1. INTRODUCTION
Rice (Oryza sativa L.) is a member of the Poaceae family, a monocotyledonous plant with a diploid genome (2n = 24). As one of the world's most important staple crops, rice has been at the forefront of modern crop development efforts. The Green Revolution in the 1960s marked a significant milestone in rice breeding [1]. Since then, advances in molecular biology and genetics, especially the advent of new breeding techniques have further accelerated rice improvement programs.
To determine the function of a gene in a plant genome, researchers have traditionally relied on gene overexpression and/or repression strategies [2]. However, these conventional methods, based on gene transformation, often involve random integration of gene constructs, leading to unpredictable results [3]. The advent of gene editing technologies has offered a more precise approach by enabling targeted insertion or modification of regulatory elements at specific genomic locations.
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) has revolutionized the field of gene editing, providing researchers with a powerful tool for precise genetic modifications. The pivotal breakthrough came in 2012 when Emmanuelle Charpentier and Jennifer Doudna demonstrated that the CRISPR/Cas9 system could be programmed to cut specific DNA sequences, laying the foundation for its use as a genome editing tool [4], a discovery that was later awarded the Nobel Prize in Chemistry in 2020. This technology has proven invaluable for studying gene function and enhancing crop traits.
While CRISPR/Cas9 has been widely adopted for gene knockouts, its application in gene upregulation through homology-directed repair (HDR) is still evolving, particularly in rice [5]. HDR-based gene editing requires the simultaneous delivery of both the CRISPR/Cas9 expression system and the DNA repair template into the cell. This process can be achieved through various methods, including Agrobacterium-mediated transformation, particle bombardment, viral vectors, and protoplast transformation. Among these, Agrobacterium-mediated gene transfer remains the most popular method in plants due to several advantages such as simplicity, high efficiency, low copy number integration, wide host range, and capacity to transfer large DNA fragments [6]. Consequently, researchers are actively developing strategies to optimize HDR-based gene editing methods, with a particular focus on improvements to Agrobacterium-mediated delivery systems [7]. This study introduces a novel single-vector system that combines the CRISPR/Cas9 expression cassette and donor DNA template for HDR. This approach aims to simplify delivery and potentially increase the efficiency of precise genetic modifications.
The natural resistance-associated macrophage protein (NRAMP) gene family in rice plays crucial roles in metal ion transport. While several NRAMP genes have been characterized in rice, the specific function of OsNRAMP7 in metal transport remains unclear. This gene has not been previously studied to elucidate its role in metal homeostasis in rice, making it an intriguing target for functional genomics research [8]. Furthermore, TBR225, a major commercial rice variety in Northern Vietnam, is known for its high yield, good quality, and early maturity. While TBR225 has been successfully gene-edited using NHEJ, no CRISPR/Cas9-mediated HDR has been reported for this variety [9,10]. Applying HDR-mediated gene editing to TBR225 faces several challenges common to rice gene editing. First, indica rice varieties like TBR225 have lower gene transfer efficiency compared to japonica varieties [10,11]. Second, HDR efficiency is very low and varies significantly between different rice varieties [5], necessitating tailored approaches for TBR225. Third, gene transfer methods are limited for TBR225, with only Agrobacterium-mediated transformation reported successful [10,11], while other high-efficiency methods have not yet been published. Therefore, there is difficulty in applying traditional multi-vector gene editing systems to hard-to-transform commercial rice varieties like TBR225.
Our research focuses on three main objectives: (i) Design a single vector system for CRISPR/Cas9-mediated HDR in rice; (ii) Demonstrate HDR for precise insertion of a constitutive promoter to enhance gene expression; (iii) Advance precision genetic modifications in the TBR225 cultivar. By targeting OsNRAMP7, we aim to showcase the potential of our approach in studying previously uncharacterized genes. The insertion of a constitutive promoter upstream of OsNRAMP7 could lead to its overexpression, potentially shedding light on its function in metal transport and homeostasis in rice. This research demonstrates the broader applicability of our HDR-based gene editing approach for functional genomics studies and genetic improvements in rice and other crop plants.
2. MATERIALS AND METHODS
2.1. Plant Growth Conditions
The rice cultivar TBR225 [9] was obtained from ThaibinhSeed Cor. (Vietnam). Rice plants were cultivated in a controlled environment net-house under the following conditions: temperature range of 28°C–32°C and relative humidity of 80%–85%.
2.2. Bacterial Strains and Culture Conditions
Escherichia coli TOP10 and Agrobacterium tumefaciens EHA105 strains were provided by the Department of Molecular Pathology, Agricultural Genetics Institute (Vietnam). E. coli TOP10 was cultured in Luria-Bertani (LB) medium at 37°C, while A. tumefaciens EHA105 was cultured in LB medium at 28°C. Both media were supplemented with appropriate antibiotics as per strain requirements.
