1. INTRODUCTION
Arsenic is leading cause of poisoning in health concern worldwide and about 6.8% reported by U.S. Geological Survey [1]. According to the water survey reported that the groundwater has significant concentration of As or F in India and Asian countries. Chronic exposure of arsenic ingestion through water and food leads to skin disorders, high blood pressure, increased incidence of cancer and diabetes in humans [2]. As per the recent research report, the level of arsenic high in aquatic field has been attained by the manmade activities, such as agricultural and industrial effluence. In the aquatic environment, the redox and methylation reactions are inter-converted inorganic arsenic (arsenite (As3+) and arsenate (As5+)) [3].
Arsenic, lead, cadmium, and chromium compounds lead to the expression of gene specific such as heat shockprotein 70 and metallothionein [4]. Activation of intracellular Ca2+ production and hypersensitivity, modulaterelease of lymphokines that altered the delayed exposure in mammals [5]. Metallothionein (Mt) is well-known metalloid and metal binding protein, which response in ion homeost and detoxification, moreover it plays an important role in fortification against metalotoxicity and oxidative stress [6]. Furthermore, the expression profile of Nrf2 (cellular resistance to oxidants regulator) depend on XMEs like glutathione peroxidase catalase, superoxide dismutase (SOD), and NAD (P) H: quinone oxidoreductase 1 which is responsible for maintaining cellular redox homeostasis. SODs are one of the major cellular defenses against oxidative stress knocked down superoxide radicals (O2−), and it is precursor in reactive O2 Radicals [7]. The activity of SOD was defined by its ability to inhibit the oxidation reaction of SOD2 hydroxylamine hydrochloride with superoxide to nitrite. In addition, SOD is one of the significant metalloenzymes and impied for the degradation of free radicals O2•− to O2 and H2O2. Arsenic derivatives induce oxidase stress by reacting with dithiol groups [8]. The protein p53 has an essential role in the transcriptional regulation of metallothionein, which shows an unusual regulatory role for p53. Besides, p53 - intracellular Ca2+ levels increased by arsenic in lymphocytes.Response to level of arsenic changes in intercellular Ca2+ regulates the formation of p53-DNA complex Instead, different mechanisms are involved in the activation of kinases in p53 responses to arsenite in the cell system [9].
In aquaculture system, the numbers of fishes are found to impairment of immune and physiological function following to exposure of heavy metals. In brown trouts (Salmo trutta), whole transcriptome analysis revealed that the down regulation of numerous factors of the complements systems are following exposure of Ni, As, Cu, Cd, Pb, Fe, and Zn [10]. Arsenic causes perturbation of defense systems in Danio rerio, Clarius batrachus, and Sparus aurata [11,12].
However, gene expression profiling of fish species in water system following As exposure has not been reported. Dawkinsia tambraparniei (Silas, 1953) is an endangered cyprinid only found in Tamiraparani River in southern Western Ghats, India [13]. Recently, D. tambraparniei population have declined due to anthropogenic activities such as mining, alterations of habitat, brooders collection for aquarium trade and environmental cruses. Due to these threats, D. tambraparniei announced as endangered in IUCN red list [14]. Effective conservation of wild stocks requires information on growth and condition. Hence, D. tambraparniei as a suitable model that we used for current experiment to investigate the alteration of the gene expression and regulate stress response by arsenic exposure using RT-PCR.
2. MATERIALS AND METHODS
2.1. Fish
D. tambraparniei (weight 10 ± 0.5 g, length 12.8 ± 05 cm SL) were collected from Thamirabarani River basin in Tamil Nadu, India. Fish were acclimated under standardized laboratory conditions. Commercial diet was supplemented in consistent temperature (28 ± 0.5°C), oxygen above 6.0 mg/L and 12 h light and dark photoperiod. The experiment followed by the guidelines of animal ethics committee, Government of India.
2.2. Chemicals Required
Arsenic (III) oxide (As2O3) (CAS Number 1327-53-3) and sodium arsenate (NaAsO2) (CAS Number 10048-95-0) purchased from Sigma-Aldrich. The gene specific primer, RNase free DNase I, cDNA synthesis PCR master mixture kit procured from ThermoFisher Scientific, India. Analytical grade chemical was used for experiment.
