ARTICLE HIGHLIGHTS
Flow cytometry (FCM) analysis is a rapid and efficient method for determining the ploidy level and DNA content in plants.
Combining fresh leaves with LB01 buffer produces clean histograms with sharper peaks and reduced noise levels.
FCM analysis classified the polyploid plants of Neolamarckia cadamba into tetraploids, mixoploids, and octoploids.
FCM showed efficacy in identifying mixoploids in N. cadamba polyploids compared to traditional methods of chromosome counting.
1. INTRODUCTION
Flow cytometry (FCM) analysis is commonly used to determine the ploidy level, and DNA content of plantlets regenerated from explants treated with varying concentrations and durations of colchicine. FCM plays an essential role in studies related to polyploidization, allowing the distinction between induced polyploids or mixoploids and plantlets with unchanged ploidy levels [1-3]. There are many advantages of using FCM over the chromosome counting method. FCM only uses a small amount of plant tissue, such as the leaf or other non-destructive parts of the plant itself. FCM does not rely on a specific stage of mitosis for analysis, allowing for greater flexibility in sample procurement regardless of harvest time or tissue type [4,5]. Furthermore, the easy and quick sample preparation for FCM enables the bulk screening of many samples in a polyploidization study. Apart from ploidy level determination, total nuclear DNA content (C-value) can also be estimated [6].
The procedure for determining ploidy level and DNA content using FCM involved three main steps: isolation of cell nuclei, fluorochrome staining, and analysis [4]. In the first step, leaf samples are finely chopped using a sharp razor blade in a buffer to release nuclear suspension. The suspension is then filtered through a nylon mesh to separate cell nuclei from debris [7]. It is crucial to use fresh, uncontaminated plant samples for FCM analysis. After filtration, the next step involves DNA staining. Depending on the excitation source of the flow cytometer, one of the following DNA-specific fluorochromes can be used: propidium iodide, ethidium bromide, 4’,6-diamidino-2-phenylindole, or Hoechst 33258. Once the staining is complete, the suspension is ready for analysis using the flow cytometer [4,8]. During the FCM analysis, the sample suspension containing cells with intact and stained chromosomes is drawn into the flow cytometer’s sheath in a narrow stream. The flow cytometer chamber is equipped with an excitation source, which can be LED or laser, emitting excitation light. This light beam intersects each stained chromosome, causing the fluorochromes to scatter light and emit fluorescence. The fluorescence intensity is then detected and quantified, forming a dominant peak that corresponds to the sample’s karyotype composition [5,9,10].
Advanced cytometers come with a high price tag because they can simultaneously quantify multiple parameters and provide real-time observations. However, in plant research, most of these parameters and functions offered by advanced cytometers are seldom utilized, as the primary focus lies in determining the ploidy level and DNA content [4]. Therefore, this study aimed to optimize and develop an efficient FCM analysis procedure for N. cadamba. This involved exploring different types of leaf tissues and lysis buffers to identify the most suitable combination. In addition, the study aimed to employ the optimized FCM procedure to determine the ploidy level and DNA content of tetraploid and octoploid N. cadamba specimens.
2. MATERIALS AND METHODS2.1. Plant Materials
The plant materials used in this study were obtained from a previous study conducted by Eng et al., [11]. Fully developed young leaves from both the control plantlets and the plantlets treated with colchicine were collected for ploidy level determination. The untreated plantlets served as the control group, while Glycine max Merr. “Polanka” was used as an external standard in the FCM analysis, following the method described by Doležel et al. [12].
2.2. FCM Analysis
To optimize the FCM study for N. cadamba, two types of leaves were assessed. Leaf samples for the FCM optimization study were obtained from ex vitro and in vitro N. cadamba plantlets. The ex vitro N. cadamba leaf samples were harvested at least 6 h before their transportation to the Advanced Technology and Breeding Center at the Malaysian palm oil board (MPOB) in Bangi, Selangor. During transportation, the harvested leaves were placed in a moistened ziplock bag. On the other hand, the in vitro plantlets were transported from UNIMAS to MPOB in culture vessels. The in vitro leaves were harvested at MPOB right before the FCM study.