2.3. gRNA Design
A fragment of OsNRAMP7 was amplified by polymerase chain reaction (PCR) from TBR225 rice genomic DNA using specific primers (NRAMP7-F: 5′-TGTTGCCTAGTGGAGTCGTG-3′ and NRAMP7-R: 5′-TGAACAGCCACAGCTTCCTC-3′) (Fig. 1a). PCR conditions were 35 cycles of 94°C/30, 55°C/30, and 72°C/40 seconds. The amplified fragment was sequenced, and gRNA target sequences for editing TBR225 OsNRAMP7 were designed based on this sequence using three bioinformatics tools: CRISPR-P v2.0 [12] for sequence and GC content determination, Mfold v2.3 [13] for secondary structure analysis, and CCTop [14] for off-target score identification.
2.4. Vector Construction
Vector construction (Fig. 1b) was performed using complementary oligonucleotides gRNA-F (5′-gttGCCATGGATTTGGAGATGGG-3′) and gRNA-R (5′-aaaCCCATCTCCAAATCCATGGC-3′) with 3-bp overhangs, synthesized by Phusa Biochem (Vietnam). These oligonucleotides were heat-denatured, annealed, phosphorylated, and cloned into the LguI site downstream of the OsU6 promoter on a modified sgRNA expression vector [15], creating the recombinant vector pEN-gRNA. A donor DNA template fragment, previously generated in the cloning vector pJET1.2/35S by Nanning GenSys Biotechnology (China), was excised and ligated into both NotI and BamHI sites on the pEN-gRNA vector. All inserted fragments were sequence-verified. The donor DNA template fragments and sgRNA expression cassette were then integrated into the gateway destination vector pCas9 [16] using Gateway LR clonase (Thermo Fisher Scientific). The resulting final construct, pCas9/gRNA-35Sx2, was validated by Sanger sequencing of the insertion junctions.
2.5. Rice Transformation
The pCas9/gRNA-35Sx2 vector was transformed into A. tumefaciens EHA105 via heat shock, and the resulting transformant was used for TBR225 rice transformation as described previously [11]. Briefly, Agrobacterium-infected calli were selected using Hygromycin-containing medium, followed by transfer to a regeneration medium to enable transgenic plant growth. Transgenic lines were screened from all regenerated plants through PCR using primer pairs Ubi-F/Cas9-R (5′-CCCTGCCTTCATACGCTATT-3′/5′-GCCTCGGCTGTCTCGCCA-3′) and HPT-F/HPT-R (5′-AAACTGTGATGGACGACACCGT-3′/5′-GTGGCGATCCTGCAAGCTCC-3′) specific for Cas9 and HPT marker gene, respectively. Transgene-free plants were selected by PCR with primer pairs Ubi-F/ Cas9-R, U6-F/gRNA-R, and HPT-F/HPT-R. OsActin was used as a control of the PCR experiment.
2.6. Mutation Analysis
To determine the insertion of the 35S promoter at the target site, two specific primer pairs were designed: NRAMP7-t-F (5′-GGCAGTAGTACAAGTGAGAGGA-3′)/NRAMP7-t-R (5′-GGGGAAGTCCGAGATCGAG-3′) and 35S-F (5′-CCCACTATCCTTCGCAA-3′)/NRAMP7-t-R. These primers amplified the genomic area containing the CRISPR/Cas9 target site and a part of the 35S promoter. Genomic DNA was extracted from transgenic T0 plant leaves using the CTAB method. PCR was conducted using 50 ng of extracted DNA as a template, with the two diagnostic primer pairs and a positive control pair (5′-AGGCGCTGCTCAACTTCCCG-3′ and 5′-CACGCACTCACCGTCGCTCA-3′) specific to OsEF1α. The amplified products from NRAMP7-t-F/NRAMP7-t-R primers were directly sequenced using the Sanger method. Sequences obtained from wild-type and transgenic plants were compared using CRISP-ID v1.1 8 to identify mutations [17]. The amplicon resulting from line #33 was cloned into pGEM-T for sequencing. The terms "heterozygous mutation" and "homozygous mutation" were used to describe transgenic plants containing 35S insertion mutations on one and both alleles, respectively.
