaccinology methods involve the comparative in silico analysis of multiple genome sequences to find conserved antigens in a population of heterogeneous pathogens. These methods can also be used to identify antigens that are distinctive to pathogenic isolates but are absent in commensal strains [6].
The primary objective of this study is to design a multi-epitope-based vaccine against A. caviae causing gastrointestinal and extra-intestinal infections such as soft-tissue infections, osteomyelitis, endocarditis, and meningitis. This vaccine candidate aims to enhance protective immunity while addressing the growing challenge of antimicrobial resistance. This study explores the core proteome of the A. caviae strain (GSH8M-1) assessed through proteome subtraction to design a multi-epitope-based vaccine construct using reverse vaccinology and immunoinformatics techniques. Suitable candidates for vaccine design were identified using in vivo techniques, and multiple computational-based bioinformatics tools were applied to predict antimicrobial resistance proteins, and their localization, and to select promiscuous epitopes (B-cell and T-cell epitopes) [7]. The epitopes were further shortlisted based on antigenicity, allergenicity, solubility, and toxicity. Highly antigenic, non-allergenic, and nontoxic epitopes were selected for vaccine prediction. Further secondary structures were predicted, the vaccine was docked with human toll-like receptor 2 (TLR-2), codon optimization was conducted, and it was evaluated through immune simulation [8]. The results will serve as trailblazer work for identifying an immunogenic vaccine model to prevent infections with A. caviae. The vaccine construct developed would provide a proactive approach to preventing A. caviae infections, particularly benefiting immunocompromised individuals. By reducing reliance on antibiotics, the vaccine helps mitigate the spread of antimicrobial resistance.
The process began with the retrieval of the complete proteome sequence (UniProt ID: UP000280168) of A. caviae from UniProtKB, comprising 4364 proteins. The cited proteome was then subjected to the Comprehensive Antibiotic Resistance Database (CARD) (RGI Resistance Gene Identifier - McMaster University) [9] for the identification of the resistance protein sequences. This was followed by subjecting the shortlisted resistance protein to surface localization using an online server PSORTb (PSORTb Subcellular Localization Prediction Tool, version 3.0) [10], which predicts the subcellular localization of proteins from Gram-positive and Gram-negative bacteria. In the analysis of subcellular localization, the focus was on retaining cytoplasmic membrane and periplasmic membrane proteins. Other cytoplasmic proteins were not deemed viable targets for vaccines but rather for drug development purposes. Surface proteins, which are exposed to the host immune system, are considered suitable vaccine candidates since they can elicit the appropriate immune responses. Virulent proteins, which can cause infections, are also viewed as promising vaccine candidates due to their potential to provoke robust immune responses. This selective approach is designed to identify vaccine candidates that are most likely to generate effective immune responses against A. caviae, thereby optimizing the efficiency of epitope-based vaccine design. To identify virulent proteins, Vaxi-DL (Vaxi-DL|Home) [11] (a web server used to evaluate the potential of protein sequences to serve as vaccine target antigens) was used. After analyzing extensively, only shortlisted target proteins underwent putative epitope predictions.
The immune epitopes and database and analysis resource (Immune Epitope Database [IEDB]) webserver was utilized to predict B-cells and Major Histocompatibility Class I (MHC-I) and MHC-II epitopes. For B-cell epitope prediction, the Bepipred linear epitope prediction 2.0 server (B Cell Tools - IEDB) [12] was utilized. In T-cell epitope prediction, IEDB-recommended (MHC-I Binding - IEDB) and (MHC-II Binding - IEDB) were used to predict MHC-I and MHC-II epitopes, respectively [13]. In both the MHC epitopes prediction phase, a reference set of MHC alleles was used, and the promiscuous epitopes were selected [14]. Epitopes were analyzed for antigenicity, allergenicity, water solubility, and toxicity to evaluate their potential to provoke an immune response, avoid allergic reactions, select water-soluble peptides, and prevent toxic responses [15], respectively. Vaxijen 2.0 (VaxiJen - ddg-pharmfac.net) [16], AllerTop (Bioinformatics tool for allergenicity prediction - ddg-pharmfac.net) [17], PepCalc (Peptide solubility calculator - PepCalc.com), and ToxIBTL (https://server.wei-group.net/ToxIBTL/Server.html) [18] conduct these analyses.
The selected epitopes were combined with the Human β-defensin adjuvant [19], which enhances the immune response evoked by the vaccine. Human β-defensin can activate immune cells and amplify immune responses [20]. Along with the adjuvant, linkers such as EAAK and GPGPG are also used [21]. They maintain structural integrity, immunogenicity, and balance of epitope presentation. The contemporary method described herein lays the groundwork for the development of a multi-epitope-based vaccine [22] with significant potential to effectively combat A. caviae infections. These methods enhance the vaccine’s efficiency, safety, and immunogenicity.
The physicochemical properties of the construct were evaluated using an online server, the ProtParam tool (Expasy - ProtParam tool) [23], which depicted molecular weight, theoretical pI, aliphatic index, hydrophobicity, and other parameters of the vaccine construct. After physicochemical analysis, the secondary structure of the vaccine model was predicted by an online server, Protein Structure Prediction Server (PSIPRED) [24]. The predicted model was subjected to validation after visualization in PyMOL software. Using ProSA-web [25] and PROCHECK [26], the Z-score and Ramachandran plots were obtained, respectively. The Z-score predicts the accuracy, and any deviation in it suggests errors, whereas the Ramachandran plot helps in stereo-chemical analysis [27].
Molecular docking analysis is a significant method in studying how the vaccine construct interacts with immune receptors. Herein, the molecular docking interactions of the vaccine construct were analyzed using the HDOCK (HDOCK Server - HUST) server [28]. Initially, the immune receptors: MHC I (1CE6), MHC II (1KG0), TLR 2 (6 NIG), and TLR 3 (7C76) were retrieved from the RCSB Protein Data Bank (RCSB PDB) and were given for docking, resulting in the evaluation of docking score. The lower the docking score, the higher the interaction between the vaccine and receptor [28,29]. Further, the server PDBsum (EMBL-EBI homepage|EMBL-EBI) [30] was utilized to interpret the binding interactions between the residues of the vaccine and receptors.
The codon optimization of the vaccine construct has been performed using the JCat tool (JCat: A novel tool to adapt codon usage of a target gene) [31]. Herein, codon optimization of the multi-epitope-based vaccine model was reverse transcribed to DNA using the JCat tool. The gene expression was optimized with Codon Adaptation Index (CAI) >0.7 and GC content >60%, and the optimized DNA sequence for in silico cloning was acquired. This optimized and transcribed DNA was virtually cloned into an expression pET28a (+) vector with restriction enzyme sites (Eco53KI and EcoRV) [32] and the cloned vaccine was obtained using SnapGene software (SnapGene | Software for everyday molecular biology).
The designed vaccine was subjected to an online web server C-Immsim (kraken.iac.rm.cnr.it) [33] to review the ability of the vaccine toward different immune responses. The evaluation involves quantifying antibody levels (xx/ml), interferons, and cytokines (ng/ml) against antigenic constructs. The measurements indicate the usefulness of epitope-based vaccines against diverse pathogens. Antibody counts reflect immune response strength, while interferons and cytokines indicate immune system activation. Such evaluations assist in identifying promising vaccine candidates and improve the design for effective immune responses against various pathogens. This computational vaccinology approach, along with experimental validations, helps in constructing a multi-epitope-based vaccine.