Two lysis buffers, LB01 and Otto’s buffers, were evaluated to optimize the DNA extraction for FCM. LB01 buffer consisted of 80 mM KCl, 20 mM NaCl, 15 mM Tris, 15 mM mercaptoethanol, 2 mM Na2EDTA, 0.5 mM spermidine, 0.1% Triton X-100, pH 7.5, 50 μg propidium iodide, and 50 μg RNase [12]. Otto’s buffer comprised 0.1 M citric acid, 0.5% Tween-20, 50 μg propidium iodide, and 50 μg RNase [13]. For each sample, approximately 1 cm2 of a fresh young leaf was chopped in 1 mL of either LB01 buffer or Otto’s buffer using a scalpel on a disposable Petri dish. The finely chopped leaf pieces were individually filtered through a 50-μm cell strainer into a 5 mL FCM tube. The filtrate of each sample was then analyzed using a FACSCalibur flow cytometer (Becton Dickinson). Histograms were obtained for each sample across 1024 channels.
2.3. Data Analysis
The FCM-generated histograms were used to determine (a) the ploidy level and (b) the DNA content of a sample. The formulas employed for these calculations are as follows:
To determine the ploidy level, the following formula by Guo et al., [14] was used:
SPL = (S (G0/G1)/R(G0/G1)) × RPL
where SPL represents the ploidy level of the colchicine-treated sample, RPL denotes the ploidy level of the control sample, S(G0/G1) indicates the mean position of the peak in the colchicine-treated sample, and R(G0/G1) signifies the mean position of the peak in the control sample.
To determine the DNA content, the formula developed by Doležel et al., [4] was employed:
SPL = (S (G0/G1)/R(G0/G1))×RPL
where SPL corresponds to the 2Cx value of the colchicine-treated sample, RPL represents the 2C value of the control sample (specifically, Glycine max Merr. “Polanka” with a value of 2.50), S(G0/G1) indicates the mean position of the colchicine-treated sample, and R(G0/G1) signifies the mean position of the peak in the control sample.
3. RESULTS AND DISCUSSION
3.1. FCM Optimization of N. cadamba
The analysis of leaf samples obtained from ex vitro N. cadamba plants using a flow cytometer [Figure 1a] revealed poor results in the FCM analysis. The low count of events, only 1515, can be attributed to the rapid degradation of DNA in leaves within hours after harvesting. The low event number count in this study indicated that a significant number of cells were not analyzed, thus failing to represent the entire cell population of the plantlet. However, when the in vitro leaf samples were analyzed, the number of event counts significantly increased to 10,980, which was 10 times higher [Figure 1b]. This discrepancy indicated that utilizing leaves from ex vitro plants harvested at least 6 h before analysis resulted in minimal viable DNA that the cytometer could detect effectively.
 | Figure 1: Flow cytometer analysis of Neolamarckia cadamba plantlets. (a). The ex vitro control plant using Otto’s buffer (event counts 1,515; CV 9.19) and (b). The in vitro control plantlet using Otto’s buffer (event counts 10,980; CV 5.20). [Click here to view] |
In this study, the plant samples from UNIMAS (Sarawak) and the flow cytometer located at MPOB (Selangor) were separated by a distance of 1000 km, and transporting plant samples were a major obstacle. To overcome this logistical hurdle, tissue-cultured N. cadamba plantlets cultivated in glass jars, which are more convenient for packing and transportation, were utilized. Since tissue-cultured plants are free from diseases, pests, and contaminants, they pose minimal phytosanitary risks. The presence of a horizontal laminar flow system in the laboratory where the cytometer is located facilitated the procurement of leaves without contamination. After analysis, the plantlets were transported back to UNIMAS for further maintenance and multiplication. Using in vitro plantlets as samples for FCM is a non-destructive method as they remain protected and viable throughout transportation and analysis. The vigor of the plantlets is therefore maintained.