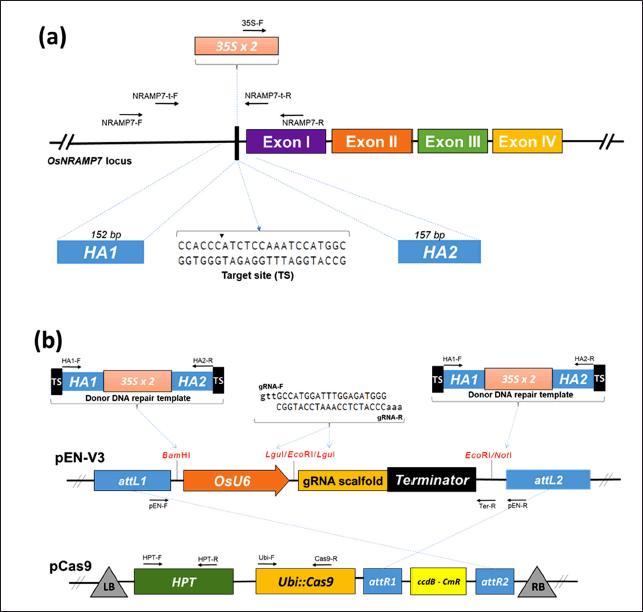 | Figure 1. CRISPR/Cas9-mediated HDR strategy for inserting the 35S promoter into the OsNRAMP7 locus of TBR225 rice. (a) Genomic structure of OsNRAMP7, showing exons and the CRISPR/Cas9 target site. The homology arms (HA1 and HA2) flanking the target site are indicated, along with their lengths and the target site sequence. (b) Schematic representation of the HDR strategy and T-DNA constructs. Donor DNA template contains the 35S promoter with a duplicated enhancer (35S x 2) flanked by homology arms (HA1 and HA2). pEN-Y3 vector carries the OsU6 promoter-driven gRNA scaffold and the donor DNA template. pCas9 vector contains the Cas9 expression cassette driven by the ZmUbiquitin promoter (Ubi::Cas9) and the hygromycin phosphotransferase (HPT) selection marker. HA: homology arm; TS: CRISPR/Cas9 target site; attL/attR: Gateway recombination sites; HPT: hygromycin phosphotransferase; ccdB-CmR: bacterial counterselection markers; LB: left border; RB: right border; 35S: cauli?ower mosaic virus 35S promoter. Arrows indicate primer positions for PCR analysis. [Click here to view] |
2.7. Gene Expression Analysis
Total RNA was isolated from rice leaves at the 3-leaf stage for gene expression analysis using GeneJET Plant RNA Purification Kit from Thermo Scientifics. The purity and concentration of extracted RNA samples were checked by UV-Vis Spectrometers (Thermo Scientifics); RNA integrity was assessed on 1.0% agarose gel. Using an oligo (dT) primer, one μg of RNA was reverse-transcribed, followed by PCR amplification with OsNRAMP7-specific primers (RT-NRMAP7-F: 5′-CGATGTCCTGAACGAGTGGC-3′ and RT-NRMAP7-R: 5′-GAAGGACTTGCGGATGGTGG-3′). PCR was performed for 33 cycles on an Eppendorf Mastercycler ep Gradient S. Target gene expression was normalized to OsEF1α [10]. PCR products were visualized by 1.0% agarose gel electrophoresis, and the resulting images were analyzed using ImageJ v1.1 software. The OsNRAMP7 band intensity was normalized to the corresponding OsEF1α band from the same cDNA sample.
3. RESULTS
3.1. Design of a Single CRISPR/Cas9 Vector System for HDR-Mediated Insertion of 35S Into TBR225 OsNRAMP7
A single vector system was designed to insert the 35S constitutive promoter into the TBR225 genome for OsNRAMP7 overexpression. This system comprises three key components: (i) a gRNA expression construct targeting the site (target site - TS) upstream of OsNRAMP7's open reading frame, (ii) a Cas9 protein expression construct driven by the ZmUbiquitin promoter on the pCas9 vector, (iii) a donor DNA repair template (NRAMP7-35S) containing the 35S promoter flanked by two homology arms and TS sequences at both the ends of fragment (Fig. 1b). This design enables simple delivery of the entire gene editing system via Agrobacterium-mediated transformation. The CRISPR/Cas9 complex creates double-strand breaks at the TS and cleaves the NRAMP7-35S fragments, which serve as donor repair templates. This process inserts the 35S promoter upstream of OsNRAMP7 around TS via the HDR mechanism, resulting in a recombinant 35S::OsNRAMP7 construct in the TBR225 genome.
3.2. Validation of Cloning Vectors
To create the TBR225 OsNRAMP7-targeting gRNA expression cassette (Fig. 1b), we treated the pEN-V3 vector system with LguI before ligating it to a 21-bp gRNA fragment. To confirm the successful insertion of gRNA, we subjected the resulting plasmid (pEN-V3/gRNA) to PCR using gRNA- and vector-specific primer pairs (Fig. 2a). We then inserted a 1.2 kb donor DNA template, excised from the cloning vector pJET1.2, into both NotI and BamHI sites of the pEN-V3/gRNA vector (Fig. 1b). We validated these recombinant vectors by restriction digestion (Fig. 2b and c). Next, we combined the recombinant construct (containing the gRNA expression cassette and two donor DNA template fragments) with the Cas9 expression cassette on the pCas9 vector via Gateway cloning reaction. Finally, we verified the presence of all required cassettes - including gRNA, Cas9, and marker gene (hygromycin phosphotransferase - HPT) expression constructs, as well as the donor DNA template - on the recombinant pCas9 vector through PCR and sequencing (Fig. 2d). The resulting vector was then used for further TBR225 rice transformation via Agrobacterium.
3.3. Generation of Transgenic TBR225 Rice Expressing CRISPR/Cas9 for OsNRAMP7 Targeting
We transformed the T-DNA containing the OsNRAMP7-targeting gRNA expression cassette and donor DNA template fragment into A. tumefaciens strain EHA105. Embryogenic calli derived from mature TBR225 embryos were then inoculated and co-cultured with the transformed Agrobacteria. We screened for transgenic T0 individuals using hygromycin selection, leveraging the HPT marker gene present on the foreign vector. The results of the Agrobacterium-mediated transformation of TBR225 rice are shown in Figure 3. The transformation efficiency of TBR225 (Table 1) reached 33.56% for resistant callus production and 4.86% for resistant plant regeneration. We obtained a total of 415 independent regenerated shoots, all of which exhibited normal root system development after 30 days of growth on the rooting medium.