The whole proteome of A. caviae (strain no. UP000280168) was retrieved from UniProt and analyzed for antimicrobial-resistant proteins. The core proteome consists of a total of 4364 proteins, among which fourteen proteins were found to be antimicrobial resistant by using the CARD and were categorized using PSORTb based on their subcellular localization. CARD analysis was utilized in selecting Antimicrobial Resistance (AMR) genes from Acinetobacter baumannii, responsible for nosocomial infections in humans [34]. Among the AMR proteins selected by CARD analysis, five were cytoplasmic membranes, one was periplasmic, three were unknown, and five were found to be in the cytoplasm as mentioned in Figure 1. The cytoplasmic membrane and periplasmic-based proteins (A0A3G9I8K5, Q68E66, A0A2U8QNR4, A0A3S5WNV0, A0A3G9I627, and A0A3G9ILN3) were selected for further processes. Periplasmic and cytoplasmic proteins often contain highly immunogenic epitopes, meaning they can elicit strong immune responses. The selected six protein sequences were evaluated by Vaxi-DL, a web-based deep learning server that evaluates the potential of protein sequences to serve as vaccine target antigens. Out of six, four were obtained to be suitable candidates for vaccine constructs based on their antigenicity, as shown in Figure 2. Another study by Dixit, Vaxi DL, was found and acknowledged for predicting antigenicity in the N-acetylmuramoyl-L-alanine amidase vaccine against immunoproliferative small intestine disease [35].
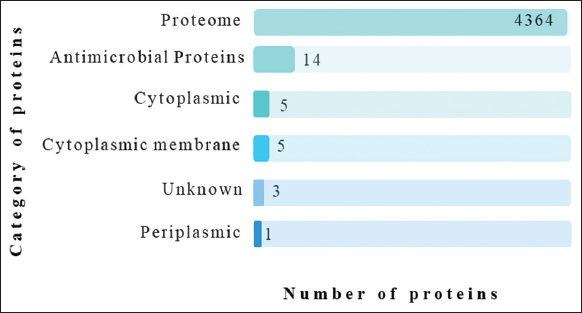 | Figure 1: Number of proteins obtained at each step of protein selection. [Click here to view] |
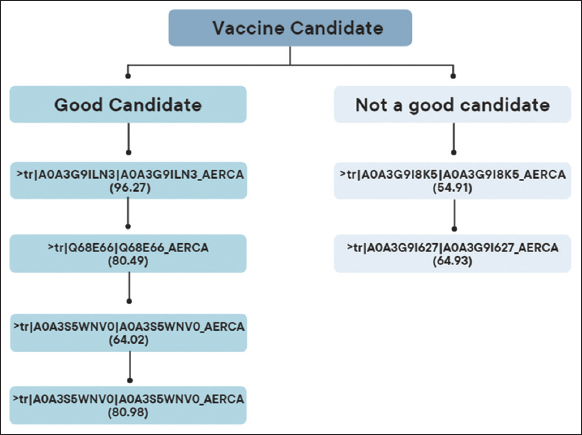 | Figure 2: Candidates for vaccine construct. [Click here to view] |
The filtered four protein sequences were selected to produce B-cell and T-cell (MHC-I and II) epitopes using the IEDB. The importance of B-cell and T-cell epitope prediction has been validated in a molecular modelling study on vaccine design against Klebsiella aerogenes [36], where they have explained how B-cell and T-cell epitopes enhance immune responses. Linear B-cell epitopes from four different protein sequences were analyzed using the IEDB B-cell epitope prediction tool; all the predicted epitopes are mentioned in Table 1. Similarly, T-cell epitopes were predicted using the IEDB T-cell Epitopes-MHC Binding Prediction tool. In T-cell epitope prediction, both MHC-I and II were considered, and only promiscuous epitopes were shortlisted and tested for antigenicity, solubility, toxicity, and allergenicity using Vaxijen, PepCalc, ToxIBTL, and AllerTop, respectively. The antigenicity score was predicted by using a threshold value of 0.5 for higher accuracy and higher specificity in VaxiJen 2.0. The ones with antigenicity near one, water-soluble, non-allergic, and non-toxic were selected for vaccine design as shown in Table 2. The work performed by Kobayashi defined how promiscuous T-cell epitopes were used for HER2/neu tumor antigen [37]. The epitopes EIKPKDYPK, PYKFAPDGF, TTLGDDAKR, FELDDKASL, LPARAARTM, and SLLPARAAR were shortlisted based on the promiscuity.
Table 1: Predicted B-cell epitope selected for vaccine candidates in Aeromonas caviae.
| Protein accession number/name | Predicted B-cell epitopes |
|---|---|
| >tr|A0A3G9ILN3|A0A3G9ILN3_AERCA Beta-lactamase | GDKAATDPLDRERAVGVSGGFELDDKASLPWLKGSAFLQFPEE VDSLEKAYRQWTPAYSPGSHRQPFDRLYHTYLNVPEQAMGHYSK EDKPIRVTPGMLAGVDNAAMQQAIDLTHQGQYAVGEMTQGA YPVSEQTLLAGNSPAMIYNTNPAAPAPAAAGPNE |
| >tr|Q68E66|Q68E66_AERCA Ethidium bromide resistance protein | SSEGFTKLAGQKLDAWPSWKSLRRP |
| >tr|A0A2U8QNR4|A0A2U8QNR4_AERCA Colistin resistance protein | IKIILYKLEDFKVLFDQNEALAEYEEKWFKGDSVGRSNLNRYL AEPIPFTTLGDDAKRDTNQSKNFSMNGYEKDTNPFTMGRKE FDDNLARNSEGCKGVCDRVPNIEIKPKDYPKFCDKNTCYPTY YKRYPDAHRQFTPDCPSSDIENCTDEELTNTDSLGALGLYLHGT PYKFAPDGFIKEKGMNMECLQKNAAANRTAIYEQELDIFKQ |
| >tr|A0A3S5WNV0|A0A3S5WNV0_AERCA MFS transporter | RRYSPLTKSAAWVDRFGRDAVGALSNTGHVAGLSVERVLLPA RAARTMTTTATLDAGRQVAELGGVGARISAQNWLQLPGTPP SASDLAPQFATMAAAATASAPTHT |
Table 2: Predicted T-cell epitope selected for vaccine candidate.
| Protein | Epitopes | Antigenicity | Allergenicity | Toxicity | Solubility | Promiscuous allele |
|---|---|---|---|---|---|---|
| MCR phosphor-ethanol-amine transferase | EIKPKDYPK | 0.8906 | Non-Allergic | Non-Toxic | Good water Solubility | 81 |
| PYKFAPDGF | 0.9841 | Non-Allergic | Non-Toxic | Good water Solubility | 81 | |
| TTLGDDAKR | 0.8022 | Non-Allergic | Non-Toxic | Good water Solubility | 81 | |
| MOX beta-lactamase | FELDDKASL | 0.5737 | Non-Allergic | Non-Toxic | Good water Solubility | 81 |
| GFELDDKASL | 0.4958 | Non-Allergic | Non-Toxic | Good water Solubility | 27 | |
| TPAYSPGSHR | 0.9076 | Non-Allergic | Non-Toxic | Good water Solubility | 27 | |
| Macrolide phosphor-transferase (MPH) | GHVAGLSVER | 1.1988 | Non-Allergic | Non-Toxic | Good water Solubility | 27 |
| LPARAARTM | 0.7479 | Non-Allergic | Non-Toxic | Good water Solubility | 81 | |
| SLLPARAAR | 0.5469 | Non-Allergic | Non-Toxic | Good water Solubility | 81 |
The design of multi-epitope-based vaccines against A. caviae has been constructed by joining the selected epitopes using a GPGPG linker. In a study by Aldakheel et al., a potential vaccine was created against Clostridium perfringens using EAAK and GPGPG linkers [38]. To enhance the immunogenicity and potency, an adjuvant (Human β-defensin) that can induce immune response was added using an EAAK linker. Epitopes were attached using the GPGPG spacer, chosen for its ability to lower binding affinity across key regions and its high glycine and proline content, which are associated with beta turns and the generation of secondary and tertiary structures, while also providing greater flexibility for the epitopes to adopt various conformations that enhance immune recognition. Studies indicate that a GPGPG spacer construct can effectively stimulate helper T Helper T lymphocytes cell (HTL) responses through either polypeptide or DNA immunization strategies, aiding in the control of infections [39,40]. Human beta-defensin adjuvant serves as an immunomodulatory agent that has been found effective in boosting the immune response and in potentiating antigen-specific immunity by activating dendritic cells and T cells. The review on the application of built-in adjuvant for epitope-based vaccines by [19] offers evidence of the usage of beta-defensin as an adjuvant. The schematic diagram of the constructed vaccine along with adjuvants and linkers is illustrated in Figure 3. Similar designing of a vaccine construct was successfully done by Naveed et al. to construct a peptide vaccine against Citrobacter freundii, which is associated with gastrointestinal and respiratory tract infection [41].