Selecting fresh, pest-free, and contaminant-free explants are vital to isolate the intact nuclei. In this study, in vitro plantlets were chosen as FCM samples as they met these criteria. This precautionary measure prevents FCM from generating inaccurate data by detecting foreign or damaged DNA. DNA in plant samples begins to degrade after being excised from the plant, but the rate of deterioration varies depending on the species and environmental conditions [15,16]. This limitation has posed a significant challenge in FCM usage, especially when fresh plant materials are logistically or geographically distant from the flow cytometer. In addition, the flow cytometer is a large and sensitive machine, making it nearly impossible to relocate easily. Consequently, logistical issues arise when conducting field studies, as the samples are often situated in remote locations.
Seeds can be used as samples in FCM since they can be stored for extended periods without DNA degradation and lack cytosols [6]. Leaves, on the other hand, contain cytosols that may hinder the staining process using fluorochromes, thereby affecting FCM readings. However, data derived from seeds should be interpreted cautiously since they exhibit genetic variation due to the combination of male and female gametes. In the present study, we encountered difficulties in the FCM analysis when using fresh leaves obtained from acclimatized N. cadamba plants, which had undergone 6 h of packing and transportation, resulting in significantly low nuclear event counts during FCM. This failure indicated two potential causes, either propidium iodide stains were ineffective in staining deteriorated DNA, or cytosol oxidation-prevented DNA staining.
Several alternative methods have been developed to overcome the limitations of using fresh materials in FCM, but they are still considered inferior. These methods involve fixation, freezing, or dehydration [17]. Chemical-based fixatives, such as acid methanol fixation [18], glycerol [19], and formaldehyde preservative [20], have been employed to preserve plant samples before FCM analysis. Compared to non-fixed materials, these fixatives can enhance fluorochrome staining and increase nucleus yield. Dehydration of plant materials is another utilized method [21,22]. However, non-fresh plant material cannot be recommended for estimating DNA content in absolute units unless the protocols undergo further investigation and demonstrate reliability [4].
Both histograms [Figure 1a and b] exhibited broad bases, indicating a high coefficient of variation (CV) and reduced data reliability. Numerous secondary peaks, representing unwanted events or noise in the DNA sample, were observed alongside the dominant peak. The ex vitro sample in Otto’s buffer showed a CV of 9.19 [Figure 1a], while the in vitro sample in the same buffer displayed a CV of 5.20 [Figure 1b]. This suggests that Otto’s buffer is unsuitable as a lysis buffer for N. cadamba leaf samples as it produced a high CV value. To enhance the FCM data and reduce the CV value, Otto’s buffer was replaced with LB01 buffer. The use of LB01 buffer produced histograms with a CV value below five [Figure 2a], while also significantly reducing excessive noise between the dominant peaks. Again, in this study, a higher CV value was obtained with the ex vitro sample. The doubling of the CV value in the ex vitro sample indicates the deterioration of leaf freshness, which affects sample quality. The evidence from both FCM studies of N. cadamba demonstrates that only fresh leaves combined with LB01 buffer can generate clean histograms with sharper peaks, fewer noises, and low CV values.