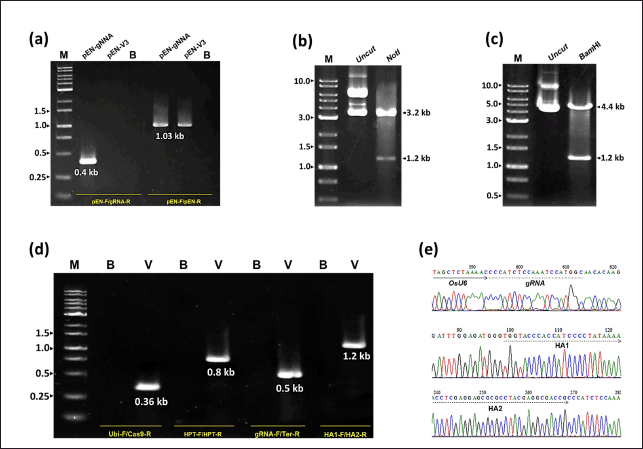 | Figure 2. Validation of recombinant vectors. (a) PCR confirmation of gRNA sequence insertion into pEN-V3 vector. (b) Restriction digestion analysis of pEN-gRNA plasmid with the first donor DNA template fragment inserted. (c) Restriction digestion analysis of pEN-gRNA plasmid with the second donor DNA template fragment inserted. (d) PCR confirmation of pCas9 vector recombined with gRNA expression cassette and two donor DNA template fragments. (e) Sequencing confirmation of pCas9 vector recombined with gRNA expression cassette and two donor DNA template fragments. M: 1.0 kb DNA ladder; B: Blank control; V: Tested plasmid (PCR template); HA: Homology arm. [Click here to view] |
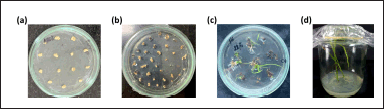 | Figure 3. Stages of TBR225 rice transformation. (a) Callus formation: Rice calli on co-cultivation medium after Agrobacterium infection. (b) Selection: Transformed calli on selection medium containing 50 mg/l hygromycin, eliminating non-transformed tissue. (c) Shoot regeneration: Initial emergence of shoots from successfully transformed and selected calli. (d) Plantlet development: regenerated plantlets in glass tubes for shoot elongation and root formation. [Click here to view] |
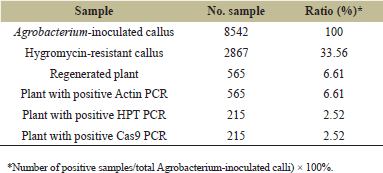 | Table 1. Result of TBR225 rice transformation. [Click here to view] |
We isolated genomic DNA from regenerated T0 plants for PCR analysis using primer pairs specific for the internal control gene OsActin, Ubiquitin::Cas9 cassette, and HPT marker gene. Our analysis revealed that 215 transformed plants contained the amplicon fragment of targeted T-DNA regions, with sizes coinciding with that of the positive control. This indicated the successful integration of the foreign T-DNA into the TBR225 genome. The selection efficiency of the antibiotic was approximately 38.05%. This finding suggests that maintaining antibiotics in the regeneration medium, although potentially reducing regeneration efficiency, ensures selective efficiency.
3.4. Insertion of 35S Promoter at Target Site
Mutation genotyping of the transgenic lines was performed using two primer pairs: NRAMP7-t-F/NRAMP7-t-R for the ~0.4-kb endogenous OsNRAMP7 fragment; 35S-F/NRAMP7-t-R for the ~0.3-kb recombinant fragment containing part of the 35S promoter and OsNRAMP7 (Fig. 4b). A ~1.2-kb recombinant OsNRAMP7 fragment was amplified in 6 lines (#33, #35, #36, #44, #52, #55), indicating exogenous DNA insertion into the TBR225 genome. The 35S-F/NRAMP7-t-R primer pair amplified the specific ~0.3 kb recombinant fragment in 5 of these 6 lines (#33, #35, #44, #52, #55), confirming correct 35S promoter insertion. In contrast, line #36 showed the amplicon with 35S-F/NRAMP7-t-F (Fig. 4b) but not 35S-F/NRAMP7-t-R, suggesting an inverse insertion of 35S promoter. Among the 6 mutant lines, 5 (#35, #36, #44, #52, #55) exhibited heterozygous insertion (two amplicons of ~0.4 and ~1.2 kb in size), while line #33 showed homozygous insertion (a unique ~1.2-kb amplicon). Sequencing of PCR products confirmed the presence of the 35S promoter upstream of the OsNRAMP7 ORF in this mutant line (Fig. 4c). These results demonstrate successful HDR-mediated insertion mutation using a combination of CRISPR/Cas9 expression construct and DNA donor template within a single transformation vector.