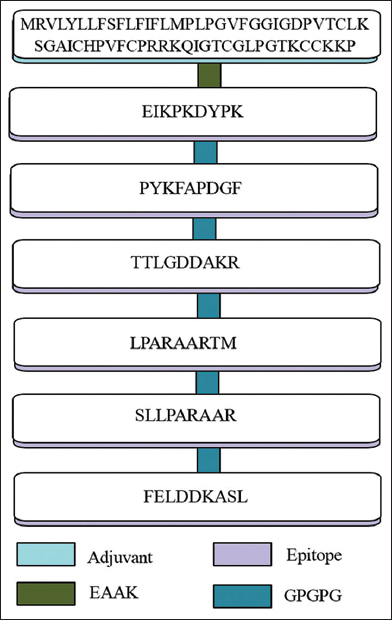 | Figure 3: Schematic diagram of multi-epitope-based vaccine. [Click here to view] |
The physiochemical properties were predicted before the construction of the 3D structure. The physicochemical properties of the predicted vaccine constructs are shown in Table 3 using the ProtParam tool. The vaccine consists of 146 amino acids with a molecular weight of 15104.80 kDa. Here, theoretical Isoelectric pH defines the pH at which the net charge of the protein is zero and was found to be 9.59. The aliphatic index refers to the relative volume occupied by the aliphatic side chains. The results have been similar to the vaccine developed by Alawam et al., against Aeromonas hydrophila [42]. It was found that almost 66.92 is occupied by alanine, valine, isoleucine, and leucine residues. The hydropathicity value indicates that the molecule is slightly hydrophilic; it has a slight tendency to interact with or be attracted to water. The antigenicity score and solubility refer to the capacity to bind specifically to binding sites and the amount of protein that dissolves in the solution, respectively. Among the six, the one with greater antigenicity (1.0553) and solubility value (0.9332) was selected for secondary structure prediction. The secondary structure of the vaccine construct has been predicted using PSIPRED and is illustrated in Figure 4. In an approach to form a multi-epitope vaccine against Pseudomonas to analyze the vaccine’s secondary structure by Dey et al., the PSIPRED web server was found to be effective [43]. The predicted (3D) model was subjected to validations. Here, the ProSA-web was utilized for analyzing Z-scores while PDBsum gave the Ramachandran plot. The Z score for the construct was found to be −6.63. This determines the overall model quality, and here groups of structures from different sources (X-ray, NMR) are distinguished by different colors. The Ramachandran plot shows the phi-psi torsion angles for all residues in the structure. About 49% of the residues fall into the most favorable regions, about 37.5% into additional allowed regions, and 13.5% are in generously allowed regions. None of the residues were in disallowed regions, indicating that the vaccine construct is stable and well-formed. A similar array of structural validation has been performed by Srijita, where they have designed an epitope-based vaccine against Clostridium difficile [44].
Table 3: Physicochemical properties of vaccine constructs.
| Vaccine construct | Total number of Amino Acids | Mol. weight (kd) | Theoretical pI | Aliphatic Index | Hydropathicity (GRAVY) | Antigenicity score | Solubility |
|---|---|---|---|---|---|---|---|
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGL PGTKCCKKPEAAKFELDDKASLGPGPGEIKPKDYPKGPGPGPYKFAPDGFGPG PGTTLGDDAKRGPGPGLPARAARTMGPGPGSLLPARAAR | 146 | 15104.80 | 9.59 | 66.92 | −0.155 | 0.9790 | 0.906174 |
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGLPG TKCCKKPEAAKEIKPKDYPKGPGPGPYKFAPDGFGPGPGTTLGDDAKRGPGP GLPARAARTMGPGPGSLLPARAARGPGPGFELDDKASL | 146 | 15104.80 | 9.59 | 66.92 | −0.155 | 1.0553 | 0.933284 |
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGLPGTK CCKKPEAAKPYKFAPDGFGPGPGTTLGDDAKRGPGPGLPARAARTMGPGPGSL LPARAARGPGPGFELDDKASLGPGPGEIKPKDYPK | 146 | 15104.80 | 9.59 | 66.92 | −0.155 | 1.0440 | 0.926247 |
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGLPGTK CCKKPEAAKTTLGDDAKRGPGPGLPARAARTMGPGPGSLLPARAARGPGPGFELDD KASLGPGPGEIKPKDYPKGPGPGPYKFAPDGF | 146 | 15104.80 | 9.59 | 66.92 | −0.155 | 0.9766 | 0.934967 |
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGLPGTKCC KKPEAAKLPARAARTMGPGPGSLLPARAARGPGPGFELDDKASLGPGPGEIKPK DYPKGPGPGPYKFAPDGFGPGPGTTLGDDAKR | 146 | 15104.80 | 9.59 | 66.92 | −0.155 | 0.9763 | 0.933050 |
| MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRKQIGTCGLPGTK CCKKPEAAKSLLPARAARGPGPGFELDDKASLGPGPGEIKPKDYPKGPGP GPYKFAPDGFGPGPGTTLGDDAKRGPGPGLPARAARTM | 146 | 15175.80 | 9.59 | 66.92 | −0.155 | 0.9997 | 0.887535 |
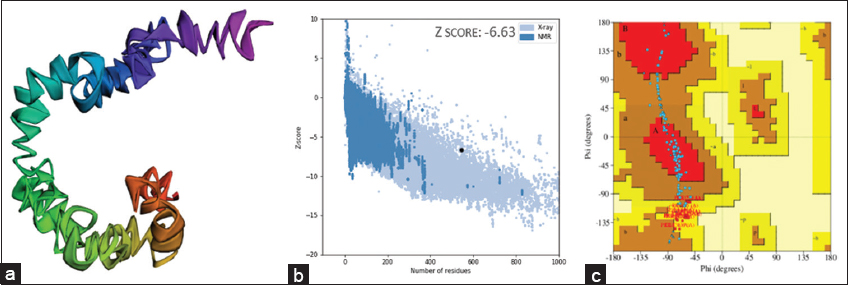 | Figure 4: (a) Predicted structure of multi-epitope-based vaccine construct, (b) Z-score graph, (c) Ramachandran plot. [Click here to view] |
The interaction of the designed vaccine construct was evaluated by docking the construct with TLR-2, TLR-3, MHC-I, and MHC-II. TLRs are known to detect microbial infection by recognizing pathogen-associated molecular patterns in microbes, leading to innate immune activation. MHC promotes antigen presentation of CD8+ and CD4+ T cells and could promote adaptive immune responses. The bond strength between receptor and vaccine was evaluated, and the one with the lowest binding affinity was selected because it provides a more stable complex, which means the ligand is more likely to remain bound to the target protein; it is even crucial for effective inhibition or activation of protein functioning, and they are most biologically relevant [45]. Among the docked complexes, TLR-2, which exhibited the lowest docking score of −450.6 kcal/mol, was selected. The docked complex of the vaccine and TLR-2 has been illustrated in Figure 5. Molecular interactions, such as hydrogen bonds, salt bridges, and hydrophobic contacts, were assessed to understand the binding strength. The conformation of the vaccine construct was also important to ensure a proper fit within the binding site. HDOCK was used for docking the TLR-2 and the construct, and it was further visualized using PyMOL. The molecular interactions were analyzed by utilizing PDBsum. Several binding interactions like hydrogen bonds, salt bridges, and non-bonding contacts were depicted in Table 4. The interactions between chains and residues are illustrated in Figure 6.