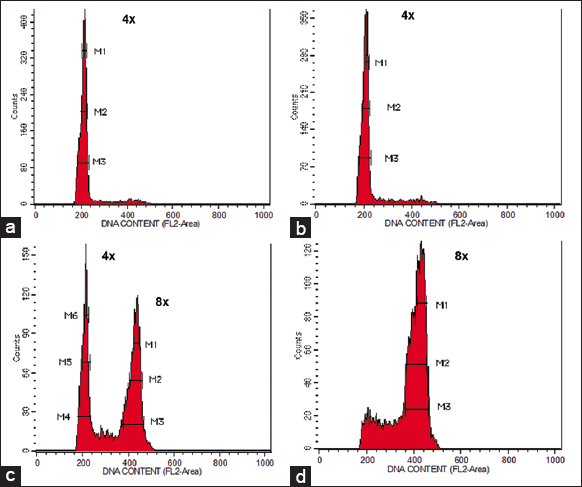 | Figure 2: Flow cytometer analysis of Neolamarckia cadamba plantlets derived from (a) nodal segment treated with distilled water (control) indicating tetraploid plant (2n = 4x), (b) nodal segment treated with colchicine indicating tetraploid plant (2n = 4x), (c) nodal segment treated with colchicine indicating mixoploid plant (2n + 4n = 4x + 8x), and (d) nodal segment treated with colchicine indicating octoploid plant (4n = 8x). [Click here to view] |
Lysis buffers were formulated to preserve nucleus integrity, prevent DNA degradation, and enhance stoichiometric staining. Commonly used non-commercial lysis buffers include Galbraith [7], LB01 [12], Otto’s [13], and Tris.MgCl2 [23]. LB01 and Otto’s buffers have been identified as the most effective [24]. Modifying lysis buffers by adjusting the pH and concentration of existing components or incorporating additional chemicals has been a common practice to enhance their effectiveness [25]. However, certain commercial buffers are customized explicitly for specific cytometer models the vendor produces, and their compositions are often undisclosed. Therefore, carefully reading the cytometer instructions and maintaining close communication with the vendor is crucial when encountering any obstacles.
Plants often contain cytosolic compounds that interfere with the ability of fluorochromes to stain DNA. DNA staining aims to achieve sufficient fluorescence intensity for detection by the cytometer. The presence of cytosols can lead to inaccurate DNA content estimation [4]. To overcome the effect of cytosols, a reducing agent called β-mercaptoethanol is added to the buffers [26]. Spermidine, an oxygen quencher, can prevent oxidative damage to DNA while maintaining its integrity [27]. Additional additives such as PVP-10 and PVP-40 can bind phenolic compounds in plants [4]. However, according to Jedrzejczyk and Sliwinska [28], the inclusion of antioxidants such as β-mercaptoethanol and PVP has not been sufficient to overcome the effects of cytosols in DNA content estimation of 11 Rosaceae woody species. In this study, the LB01 buffer was supplemented with β-mercaptoethanol and spermidine to ensure reliable and accurate DNA content estimation of N. cadamba.
Propidium iodide was used as the fluorochrome in this study to determine the ploidy level and DNA content of regenerants in N. cadamba. The obtained DNA content of N. cadamba in this study aligns with the data reported by Ohri et al., [29]. The successful determination of N. cadamba DNA content can be attributed to the use of propidium iodide as the fluorochrome, as this stain intercalates into double-helix DNA without selectively binding to specific DNA bases. The inclusion of RNase is necessary to remove RNA and prevent the fluorochrome from binding to RNA.
The choice of DNA stains depends on the excitation source used in the cytometer. In this study, laser excitation was employed, specifically an argon ion laser (15 mW) at 488 nm. Intercalating fluorochromes are more commonly used compared to base-specific fluorochromes. In a survey of three articles by Bennett and Leitch [30,31] and Bennett et al., [32], it was found that out of 892 data points for DNA content, 789 were obtained using intercalating fluorochromes, with propidium iodide being used in 689 out of the 789 cases. On the other hand, DAPI and Hoechst 33258 are used to stain DNA in flow cytometers equipped with an arc lamp. One advantage of using these stains is that RNase is not required [4]. However, these stains preferentially bind to the A-T base pair in DNA, and they are less accurate in estimating DNA content. As a result, DNA content estimation cannot be interpreted as absolute units but as relative units to the control. Therefore, when absolute units of DNA content estimation are required for a study, it is advisable to avoid using a flow cytometer equipped with an arc lamp that utilizes DAPI and Hoechst 33258 as stains, as the estimation will be in relative units compared to the control.