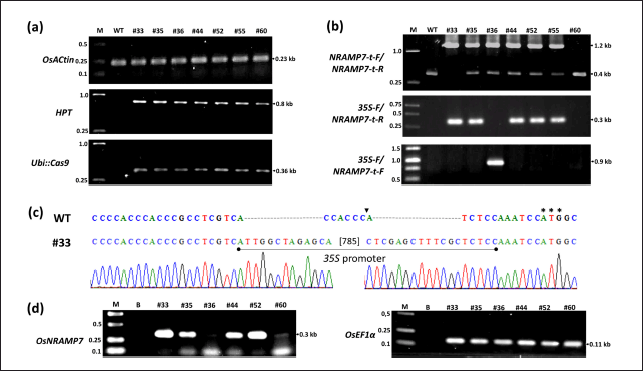 | Figure 4. Molecular characterization of T0 transgenic TBR225 rice lines. (a) PCR analysis of transgene integration. Top: OsActin (endogenous control); middle: HPT (selectable marker gene); bottom: Ubi::Cas9 cassette. (b) 35S promoter insertion detection. PCR analysis using three specific primer pairs to confirm 35S promoter integration. (c) CRISPR/Cas9 editing confirmation. Sequence alignment of transgenic line #33 and wild-type (WT) around the target site. Arrow: intended CRISPR/Cas9 target site; solid line: 35S promoter sequence; dashes (-): gaps in the alignment; asterisks (***): start codon (ATG). (d) OsNRAMP7 expression analysis. RT-PCR results for edited lines (#33, #35, #36, #44, #52) and non-edited line (#60). OsEF1α: internal control. M: 1.0 kb ladder, B: blank control; WT: wild-type plant. Primer sequences used for all analyses are provided in Table S1. [Click here to view] |
 | Table S1. Sequence of oligonucleotides used in the research. [Click here to view] |
3.5. Expression Analysis of 35S-Inserted OsNRAMP7 Gene
To evaluate the effectiveness of CRISPR/Cas9-induced mutation, we analyzed the expression of the recombinant OsNRAMP7 gene in six T0 edited lines using RT-PCR. We included two control lines: TBR225 transgenic line (#60) without insertion mutation and an edited line (#35) carrying an inverse 35S promoter insertion. The RT-PCR results revealed that all edited lines with correct 35S promoter insertion exhibited significantly higher OsNRAMP7 expression levels compared to the control line #60 (Fig. 4d). Notably, there was no significant difference in OsNRAMP7 mRNA expression between lines #60 (no insertion) and #35 (predicted inverse insertion). These findings suggested that the CRISPR/Cas9-induced insertion of the 35S constitutive promoter at the 5′-UTR site successfully increased the transcription of the target gene OsNRAMP7. This demonstrates the effectiveness of our gene editing approach in enhancing OsNRAMP7 expression in the edited rice lines.
3.6. Obtaining Homozygous Transgene-Free OsNRAMP7-Overexpressing TBR225 Line
To confirm the overexpression of the target gene in edited TBR225 rice plants resulting from the insertion of the 35S promoter, we genotyped T1 individuals propagated from T0 line #33. Among 20 tested plants, 8 showed negative PCR results with three primer pairs specific for gRNA, Cas9, and HPT expression cassettes, while they showed positive results in control PCR of OsActin, proving the absence of transgenes in their genome (Fig. 5a). This T-DNA segregation ratio is consistent with Mendel's 3:1 genetic segregation ratio (χ2 < 0.381). Moreover, we obtained specific amplicons of ~0.4 and ~1.2 kb with primer pairs 35S-F/NRAMP7-t-R and NRAMP7-t-F/NRAMP7-t-R, respectively, from all T1 plants (Fig. 5b). Notably, we observed an overexpression of OsNRAMP7 in T1 progenies similar to their T0 parent line (Fig. 5c). These results indicated that the homozygous 35S-insertion mutation was stably transmitted to the next generation, resulting in the overexpression of the target gene in T1 transgene-free edited TBR225 rice.
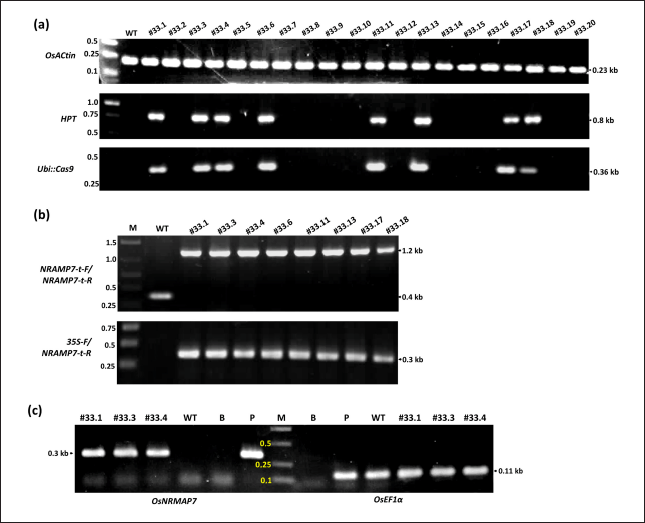 | Figure 5. Molecular characterization of the T1 progeny of line #33. (a) Transgene segregation analysis in T1 population derived from the T0 line #33. Top: OsActin (endogenous control); middle: HPT (selectable marker gene); bottom: Ubi::Cas9 cassette. (b) 35S promoter insertion confirmation. PCR analysis using two specific primer pairs to validate correct 35S promoter integration. (c) OsNRAMP7 expression analysis. RT-PCR results for edited lines (#33.1, #33.2, #33.3) and wild-type (WT). OsEF1α: internal control. M: 1.0 kb ladder, B: blank control; P: positive control (genomic DNA as PCR template); WT: wild-type plant; #33.1 - #33.20: T1 plants derived from the T0 line #33. [Click here to view] |
4. DISCUSSION
Promoter editing for precise control of target gene expression represents an advanced approach in plant biotechnology, with significant implications for rice and other crops [18]. In contrast to conventional gene overexpression methods, promoter editing minimizes unintended consequences by avoiding the production of unnecessary proteins associated with marker or reporter genes and preventing genome disruption due to random transgene insertion. This precision approach reduces energy waste, mitigates potential negative impacts on plant growth, and addresses biosafety concerns [19,20].