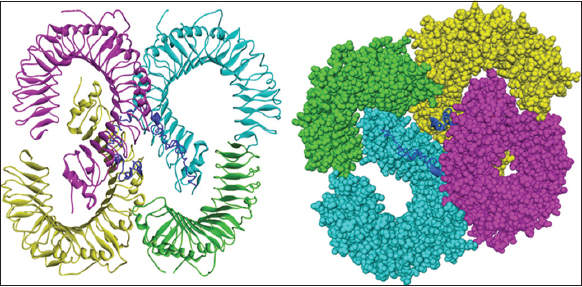 | Figure 5: Schematic diagram of docked complex (vaccine construct with toll-like receptor 2). [Click here to view] |
Table 4: No. of different bonds linked to each other.
| Chains | No. of interface residue | Interface area (A2) | No. of salt bridges | No. of hydrogen bonds | No. of non-bonded contacts |
|---|---|---|---|---|---|
| {A}{B} | 15:13 | 832:843 | 2 | 4 | 74 |
| {A}{C} | 7:9 | 516:510 | - | 1 | 31 |
| {A}{E | 16:33 | 950:909 | - | 7 | 197 |
| {B}{C} | 8:8 | 425:442 | - | 1 | 30 |
| {B}{D} | 5:6 | 283;264 | - | - | 20 |
| {B}{E} | 6:13 | 485:422 | - | - | 83 |
| {C}{D} | 12:13 | 778:762 | 2 | 5 | 69 |
| {C}{E} | 8:22 | 644:548 | - | 4 | 109 |
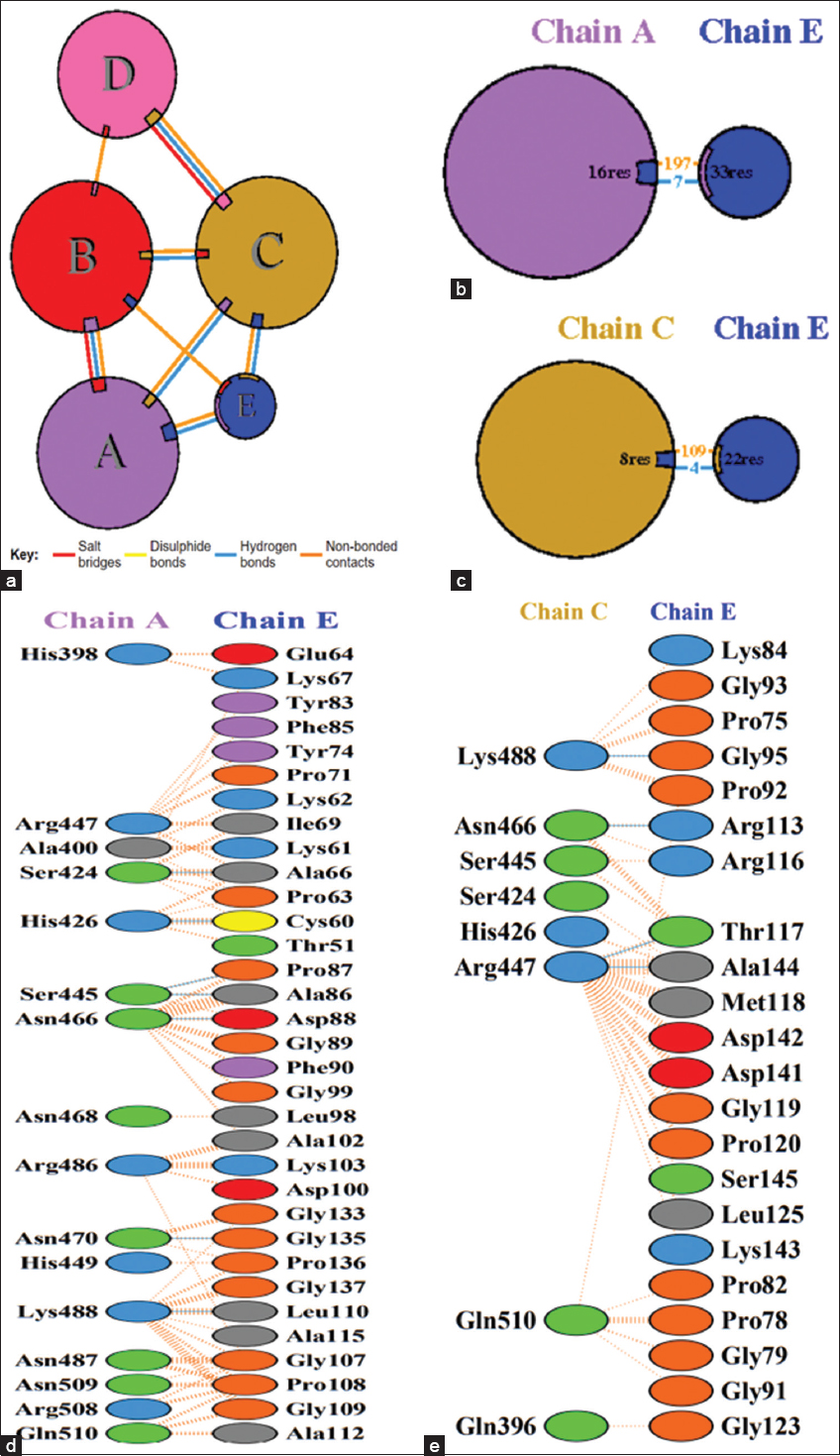 | Figure 6: Interactions between vaccine construct and receptor. (a) Bonds formed between the chains of toll-like receptor 2 and vaccine construct. (b) Chain A and vaccine construct interaction. (c) Chain C and vaccine construct interaction. (d) Interaction between the residue of chain C and the construct (e) Interaction between the residues of chain A and the construct. [Click here to view] |
Computational cloning was executed via the JCat tool and the codon optimization tool. Codon Optimization is the process of the optimization of a specific sequence of the host to obtain maximum expression in the host. It enhances protein expression by adjusting the gene sequence to match the target organism’s preferred codons. This includes replacing less favored codons, optimizing mRNA stability through UTR modifications, enhancing translation efficiency with optimal ribosome binding sites, and minimizing immune recognition by altering surface residues. The optimized gene is checked for immunogenicity and expression levels with the help of bioinformatics tools, which improve protein yield and lower immunological responses. The experiments performed in a recent work [46] elucidated how codon optimization enhances the expression of a heterologous gene in Escherichia coli. The “codon adaptation index (CAI)” and GC content values represent the effective codon usage of the model vaccine construct as shown in Figure 7. The mentioned vaccine construct was given for improved DNA sequencing, which was further used for processing. By utilizing the JCat server, the vaccine design was also subjected to a process for obtaining in silico clones in expression vector pET28a (+). This was performed after obtaining the optimal CAI score of more than 0.7 and GC content >60% for vaccine sequence (DNA). A T-cell-based multi-epitope vaccine was developed against leishmaniasis, a common parasitic infection, where in the final step, the two proposed vaccine candidates were ligated into the pET28a(+) vector for subsequent vaccine production [47]. The optimized DNA sequence for the vaccine construct was then virtually cloned into the expression pET28a (+) vector using restriction enzymes (Eco53KI and EcoRV) at their respective restriction sites. Finally, using SnapGene software, the virtually cloned vaccine with inserted optimized (DNA) sequences was created, as shown in Figure 8.