3.2. Ploidy Level and DNA Content Determination of N. cadamba
The histogram generated by FCM analysis revealed distinct patterns in DNA content for different plant types. Both control and tetraploid plants exhibited a single dominant peak, with the mean position corresponding to 4x [Figure 2a and b]. In contrast, mixoploid plants showed two separate dominant peaks in the histogram. The first peak corresponded to 4x, similar to tetraploids, while the second peak corresponded to 8x [Figure 2c]. Octoploid plants displayed a single dominant peak at 8x [Figure 2d]. Notably, the mean position of octoploid cells was twice that of tetraploid cells.
FCM is a valuable tool for detecting mixoploids and quickly analyzing large cell populations. Previous studies on polyploidization, conducted without using FCM, reported high percentages of polyploidization without detecting mixoploids. For instance, percentages of 41.7% for Vitis sp. shoot tips [33], 42.3% for Gossypium arboreum seeds [34], and 100% for Pogostemon cablin leaves [35] were determined through the chromosome counting method. To avoid mixing mixoploids with induced polyploids or the original ploidy category, FCM should be employed in these studies to identify mixoploids accurately.
Mixoploids, consisting of cells with different ploidy levels, represent genetic variants. However, the extent to which the composition of mixoploids changes over time in terms of the proportions of original cell ploidy and polyploid cell ploidy remains uncertain. According to Regalado et al., [36], mixoploid plants are unstable due to competition between the original cells and the polyploid cells, leading to the elimination of the latter. As a result, the mixoploid status reverts back to its original ploidy level. Due to the complexities involved, mixoploids have often been excluded from further characterization or study on their detection [3,37-39]. While generating biodiversity and genotype variation through polyploidization is well-known, the study of mixoploids has been limited, resulting in a scarcity of published literature on the subject. There is currently a lack of empirical evidence regarding the unfavorable aspects of mixoploidy in plant breeding. Therefore, this area deserves more attention and further investigation in the future, including aspects such as productivity and growth, fertility testing through the crossing, adaptation to different environments, and the extent of genotype stability.
The analysis of FCM data revealed that N. cadamba tetraploids (4C DNA) had an estimated DNA content of 2.59 ± 0.09 pg, while octoploids showed a twofold increase in DNA content (8C DNA) at 5.35 ± 0.24 pg. These findings are consistent with the results reported by Ohri et al., [29]. The estimated 4C DNA content for Anthocephalus cadamba (synonyms: N. cadamba), according to Ohri et al., [29], is 2.77 ± 0.27 pg using the M86 Vickers microdensitometer. These data have been published and archived in the C-value Kew Garden database (http://www.kew.org/cval/homepage.html), where the values are reported as estimated 1C DNA, equivalent to 0.69 pg. These findings confirm that N. cadamba, the species under investigation in our study, is a tetraploid. Based on these data, our estimated 4C DNA content of N. cadamba falls within the published range at 2.59 ± 0.09 pg. In addition, our study observed that the estimated 8C DNA content of octoploids was 5.35 ± 0.24 pg. In our experimental setup, the use of colchicine, a chromosome-doubling agent, resulted in doubling the DNA content of N. cadamba octoploids. Estimated DNA content serves various purposes beyond indicating changes in ploidy levels, including molecular investigations, evolution, phylogenetics, phenotypic studies, phenological analyses, ecological and environmental indicators, as well as paleobiological trends [32].
4. CONCLUSION
This study demonstrated the importance of using fresh leaf samples from in vitro plantlets of N. cadamba and lysis buffer LB01 to ensure reliable and consistent results in FCM analysis. The findings further proved that FCM is a valuable tool for characterizing polyploid plants following colchicine treatment. In addition, FCM enables the identification of mixoploids, a task that cannot be accomplished using traditional methods such as chromosome counting.