A straightforward strategy involves the direct insertion of an endogenous or exogenous promoter upstream of the target gene's open reading frame, utilizing either HDR [21] or NHEJ [22] mechanisms. These inserted promoters can be either constitutive or inducible, offering flexibility in gene expression control. Notable examples of this approach include the work by Cermak et al. [23] who inserted a constitutive 35S promoter upstream of the SlANT1 gene in tomato using homologous recombination, resulting in overexpression of the target gene and increased pigment production [23]. Similarly, researchers employed CRISPR-Cas9 technology to insert the native maize GOS2 promoter into the 5′-untranslated region of the ARGOS8 gene, creating a new ARGOS8 variant with significantly elevated transcript levels compared to the native allele [21]. In our study, we achieved substantial enhancement of OsNRAMP7 expression in a transgene-free rice line with 35S promoter integration. This result, corroborated by similar findings from other researchers, demonstrates the effectiveness of the precise promoter insertion strategy in modulating gene expression, offering a promising alternative to conventional methods using recombinant gene overexpression T-DNA vectors for plant genetic engineering.
While gene editing via the HDR mechanism is not novel, researchers continue to refine this system, particularly in rice. Previous studies predominantly employed multi-vector systems, utilizing at least two separate vectors to deliver sgRNA, Cas9 complex, and template DNA constructs into cells [22,24]. This approach offers flexibility but complicates the transformation process. Virus-based systems have been developed to address transformation challenges [25], but they suffer from limited cargo capacity. Recent advancements include the use of RNA templates combined with sgRNA expression constructs in single-vector systems, enabling efficient induction of nucleotide substitution mutations in rice [26,27]. Barone et al. [28] pioneered a selectable marker-free intra-genomic gene targeting method in maize, where the donor DNA template, along with the Cas9/gRNA expression cassettes, were pre-integrated into the T-DNA, and subsequently, the donor DNA was excised by the Cas9/gRNA complex. We have successfully adapted this strategy for rice, with a key modification: our T-DNA contains two donor DNA template sequences instead of one, potentially enhancing gene editing efficiency. This single-vector system aims to overcome the challenges faced by indica rice varieties with low gene transformation efficiency, such as TBR225, because it requires only one step instead of several steps in the gene transfer process, compared to multi-vector and virus-based systems.
A significant challenge in gene editing systems is the prolonged activity of CRISPR/Cas9, which can result in more extensive genetic modifications than initially intended. This can complicate the interpretation of experimental results and potentially affect the outcomes of gene therapy approaches [29,30]. Our study introduces a novel gRNA expression structure design, positioned between two donor DNA template fragments (Fig. 1b). While not experimentally verified in this study, this design aims to mitigate the issue of prolonged CRISPR/Cas9 activity. The rationale is that if both template fragments are removed, the gRNA expression construct would also be excised from the rice chromosome. This approach builds upon a similar strategy employed by Tan et al. [31], who successfully developed a CRISPR/Cas9-mediated HDR auto-excision system for the efficient removal of marker genes from transgenic plants. The primary challenge with this method is ensuring the CRISPR/Cas9 complex functions effectively to accurately excise the donor DNA template fragment. Additionally, low copy numbers of the donor DNA template fragment can reduce the efficiency of the HDR process [32]. However, these issues can be addressed through in vivo or in vitro validation of the designed sgRNA and by increasing the number of donor DNA template fragments inserted into the T-DNA vector. Our results demonstrated success using a single vector carrying one well-designed gRNA and two donor DNA template copies, offering a promising solution to enhance gene editing precision and efficiency in rice.