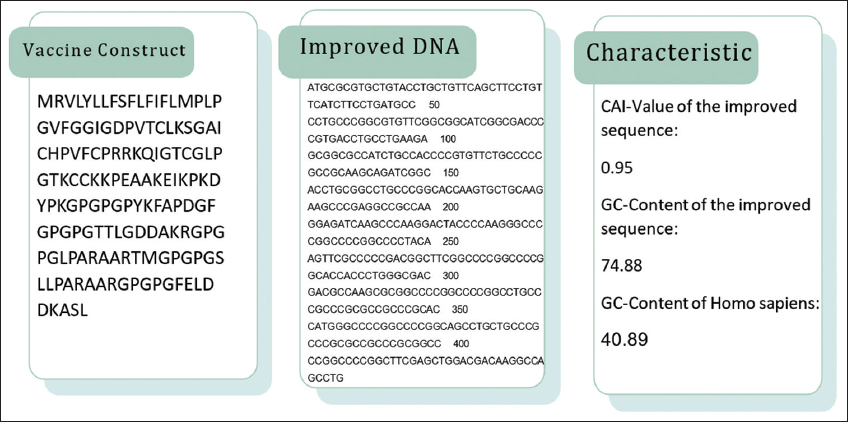 | Figure 7: Codon optimization using the JCat tool. [Click here to view] |
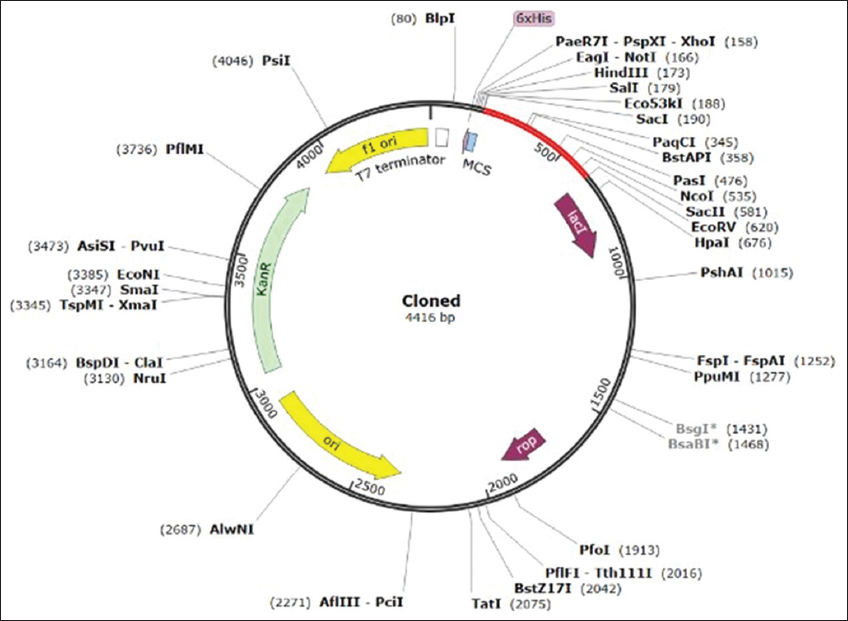 | Figure 8: Cloning of a designed vaccine into a pET28a (+) vector. The inserted DNA segment is shown in red, while the plasmid is in black. [Click here to view] |
The C-Immsim is used to analyse the interaction of vaccines with the immune system. The results demonstrated that the vaccine triggered a potent immune response. Zheng et al. ensembled the vaccine against the Pegivirus, earlier known as the Hepatitis G virus, where the vaccine formed was further validated through immune simulation [48]. Through C-Immsim, it has been analysed that the vaccine elicited strong immune response, including the highest production of Immunoglobulin (Ig)M and IgG antibodies around 15 days, followed by other Igs, indicating a strong defence against the targeted pathogen as shown in Figure 9. It activates both HTL and cytotoxic T lymphocytes to enhance cellular immunity. In addition, the vaccine is anticipated to stimulate the release of cytokines, such as interleukins and interferon-gamma, suggesting an active immune system response. While these simulation results are promising, real-world experiments are necessary to confirm the vaccine’s effectiveness against A. caviae. Thus, the study indicates that the constructed vaccine can elicit a broad range of strong immune responses [49]. Immune simulations are essential for evaluating the vaccine’s efficacy by modelling immune responses in a controlled environment. They provide information about humoral and cellular immunity by measuring antibody levels, evaluating T-cell activation, and assessing cytokine production. Furthermore, simulations forecast the development of immunological memory, which shows how effectively the immune system will react to infections in the future. Before beginning experimental trials, researchers can improve the vaccine’s design and guarantee its efficacy and safety by examining these results.
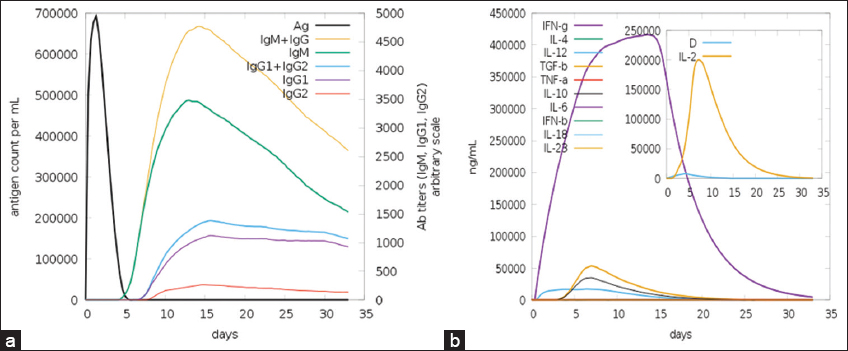 | Figure 9: Host immune system stimulation in response to the designed vaccine. (a) production of antibodies against the vaccine antigen, (b) cytokines, and interleukin concentration against the vaccine antigen. [Click here to view] |
The emergence of antimicrobial-resistant A. caviae has become a life-threatening issue globally, raising the need for developing a novel vaccine against the bacterial disease. This study focused on designing an epitope-based vaccine construct using reverse vaccinology and immunoinformatics approaches that will elicit an immune response against A. caviae. Potential antigenic epitopes were selected based on the bacterium’s known resistance to antibiotics like penicillin, carbenicillin, ampicillin, and ticarcillin, which complicate treatment options for infections. The constructed vaccine aims to provoke an active immunological response against A. caviae infections, such as osteomyelitis, soft tissue infection, endocarditis, and meningitis, addressing the urgent public health threat posed by increasing extra-intestinal infections in immunocompromised individuals and patients with severe underlying diseases. Potential antigenic epitopes against the bacteria were selected, and the construct was formulated with the aid of various bioinformatics tools, it is suggested that the vaccine construct provokes an active immunological response in the host. Furthermore, the validation of these in silico predictions can be done using wet laboratory experiments, and it is hoped that this work may help to develop the interest of biotechnologists in the field of bioinformatics and vaccine technology. Even though the study showed favorable outcomes, there are still a few methodological limitations that can be overcome in the future. The predicted results may not always match the biological outcomes, as it is difficult for computational models to capture the complexity of the immune responses in vivo. Factors such as protein folding and posttranslational modifications could also affect the immunogenicity of the predicted epitopes. This might be overcome in the future by developing advanced servers or tools with more advanced algorithms to validate the predictions. Furthermore, substantial experimental support is required for the optimal combination of epitopes in the vaccine construction. The next steps in the development of this vaccine involve several phases, including vaccine formulation using recombinant technology, purification and confirmation of the recombinant protein, and conducting animal studies. Furthermore, toxicity tests, efficacy as well and assessments for predicting humoral response in immunised mice, focusing on the measurement of antibody isotypes, mucosal responses, and cytokine responses should be included. Efficacy testing will assess the protective effect of the vaccine against A. caviae infections, while histopathological assays will evaluate tissue responses and any potential pathological changes resulting from vaccination. Ultimately, it is essential to validate the vaccine construct experimentally both in vitro and in vivo to ensure its effectiveness and safety before advancing to clinical trials.