2.3. Experimental Setup
The experimental fishes divided into control (without test solution) and treatment group, each group contains 08 fishes. As2O3 (22.60 ppm) and NaAsO2 (31.65 ppm) dissolved in distilled water as test solution. In the experimental study, the final LC50 concentrations used based on previous experiment [15]. The test solutions added in treatment group and the experiment were followed for 42 days. Three experimental samples were collected in each group, anaesthetized for 5 min by clove oil and the brain, heart, and liver tissues were stored at −80°C for future use.
2.4. Synthesis of cDNA
Initially, RNA extraction performed by Trizol (HiMedia, India) according to manufacturer’s procedure. Quality of RNA samples assessed by agarose-formaldehyde gel electrophoresis method (agarose 1% w/v; formaldehyde 16%). Quantity and quality of experimental RNA samples analyzed by Nano-drop Spectrophotometer (ThermoFisher, USA) absorbance at A 260/280 ratio of 1.8–2.0. 1 μg of RNA subjected to cDNA synthesis conducted in 20 μl reactions, containing 1 mg of RNA, 2 μl of oligo dT primers, 2 μl of dNTPs, 400 mL of 2× HiScript mix, 0.5 μl of reverse transcriptase. The cDNA synthesis following to the manufacturer’s procedure, samples were stored at −20°C.
2.5. Gene Expression Analysis by Semi-Quantitative PCR
The primers Mt, p53, SOD, and β-actin [Table 1] used for amplification of gene expression by PCR analyzer. PCR reaction conducted in 20 µl volume having of master mix (10 µl), cDNA (1 µl) and primer (0.5 µl of forward and reverse primers). Amplification was carried out following PCR program: 3 min at 95°C then 30–35 cycles of 95°C for 1 min, annealing 55–58°C for 30 s and extension at 72°C for 1 min followed by a final extension at 72°C for 7–10 min.
Table 1: Oligonucleotide primers used to analysis of expression by semi-quantitative PCR.
| Primer name | Primers usage | Nucleotide sequences | Size |
|---|---|---|---|
| β-actin | Sense | 5’ TCATCACCATTGGCAATGAGC 3’ | 250 |
| β-actin | Antisense | 5’ CACTGTGTTGGCGTACAGGT 3’ | |
| Mt | Sense | 5’ ATGGATCCNTGCGAWTGC 3’ | 190 |
| Mt | Antisense | 5’ CANTGACAGCAGCTGGAGC 3’ | |
| p53 | Sense | 5’ GGGCAATCAGCGAGCAAA 3’ | 180 |
| p53 | Antisense | 5’ GGGCAATCAGCGAGCAAA 3’ | |
| SOD | Sense | 5’ TGAGACACGTCGGAGACC 3’ | 230 |
| SOD | Antisense | 5’ TGCCGATCACTCCACAGG 3’ |
SOD: Superoxide dismutase.
2.6. Statistical Analysis
The expression patterns mentioned in graphical representation by GelAnalyzer software. Expression values analysis carried out by Image J 1.8 software and statistical significance established at P < 0.05.
3. RESULTS AND DISCUSSION
Genes such as SOD, p53, Mt, and β-Actin were analyzed for expression studies. Fragments of 230bp, 180bp, 190bp, and 250bp were amplified for SOD, p53, Mt, and β-actin, respectively. When compared with expression pattern, in As2O3 ide treated fish the heart shows Mt expressed laterally at 42 days while in NaAsO2 group the expression fold was higher in 28 days itself. The case was happened vice versa in brain where the As2O3 treated group displays Mt expression at 28 days and prolonged to 42 days. In NaAsO2 group Mt lacks expression in brain. Significantly, no changes found in liver for most genes after 7 days of arsenic exposure. While high concentrations of As2O3 and NaAsO2 accumulated in the liver, along with the gills, only the Mt gene was upregulated after 28 days of exposure [Figures 1 and 2].