The efficiency of HDR in gene editing remains a significant challenge, particularly in rice [5]. The low efficiency of HDR-mediated gene insertion in rice is further illustrated by several studies. For instance, Dong et al. [22] attempted to insert a 5.2 kb carotenoid biosynthesis cassette into the Kitaake rice genome using CRISPR/Cas9. While they obtained 8 out of 55 regenerated plants carrying the insertion fragment, only one plant contained the DNA insertion with the correct orientation. Notably, subsequent analyses suggested that the insertion process occurred through NHEJ rather than HDR, highlighting the cellular preference for this repair pathway [22]. Li et al. [33] reported a modest success rate of 2.2% for targeted gene insertions. Another study focusing on two rice varieties, Nangeng 9108 and BL3045, initially showed promising results with an HDR-mediated insertion efficiency of 20.9%. However, the percentage of transgenic plants carrying the correct insertion was significantly lower, reaching only 3.8% [34]. In this study, our work on TBR225 rice yielded a mere 2.79% efficiency for 35S promoter insertion via HDR, contrasting sharply with previous studies reporting a 90% indel mutation rate through NHEJ in the same variety [10]. These findings underscore that HDR efficiency in rice depends on multiple factors, including CRISPR/Cas system components, donor template and sgRNA design, delivery method, target sequence, and rice genotype. The preference of cells to repair double-strand breaks through NHEJ rather than HDR further complicates the process. To enhance HDR efficiency in rice and overcome the cellular preference for NHEJ, several strategies could be explored [5]. These include cell cycle synchronization to target S and G2 phases, suppression of the NHEJ pathway, optimization of donor DNA design, use of HDR enhancers, exploration of Cas9 variants and fusion proteins, improved delivery methods, temperature modulation, and use of tissue-specific promoters. These approaches aim to increase the precision of gene editing, reduce off-target effects, improve integration rates of large DNA fragments, and potentially lower regeneration times. Implementing these methods could significantly improve the overall success rate of targeted gene insertions in rice, although their effectiveness may vary depending on the specific target gene and tissue type. Despite the challenges, we successfully demonstrated the potential for introducing a new promoter to control target gene expression, as evidenced by the increased expression in rice line #33, even with the presence of indel mutations. This variability in success rates highlights the complexity of achieving efficient and precise DNA insertions in rice using HDR-mediated gene editing, emphasizing the need for continued research and optimization in this field.
As mentioned above, the length of homology arms and donor DNA template being particularly crucial which can influence the efficiency of HDR-mediated gene editing. Previous studies have successfully used homology arms of varying lengths, ranging from a few dozen to over 1,000 base pairs (bp), with donor DNA templates extending to several thousand bp [5]. Longer homology arms generally increase HDR efficiency up to a certain point [20]. However, the size of the T-DNA can affect its integration into the plant genome, with larger inserts potentially having lower integration rates [35]. Therefore, in our study, we designed homology arms of approximately 150 bp to minimize the size of the single T-DNA vector, facilitating efficient gene transfer via Agrobacterium-mediated transformation. While this approach proved successful, the insertion efficiency was not high. This occurrence may be related to the design of homology arms [5], highlighting the complexity of gene editing outcomes and the need for further research in this area.
Our findings underscore the challenges and intricacies involved in HDR-mediated gene editing, particularly in TBR225 rice. They also emphasize the importance of optimizing various parameters, such as homology arm design and donor DNA template length, to improve editing efficiency and accuracy.
5. CONCLUSION
This study successfully demonstrated precise genetic modification of TBR225 rice using CRISPR/Cas9-mediated HDR. We implemented a single vector system to insert the 35S promoter upstream of OsNRAMP7 in this Vietnamese rice variety, generating and validating transgenic rice lines. Results confirmed successful 35S promoter insertion, leading to significant OsNRAMP7 overexpression in edited lines, including homozygous transgene-free OsNRAMP7-overexpressing TBR225 lines. Our HDR-based editing approach for TBR225 rice enables precise genetic modification in one generation, unlike traditional breeding's multi-generation process. It creates transgene-free lines while avoiding undesired genetic material introduction. This research advances precise genome editing in economically important rice varieties, providing a valuable resource for studying the role of OsNRAMP7 in rice biology, particularly metal ion transport. Our study lays the groundwork for enhancing traits like stress tolerance and nutrient use efficiency in rice, opening new avenues for crop improvement and functional genomics studies in Vietnamese rice varieties, potentially contributing to food security and sustainable agriculture.
6. ACKNOWLEDGMENTS
The authors extend their gratitude to the Vietnam Ministry of Science and Technology for funding the project "Research on micronutrient biofortification in rice using biotechnology" (Code: N?T/CN/21/24). The authors are thankful to their colleagues at the Institute of Agricultural Genetics for their assistance with rice transformation experiments, and to ThaiBinh Seeds Corporation for providing the rice accession. These contributions were instrumental to the authors' research success.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data are available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Khush GS. Green revolution: the way forward. Nat Rev Genet 2001;2(10):815–22; CrossRef