5. ACKNOWLEDGMENTS
The funding for this study was provided by the Sarawak Timber Association (STA) to Universiti Malaysia Sarawak under Grants No. GL/F07/STA01/2019 and GL/F07/STA/2020. The authors would like to sincerely thank WTK for their generous contribution to the plant materials used in this research.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors requirements/guidelines.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
Ethics approval was not required for this study, as it did not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
All data obtained or analyzed in this study are included in this manuscript.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Bicknell R, Boase M, Morgan E. Developing polyploid genotypes of Leptospermum scoparium. N Z J Crop Hortic Sci 2019;47:273-81. [CrossRef]
2. Huy NP, Tam DT, Luan VQ, Tung HT, Hien VT, Ngan HT, et al. In vitro polyploid induction of Paphiopedilum villosum using colchicine. Sci Hortic 2019;252:283-90. [CrossRef]
3. Julião SA, Ribeiro CD, Lopes JM, Matos EM, Reis AC, Peixoto PH, et al. Induction of synthetic polyploids and assessment of genomic stability in Lippia alba. Front Plant Sci 2020;11:292. [CrossRef]
4. Doležel J, Greilhuber J, Suda J. Flow cytometry with plants:An overview. In:Flow Cytometry with Plant Cells Analysis of Genes, Chromosomes and Genomes. Germany:Wiley-VCH Verlag GmbH and Co.;2007. 41-65. [CrossRef]
5. D'Hondt L, Hofte M, van Bockstaele E, Leus L. Applications of flow cytometry in plant pathology for genome size determination, detection and physiological status. Mol Plant Pathol 2011;12:815-28. [CrossRef]
6. Sliwinska E, Zielinska E, Jedrzejczyk I. Are seeds suitable for flow cytometric estimation of plant genome size?Cytometry 2005;64:72-9. [CrossRef]
7. Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983;220:1049-51. [CrossRef]
8. Dolezel J, Greihuber J, Lucretti S, Meister A, Lysak MA, Nardi L, et al. Plant genome size estimation by flow cytometry:Interlaboratory comparison. Ann Bot 1998;82:17-26. [CrossRef]
9. Bakke AC. The principles of flow cytometry. Lab Med 2001;32:207-11. [CrossRef]
10. Doležel J, Vrána J, Šafá?J, BartošJ, KubalákováM, ŠimkováH. Chromosomes in the flow to simplify genome analysis. Funct Integr Genom 2012;12:397-416. [CrossRef]
11. Eng WH, Ho WS, Ling KH. Effects of colchicine treatment on morphological variations of Neolamarckia cadamba. Int J Agric Technol 2021;17:47-66.
12. Doležel J, BinarováP, Lucretti S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 1989;31:113-20. [CrossRef]
13. Otto F. Preparation and staining cells for high resolution DNA analysis. In:Radbruch A, editor. A Flow Cytometry and Cell Sorting. Germany:Springer Verlag;1992. 101-4. [CrossRef]
14. Guo WW, Liang WJ, Xie KD, Xia QM, Fu J, Guo DY, et al. Exploitation of polyploids from 39 Citrus seedling populations. Acta Hortic 2016;1135:11-6. [CrossRef]
15. Sahu SK, Thangaraj M, Kathiresan K. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol 2012;2012:205049. [CrossRef]
16. Guo Y, Yang G, Chen Y, Li D, Guo Z. A comparison of different methods for preserving plant molecular materials and the effect of degraded DNA on ddRAD sequencing. Plant Divers 2018;40:106-16. [CrossRef]
17. Loureiro J, Doležel J, Greilhuber J, Santos C, Suda J. Plant flow cytometry-Far beyond the stone age. Cytometry 2008;73:579-80. [CrossRef]
18. Silva TC, Abreu IS, Carvalho CR. Improved and reproducible flow cytometry methodology for nuclei isolation from single root meristem. J Bot 2010;2010:320609. [CrossRef]