We, the authors, acknowledge the Department of Biotechnology, SRMIST, and the facilities provided by the college for the successful completion of work.
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
There is no funding to report.
The authors report no financial or any other conflicts of interest in this work.
This study does not involve experiments on animals or human subjects.
All the data is available with the authors and shall be provided upon request.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
1. Lawrence R, Jeyakumar E. Antimicrobial resistance:A cause for global concern. BMC Proc 2013;7:S1. [CrossRef]
2. McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis 2002;34 Suppl 3:S93-106. [CrossRef]
3. Song Y, Wang LF, Zhou K, Liu S, Guo L, Ye LY, et al. Epidemiological characteristics, virulence potential, antimicrobial resistance profiles, and phylogenetic analysis of Aeromonas caviae isolated from extra-intestinal infections. Front Cell Infect Microbiol 2023;13:1084352. [CrossRef]
4. Batra P, Mathur P, Misra MC. Aeromonas spp.:An emerging nosocomial pathogen. J Lab Physicians 2016;8:1-4. [CrossRef]
5. Pereira C, Duarte J, Costa P, Braz M, Almeida A. Bacteriophages in the control of Aeromonas sp. in aquaculture systems:An integrative view. Antibiotics (Basel) 2022;11:163. [CrossRef]
6. Stabler RA, Marsden GL, Witney AA, Li Y, Bentley SD, Tang CM, et al. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology (Reading) 2005;151:2907-22. [CrossRef]
7. Solanki V, Tiwari M, Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci Rep 2019;9:5240. [CrossRef]
8. Rahmani A, Baee M, Saleki K, Moradi S, Nouri HR. Applying high throughput and comprehensive immunoinformatics approaches to design a trivalent subunit vaccine for induction of immune response against emerging human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2. J Biomol Struct Dyn 2022;40:6097-113. [CrossRef]
9. Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020:Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020;48:D517-25. [CrossRef]
10. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0:Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010;26:1608-15. [CrossRef]
11. Rawal K, Sinha R, Nath SK, Preeti P, Kumari P, Gupta S, et al. Vaxi DL:A web-based deep learning server to identify potential vaccine candidates. Comput Biol Med 2022;145:105401. [CrossRef]
12. Galanis KA, Nastou KC, Papandreou NC, Petichakis GN, Pigis DG, Iconomidou VA. Linear B-cell epitope prediction for in silico vaccine design:A performance review of methods available via command-line interface. Int J Mol Sci 2021;22:3210. [CrossRef]
13. Janahi EM, Dhasmana A, Srivastava V, Sarangi AN, Raza S, Arif JM, et al. In silico CD4+, CD8+T-cell and B-cell immunity associated immunogenic epitope prediction and HLA distribution analysis of Zika virus. EXCLI J 2017;16:63-72.
14. Ong E, He Y, Yang Z. Epitope promiscuity and population coverage of Mycobacterium tuberculosis protein antigens in current subunit vaccines under development. Infect Genet Evol 2020;80:104186. [CrossRef]
15. Karkashan A. Immunoinformatics assisted profiling of west Nile virus proteome to determine immunodominant epitopes for the development of next-generation multi-peptide vaccine. Front Immunol 2024;15:1395870. [CrossRef]
16. Doytchinova IA, Flower DR. VaxiJen:A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 2007;8:4. [CrossRef]
17. Dimitrov I, Flower DR, Doytchinova I. AllerTOP - a server for in silico prediction of allergens. BMC Bioinformatics 2013;14 Suppl 6:S4. [CrossRef]
18. Wei L, Ye X, Sakurai T, Mu Z, Wei L. ToxIBTL:Prediction of peptide toxicity based on information bottleneck and transfer learning. Bioinformatics 2022;38:1514-24. [CrossRef]
19. Lei Y, Zhao F, Shao J, Li Y, Li S, Chang H, et al. Application of built-in adjuvants for epitope-based vaccines. PeerJ 2019;6:e6185. [CrossRef]
20. Cie?lik M, Bagi?ska N, Górski A, Jo?czyk-Matysiak E. Human b-defensin 2 and Its postulated role in modulation of the immune response. Cells 2021;10:2991. [CrossRef]
21. Nguyen TL, Kim H. Immunoinformatics and computational approaches driven designing a novel vaccine candidate against Powassan virus. Sci Rep 2024;14:5999. [CrossRef]
22. Shams M, Nourmohammadi H, Majidiani H, Shariatzadeh SA, Asghari A, Fatollahzadeh M, et al. Engineering a multi-epitope vaccine candidate against Leishmania infantum using comprehensive immunoinformatics methods. Biologia (Bratisl) 2022;77:277-89. [CrossRef]
23. Ramalingam PS, Arumugam S. Reverse vaccinology and immunoinformatics approaches to design multi-epitope based vaccine against oncogenic KRAS. Med Oncol 2023;40:283. [CrossRef]
24. Buchan DW, Jones DT. The PSIPRED protein analysis workbench:20 years on. Nucleic Acids Res 2019;47:W402-7. [CrossRef]
25. Prajapat R, Marwal A, Gaur RK. Recognition of errors in the refinement and validation of three-dimensional structures of AC1 proteins of Begomovirus strains by using ProSA-web. J Viruses 2014;2014:752656. [CrossRef]
26. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck:A program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993;26:283-91. [CrossRef]
27. DeVore GR. Computing the Z score and centiles for cross-sectional analysis:A practical approach. J Ultrasound Med 2017;36:459-73. [CrossRef]
28. Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK:A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017;45:W365-73. [CrossRef]
29. Gupta AK, Khan MS, Choudhury S, Mukhopadhyay A, Sakshi, Rastogi A, et al. CoronaVR:A computational resource and analysis of epitopes and therapeutics for severe acute respiratory syndrome coronavirus-2. Front Microbiol 2020;11:1858. [CrossRef]
30. Laskowski RA. PDBsum:Summaries and analyses of PDB structures. Nucleic Acids Res 2001;29:221-2. [CrossRef]
31. Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, et al. JCat:A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 2005;33:W526-31. [CrossRef]
32. Naas AE. Optimzing the 5'-End of Coding Sequences in Recombinant mRNA to Achieve High-Level Expression in the Bacterium Escherichia coli. Trondheim:Institutt for Bioteknologi;2012.