2. Capriotti L, Baraldi E, Mezzetti B, Limera C, Sabbadini S. Biotechnological approaches: gene overexpression, gene silencing, and genome editing to control fungal and Oomycete diseases in Grapevine. Int J Mol Sci 2020;21(16):5701; CrossRef
3. Ladics GS, Bartholomaeus A, Bregitzer P, Doerrer NG, Gray A, Holzhauser T, et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res 2015;24(4):587–603; CrossRef
4. Jinek M, Chylinski K, Fonfara I, Hauer Michael, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337(6096):816–21; CrossRef
5. Chen J, Li S, He Y, Li J, Xia L. An update on precision genome editing by homology-directed repair in plants. Plant Physiol 2022;188(4):1780–94; CrossRef
6. Laforest LC, Nadakuduti SS. Advances in delivery mechanisms of CRISPR gene-editing reagents in plants. Front Genome Ed 2022;4:830178; CrossRef
7. Atia M, Jiang W, Sedeek K, Butt H, Mahfouz M. Crop bioengineering via gene editing: reshaping the future of agriculture. Plant Cell Rep 2024;43(4):98; CrossRef
8. Li J, Liu Y, Kong L, Xu E, Zou Y, Zhang P, et al. An intracellular transporter OsNRAMP7 is required for distribution and accumulation of iron into rice grains. Plant Sci 2023;336:111831; CrossRef
9. Manh Bao T, Thi Hop T, Thi Tiec T, Thi Nhung N, Van Hoan N. Results of breeding Tbr225 rice cultivar. Vietnam J Agri Sci 2016;14(9):1360–67; Available via http://www1.vnua.edu.vn/tapchi/Upload/15112016-so%208.7.pdf
10. Duy PN, Lan DT, Pham Thu H, Thi Thu HP, Nguyen Thanh H, Pham NP, et al. Improved bacterial leaf blight disease resistance in the major Elite Vietnamese rice cultivar TBR225 via editing of the OsSWEET14 promoter. PLoS One 2021;16(9):e0255470; CrossRef
11. Pham TH, Tran LD, Nguyen VC, Pham TV, Do TH, Pham XH, et al. A protocol for Agrobateruim–mediated transformation into a TBR225 rice variety. Vietnam J Agric Rural Dev 2021;18:66–73. Available via https://sti.vista.gov.vn/tw/Lists/TaiLieuKHCN/Attachments/330558/CVv201S182021066.pdf
12. Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL. CRISPR-P 2.0: an improved Crispr/Cas9 tool for genome editing in plants. Mol Plant 2017;10(3):530–32; CrossRef
13. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31(13):3406–15; CrossRef
14. Stemmer M, Thumberger T, Keyer MD, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 2015;10(4):e0124633; CrossRef
15. Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 2014;42(17):10903–14; CrossRef
16. Herbert L, Meunier AC, Bes M, Vernet A, Portefaix M, Durandet F, et al. Beyond seek and destroy: how to generate allelic series using genome editing tools. Rice 2020;13(1):5; CrossRef
17. Dehairs J, Talebi A, Cherifi Y, Swinnen JV. CRISP-ID: decoding CRISPR mediated indels by sanger sequencing. Sci Rep 2016;6:28973; CrossRef
18. Tang X, Zhang Y. Beyond knockouts: fine-tuning regulation of gene expression in plants with CRISPR-cas-based promoter editing. New Phytol 2023;239:868–8; CrossRef
19. Miki B, McHugh S. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J Biotechnol 2004;107(3):193–232; CrossRef
20. Chen X, Du J, Yun S, Xue C, Yao Y, Rao S. Recent advances in CRISPR-Cas9-based genome insertion technologies. Mol Ther Nucleic Acids 2024;35(1):102138; CrossRef
21. Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, et al. ARGOS8 variants generated by Crispr-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 2017;15(2):207–16; CrossRef
22. Dong OX, Yu S, Jain R, Zhang N, Duong PQ, Butler C, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun 2020;11(1):1178; CrossRef
23. Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol 2015;16:232; CrossRef
24. Endo M, Mikami M, Toki S. Biallelic gene targeting in rice. Plant Physiol 2016;170(2):667–77; CrossRef
25. Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK. Gene targeting by homology-directed repair in rice using a Geminivirus-based CRISPR/Cas9 system. Mol Plant 2017;10(7):1007–10; CrossRef
26. Zafar K, Khan MZ, Amin I, Mukhtar Z, Zafar M, Mansoor S. Employing template-directed CRISPR-based editing of the OsALS gene to create herbicide tolerance in Basmati rice. AoB Plants 2023;15(2):plac059; CrossRef
27. Butt H, Eid A, Ali Z, Atia MAM, Mokhtar MM, Hassan N, et al. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front Plant Sci 2017;8:1441; CrossRef
28. Barone P, Wu E, Lenderts B, Anand A, Gordon-Kamm W, Svitashev S, et al. Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol Plant 2020;13(8):1219–27; CrossRef
29. Chu P, Agapito-Tenfen SZ. Unintended genomic outcomes in current and next generation GM techniques: a systematic review. Plants (Basel) 2022;11(21):2997; CrossRef
30. Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J 2017;15:257–68; CrossRef
31. Tan J, Wang Y, Chen S, Lin Z, Zhao Y, Xue Y, et al. An efficient marker gene excision strategy based on CRISPR/Cas9-mediated homology-directed repair in rice. Int J Mol Sci 2022;23(3):1588; CrossRef
32. Riesenberg S, Kanis P, Macak D, Wollny D, Düsterhöft D, Kowalewski J, et al. Efficient high-precision homology-directed repair-dependent genome editing by HDRobust. Nat Methods 2023;20(9):1388–99; CrossRef
33. Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, et al. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants 2016;2:16139; CrossRef
34. Xu Y, Wang FQ, Chen ZH, Wang J, Li WQ, Fan F, et al. Intron-targeted gene insertion in rice using CRISPR/Cas9: a case study of the Pi-ta gene. Crop J 2020;8:424–31; CrossRef
35. Liu S, Wang K, Geng S, Hossain M, Ye X, Li A, et al. Enemies at peace: recent progress in Agrobacterium-mediated cereal transformation. Crop J 2024;12(2):321–9; CrossRef