19. Kolá?F, Lu?anováM, T?šitel J, Loureiro J, Suda J. Glycerol-treated nuclear suspensions - an efficient preservation method for flow cytometric analysis of plant samples. Chromosome Res 2012;20:303-15. [CrossRef]
20. Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annu Rev Ecol Evol Syst 2007;38:847-76. [CrossRef]
21. Suda J, KrahulcováA, Trávnícek P, Krahulec F. Ploidy level versus DNA ploidy level:An appeal for consistent terminology. Taxon 2006;55:447-50. [CrossRef]
22. Bainard JD, Husband BC, Baldwin SJ, Fazekas AJ, Gregory TR, Newmaster SG, et al. The effects of rapid desiccation on estimates of plant genome size. Chromosome Res 2011;19:825-42. [CrossRef]
23. Pfosser M, Amon A, Lelley T, Heberle-Bors E. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disornic and ditelosornic wheat-rye addition lines. Cytometry 1995;21:387-93. [CrossRef]
24. Loureiro J, Rodriguez E, Doležel J, Santos C. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann Bot 2006;98:679-89. [CrossRef]
25. Galbraith DW, Bartos J, Dolezel J. Flow cytometry and cell sorting in plant biotechnology. In:Sklar LA, editor. Flow Cytometry in Biotechnology. Oxford:Oxford University Press;2005. 291-322. [CrossRef]
26. Greihuber J, Temsch EM, Loureiro JC. Nuclear DNA content measurement. In:Doležel J, editor. Flow Cytometry with Plant Cells Analysis of Genes, Chromosomes and Genomes. Germany:Wiley-VCH Verlag GmbH and Co.;2007. 67-102. [CrossRef]
27. Khan AU, Mei H, Wilson T. A proposed function for spermine and spermidine:Protection of replicating DNA against damage by singlet oxygen. Proc Natl Acad Sci U S A 1992;89:11426-7. [CrossRef]
28. Jedrzejczyk I, Sliwinska E. Leaves and seeds as materials for flow cytometric estimation of the genome size of 11 Rosaceae woody species containing DNA-staining inhibitors. J Bot 2010;2010:930895. [CrossRef]
29. Ohri D, Bhargava A, Chatterjee A. Nuclear DNA amounts in 112 species of tropical hardwood --new estimates. Plant Biol 2004;6:555-61. [CrossRef]
30. Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Ann Bot 1995;76:113-76. [CrossRef]
31. Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms, 583 new estimates. Ann Bot 1997;80:169-96. [CrossRef]
32. Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in Angiosperms and their modern uses, 807 new estimates. Ann Bot 2000;86:859-909. [CrossRef]
33. Sinski I, Bosco DD, Pierozzi NI, Maia JD, Ritschel PS, Quecini V. Improving in vitro induction of autopolyploidy in grapevine seedless cultivars. Euphytica 2014;196:299-311. [CrossRef]
34. Yang N, Rong E, Li Q, Dong J, Du T, Zhao X, et al. Tetraploid induction and identification of Gossypium arboreum. Agric Sci 2015;6:436-44. [CrossRef]
35. Widoretno W. In vitro induction and characterisation of tetraploid Patchouli (Pogostemon cablin Benth.) plant. Plant Cell Tissue Organ Cult 2016;125:261-7. [CrossRef]
36. Regalado JJ, Carmona-Martín E, Querol V, Veléz CG, Encina CL, Pitta-Alvare SI. Production of compact petunias through polyploidisation. Plant Cell Tissue Organ Cult 2017;129:61-71. [CrossRef]
37. Rao S, Kang X, Li J, Chen J. Induction, identification and characterisation of tetraploidy in Lycium ruthenicum. Breed Sci 2019;69:160-8. [CrossRef]
38. Li S, Lin Y, Pei H, Zhang J, Zhang J, Luo J. Variations in colchicineinduced autotetraploid plants of Lilium davidii var. unicolor. Plant Cell Tissue Organ Cult 2020;141:479-88. [CrossRef]
39. Niazian M, Nalousi AM. Artificial polyploidy induction for improvement of ornamental and medicinal plants. Plant Cell Tissue Organ Cult 2020;142:447-69. [CrossRef]