33. Chatterjee R, Sahoo P, Mahapatra SR, Dey J, Ghosh M, Kushwaha GS, et al. Development of a conserved chimeric vaccine for induction of strong immune response against Staphylococcus aureus using immunoinformatics approaches. Vaccines (Basel) 2021;9:1038. [CrossRef]
34. Wareth G, Brandt C, Sprague LD, Neubauer H, Pletz MW. WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective. BMC Microbiol 2021;21:210. [CrossRef]
35. Sabikhi Y, Surabhi, Dixit P, Sharma T, Sharma P, Rai M, et al. N-Acetylmuramoyl-L-Alanine Amidase:A Competent Vaccine Candidate against. United States:IPSID;2022. [CrossRef]
36. Alzarea SI. Identification and construction of a multi-epitopes vaccine design against Klebsiella aerogenes:Molecular modeling study. Sci Rep 2022;12:14402. [CrossRef]
37. Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res 2000;60:5228-36.
38. Aldakheel FM, Abrar A, Munir S, Aslam S, Allemailem KS, Khurshid M, et al. Proteome-wide mapping and reverse vaccinology approaches to design a multi-epitope vaccine against Clostridium perfringens. Vaccines (Basel) 2021;9:1079. [CrossRef]
39. Khan S, Irfan M, Hameed AR, Ullah A, Abideen SA, Ahmad S, et al. Vaccinomics to design a multi-epitope-based vaccine against monkeypox virus using surface-associated proteins. J Biomol Struct Dyn 2023;41:10859-68. [CrossRef]
40. Asif Rasheed M, Awais M, Aldhahrani A, Althobaiti F, Alhazmi A, Sattar S, et al. Designing a highly immunogenic multi epitope based subunit vaccine against Bacillus cereus. Saudi J Biol Sci 2021;28:4859-66. [CrossRef]
41. Naveed M, Hassan JU, Ahmad M, Naeem N, Mughal MS, Rabaan AA, et al. Designing mRNA- and peptide-based vaccine construct against emerging multidrug-resistant Citrobacter freundii:A computational-based subtractive proteomics approach. Medicina (Kaunas) 2022;58:1356. [CrossRef]
42. Alawam AS, Alwethaynani MS. Construction of an aerolysin-based multi-epitope vaccine against Aeromonas hydrophila:An in silico machine learning and artificial intelligence-supported approach. Front Immunol 2024;15:1369890. [CrossRef]
43. Dey J, Mahapatra SR, Patnaik S, Lata S, Kushwaha GS, Panda RK, et al. Molecular characterization and designing of a novel multiepitope vaccine construct against Pseudomonas aeruginosa. Int J Pept Res Ther 2022;28:49. [CrossRef]
44. Basak S, Deb D, Narsaria U, Kar T, Castiglione F, Sanyal I, et al. In silico designing of vaccine candidate against Clostridium difficile. Sci Rep 2021;11:14215. [CrossRef]
45. Kumar P, Kumar P, Shrivastava A, Dar MA, Lokhande KB, Singh N, et al. Immunoinformatics-based multi-epitope containing fused polypeptide vaccine design against visceral leishmaniasis with high immunogenicity and TLR binding. Int J Biol Macromol 2023;253:127567. [CrossRef]
46. Tian J, Yan Y, Yue Q, Liu X, Chu X, Wu N, et al. Predicting synonymous codon usage and optimizing the heterologous gene for expression in E. coli. Sci Rep 2017;7:9926. [CrossRef]
47. Basmenj ER, Arastonejad M, Mamizadeh M, Alem M, KhalatbariLimaki M, Ghiabi S, et al. Engineering and design of promising T-cell-based multi-epitope vaccine candidates against leishmaniasis. Sci Rep 2023;13:19421. [CrossRef]
48. Zheng B, Suleman M, Zafar Z, Ali SS, Nasir SN, Namra, et al. Towards an ensemble vaccine against the pegivirus using computational modelling approaches and its validation through in silico cloning and immune simulation. Vaccines (Basel) 2021;9:818. [CrossRef]
49. Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine 2015;33:5271-81. [CrossRef]
1. Lawrence R, Jeyakumar E. Antimicrobial resistance: A cause for global concern. BMC Proc 2013;7:S1. https://doi.org/10.1186/1753-6561-7-S3-S1
2. McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis 2002;34 Suppl 3:S93-106. https://doi.org/10.1086/340246
3. Song Y, Wang LF, Zhou K, Liu S, Guo L, Ye LY, et al. Epidemiological characteristics, virulence potential, antimicrobial resistance profiles, and phylogenetic analysis of Aeromonas caviae isolated from extra-intestinal infections. Front Cell Infect Microbiol 2023;13:1084352. https://doi.org/10.3389/fcimb.2023.1084352
4. Batra P, Mathur P, Misra MC. Aeromonas spp.: An emerging nosocomial pathogen. J Lab Physicians 2016;8:1-4. https://doi.org/10.4103/0974-2727.176234
5. Pereira C, Duarte J, Costa P, Braz M, Almeida A. Bacteriophages in the control of Aeromonas sp. in aquaculture systems: An integrative view. Antibiotics (Basel) 2022;11:163. https://doi.org/10.3390/antibiotics11020163
6. Stabler RA, Marsden GL, Witney AA, Li Y, Bentley SD, Tang CM, et al. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology (Reading) 2005;151:2907-22. https://doi.org/10.1099/mic.0.28099-0
7. Solanki V, Tiwari M, Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci Rep 2019;9:5240. https://doi.org/10.1038/s41598-019-41496-4
8. Rahmani A, Baee M, Saleki K, Moradi S, Nouri HR. Applying high throughput and comprehensive immunoinformatics approaches to design a trivalent subunit vaccine for induction of immune response against emerging human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2. J Biomol Struct Dyn 2022;40:6097-113. https://doi.org/10.1080/07391102.2021.1876774
9. Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020;48:D517-25. https://doi.org/10.1093/nar/gkz935
10. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010;26:1608-15. https://doi.org/10.1093/bioinformatics/btq249
11. Rawal K, Sinha R, Nath SK, Preeti P, Kumari P, Gupta S, et al. Vaxi DL: A web-based deep learning server to identify potential vaccine candidates. Comput Biol Med 2022;145:105401. https://doi.org/10.1016/j.compbiomed.2022.105401
12. Galanis KA, Nastou KC, Papandreou NC, Petichakis GN, Pigis DG, Iconomidou VA. Linear B-cell epitope prediction for in silico vaccine design: A performance review of methods available via command-line interface. Int J Mol Sci 2021;22:3210. https://doi.org/10.3390/ijms22063210
13. Janahi EM, Dhasmana A, Srivastava V, Sarangi AN, Raza S, Arif JM, et al. In silico CD4+, CD8+ T-cell and B-cell immunity associated immunogenic epitope prediction and HLA distribution analysis of Zika virus. EXCLI J 2017;16:63-72.
14. Ong E, He Y, Yang Z. Epitope promiscuity and population coverage of Mycobacterium tuberculosis protein antigens in current subunit vaccines under development. Infect Genet Evol 2020;80:104186. https://doi.org/10.1016/j.meegid.2020.104186
15. Karkashan A. Immunoinformatics assisted profiling of west Nile virus proteome to determine immunodominant epitopes for the development of next-generation multi-peptide vaccine. Front Immunol 2024;15:1395870. https://doi.org/10.3389/fimmu.2024.1395870
16. Doytchinova IA, Flower DR. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 2007;8:4. https://doi.org/10.1186/1471-2105-8-4
17. Dimitrov I, Flower DR, Doytchinova I. AllerTOP - a server for in silico prediction of allergens. BMC Bioinformatics 2013;14 Suppl 6:S4. https://doi.org/10.1186/1471-2105-14-S6-S4
18. Wei L, Ye X, Sakurai T, Mu Z, Wei L. ToxIBTL: Prediction of peptide toxicity based on information bottleneck and transfer learning. Bioinformatics 2022;38:1514-24. https://doi.org/10.1093/bioinformatics/btac006
19. Lei Y, Zhao F, Shao J, Li Y, Li S, Chang H, et al. Application of built-in adjuvants for epitope-based vaccines. PeerJ 2019;6:e6185. https://doi.org/10.7717/peerj.6185
20. Cie?lik M, Bagi?ska N, Górski A, Jo?czyk-Matysiak E. Human β-defensin 2 and Its postulated role in modulation of the immune response. Cells 2021;10:2991. https://doi.org/10.3390/cells10112991
21. Nguyen TL, Kim H. Immunoinformatics and computational approaches driven designing a novel vaccine candidate against Powassan virus. Sci Rep 2024;14:5999. https://doi.org/10.1038/s41598-024-56554-9
22. Shams M, Nourmohammadi H, Majidiani H, Shariatzadeh SA, Asghari A, Fatollahzadeh M, et al. Engineering a multi-epitope vaccine candidate against Leishmania infantum using comprehensive immunoinformatics methods. Biologia (Bratisl) 2022;77:277-89. https://doi.org/10.1007/s11756-021-00934-3
23. Ramalingam PS, Arumugam S. Reverse vaccinology and immunoinformatics approaches to design multi-epitope based vaccine against oncogenic KRAS. Med Oncol 2023;40:283. https://doi.org/10.1007/s12032-023-02160-0
24. Buchan DW, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 2019;47:W402-7. https://doi.org/10.1093/nar/gkz297
25. Prajapat R, Marwal A, Gaur RK. Recognition of errors in the refinement and validation of three-dimensional structures of AC1 proteins of Begomovirus strains by using ProSA-web. J Viruses 2014;2014:752656. https://doi.org/10.1155/2014/752656
26. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck: A program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993;26:283-91. https://doi.org/10.1107/S0021889892009944
27. DeVore GR. Computing the Z score and centiles for cross-sectional analysis: A practical approach. J Ultrasound Med 2017;36:459-73. https://doi.org/10.7863/ultra.16.03025
28. Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017;45:W365-73. https://doi.org/10.1093/nar/gkx407
29. Gupta AK, Khan MS, Choudhury S, Mukhopadhyay A, Sakshi, Rastogi A, et al. CoronaVR: A computational resource and analysis of epitopes and therapeutics for severe acute respiratory syndrome coronavirus-2. Front Microbiol 2020;11:1858. https://doi.org/10.3389/fmicb.2020.01858
30. Laskowski RA. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res 2001;29:221-2. https://doi.org/10.1093/nar/29.1.221
31. Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, et al. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 2005;33:W526-31. https://doi.org/10.1093/nar/gki376
32. Naas AE. Optimzing the 5’-End of Coding Sequences in Recombinant mRNA to Achieve High-Level Expression in the Bacterium Escherichia coli. Trondheim: Institutt for Bioteknologi; 2012.
33. Chatterjee R, Sahoo P, Mahapatra SR, Dey J, Ghosh M, Kushwaha GS, et al. Development of a conserved chimeric vaccine for induction of strong immune response against Staphylococcus aureus using immunoinformatics approaches. Vaccines (Basel) 2021;9:1038. https://doi.org/10.3390/vaccines9091038
34. Wareth G, Brandt C, Sprague LD, Neubauer H, Pletz MW. WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective. BMC Microbiol 2021;21:210. https://doi.org/10.1186/s12866-021-02270-7
35. Sabikhi Y, Surabhi, Dixit P, Sharma T, Sharma P, Rai M, et al. N-Acetylmuramoyl-L-Alanine Amidase: A Competent Vaccine Candidate against. United States: IPSID; 2022. https://doi.org/10.31219/osf.io/958gs
36. Alzarea SI. Identification and construction of a multi-epitopes vaccine design against Klebsiella aerogenes: Molecular modeling study. Sci Rep 2022;12:14402. https://doi.org/10.1038/s41598-022-18610-0
37. Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res 2000;60:5228-36.
38. Aldakheel FM, Abrar A, Munir S, Aslam S, Allemailem KS, Khurshid M, et al. Proteome-wide mapping and reverse vaccinology approaches to design a multi-epitope vaccine against Clostridium perfringens. Vaccines (Basel) 2021;9:1079. https://doi.org/10.3390/vaccines9101079
39. Khan S, Irfan M, Hameed AR, Ullah A, Abideen SA, Ahmad S, et al. Vaccinomics to design a multi-epitope-based vaccine against monkeypox virus using surface-associated proteins. J Biomol Struct Dyn 2023;41:10859-68. https://doi.org/10.1080/07391102.2022.2158942
40. Asif Rasheed M, Awais M, Aldhahrani A, Althobaiti F, Alhazmi A, Sattar S, et al. Designing a highly immunogenic multi epitope based subunit vaccine against Bacillus cereus. Saudi J Biol Sci 2021;28:4859-66. https://doi.org/10.1016/j.sjbs.2021.06.082
41. Naveed M, Hassan JU, Ahmad M, Naeem N, Mughal MS, Rabaan AA, et al. Designing mRNA- and peptide-based vaccine construct against emerging multidrug-resistant Citrobacter freundii: A computational-based subtractive proteomics approach. Medicina (Kaunas) 2022;58:1356. https://doi.org/10.3390/medicina58101356
42. Alawam AS, Alwethaynani MS. Construction of an aerolysin-based multi-epitope vaccine against Aeromonas hydrophila: An in silico machine learning and artificial intelligence-supported approach. Front Immunol 2024;15:1369890. https://doi.org/10.3389/fimmu.2024.1369890
43. Dey J, Mahapatra SR, Patnaik S, Lata S, Kushwaha GS, Panda RK, et al. Molecular characterization and designing of a novel multiepitope vaccine construct against Pseudomonas aeruginosa. Int J Pept Res Ther 2022;28:49. https://doi.org/10.1007/s10989-021-10356-z
44. Basak S, Deb D, Narsaria U, Kar T, Castiglione F, Sanyal I, et al. In silico designing of vaccine candidate against Clostridium difficile. Sci Rep 2021;11:14215. https://doi.org/10.1038/s41598-021-93305-6
45. Kumar P, Kumar P, Shrivastava A, Dar MA, Lokhande KB, Singh N, et al. Immunoinformatics-based multi-epitope containing fused polypeptide vaccine design against visceral leishmaniasis with high immunogenicity and TLR binding. Int J Biol Macromol 2023;253:127567. https://doi.org/10.1016/j.ijbiomac.2023.127567
46. Tian J, Yan Y, Yue Q, Liu X, Chu X, Wu N, et al. Predicting synonymous codon usage and optimizing the heterologous gene for expression in E. coli. Sci Rep 2017;7:9926. https://doi.org/10.1038/s41598-017-10546-0
47. Basmenj ER, Arastonejad M, Mamizadeh M, Alem M, KhalatbariLimaki M, Ghiabi S, et al. Engineering and design of promising T-cell-based multi-epitope vaccine candidates against leishmaniasis. Sci Rep 2023;13:19421. https://doi.org/10.1038/s41598-023-46408-1
48. Zheng B, Suleman M, Zafar Z, Ali SS, Nasir SN, Namra, et al. Towards an ensemble vaccine against the pegivirus using computational modelling approaches and its validation through in silico cloning and immune simulation. Vaccines (Basel) 2021;9:818. https://doi.org/10.3390/vaccines9080818
49. Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine 2015;33:5271-81. https://doi.org/10.1016/j.vaccine.2015.06.117
Year
Month