 | Figure 1: Expression of genes coding for Mt isoforms in various organs of Dawkinsia tambraparniei relative to different days with sodium arsenite (NaAsO2) and arsenic trioxide (As2O3). [Click here to view] |
 | Figure 2: Effect of As2O3 (A) and NaAsO2 (S) on heart. Analysis of Mt, p53, SOD genes expression was performed on heart by means of semi quantitative reverse transcriptase PCR. Amplification was done for 7 days to 42 days samples with respective genes (C for control). [Click here to view] |
Our study depicted that after 14 days of exposure to As2O3 drastically increase the expression of mRNA of p53 gene in comparison with control brain tissue; whereas the significantly elevated expression of mRNA of p53 gene was found in the NaAsO2 exposed heart and liver in comparison with the As2O3 group [Figure 3]. There are no significant expression observed in SOD after 7 days of brain exposure to both As2O3 and NaAsO2. Up-regulated p53 and Mt genes only expressed at 28 days by predictable marker of heavy metal pollution, but there were no significant differences observed in between the two arsenical exposure environments. Nevertheless, genes that respond to oxidative stress SOD not affected by these arsenic derivatives within brain, heart, and liver cells [Figure 4].
 | Figure 3: Expression of genes coding for p53 in various organs of D. tambraparaniei relative to different days with NaAsO2 and As2O3. [Click here to view] |
 | Figure 4: Expression of genes coding for superoxide dismutase (SOD) isoforms in various organs of D. tambraparaniei relative to different days with NaAsO2 and As2O3. [Click here to view] |
Our results show that p53 expression is substantially similar in heart, liver, and brain exposed to both As2O3 and NaAsO2. Thus, it indicates that Mt gene plays an important role in elimination of arsenical derivatives. In brain at 28th day of exposure with As2O3, there is increase expression of p53 was seen. Individually delayed division of cells as well as decreases the time taken for G1-cell cycle arrested and damage of DNA/mitochondrial membrane have been generally stated that elevated expressions of p53 in exposed cells to As2O3 as an agents for genotoxicity [Figure 3] [16].
Tissue specific Mt transcript was highest in liver, followed by brain and heart but displaying relatively low expression in heart and brain depending on the dose days. The tissue distribution patterns at the basal level of Mt in D. tambraparniei were similar to that of river puffer fish Mt, which exhibits higher expression in the liver, and minimum expression in the kidney and brain and in the arsenic induced cases the same happens i.e., expression at maximum in liver and minimum in heart and brain [17].
In rainbow trout similar accumulation pattern was observed in gills and kidney when cadmium was induced in time course manner [18]. Likewise, higher expression of Mt observed in liver compared to gills and kidney in carp and rainbow trout [19,20]. In the case of Kryptolebias marmoratus (mangrove rivulus), the highest Mt mRNA expression was found on brain region [21]. These reports showed that the tissue distribution of Mt mRNA expression has species specificity. In this report, the liver was found to be the major site of Mt expression in D. tambraparniei. Taken together, the true mode of action of arsenic derivatives in heart and brain cannot predicted with partial series expression patterns of mRNA, but we hypothesize that aqueous arsenic is a strong modulator of Mt transcripts in both heart and brain [Figures 2 and 5].
 | Figure 5: Effect of As2O3 (A) and NaAsO2 (S) on brain. Analysis of Mt, p53, SOD genes expression was performed on heart by means of semi quantitative reverse transcriptase PCR. Amplification was done for 7 days to 42 days samples with respective genes (C for control). [Click here to view] |
Based on these findings it is well known that the expression of Mt depends on arsenic accumulation on different organs of fish. Harrison and Klaverkamp (1989) observed that Cd was maximally accumulated in organs such as kidney, and later gills similar to the liver but not as evident in the muscles of rainbow trout [22]. Mt expression induction by these arsenic derivatives has been noted in other fish species such as mullet, carp, and rainbow trout, not only in liver but also in other tissues such as brain and heart [23-25].
It is known that when fishes were exposed to arsenic compound results in increase p53 expression due to damage in mitochondrial membrane and damage in DNA, finally ROS. According to early report, PLHC-1 and primary trout cells that p53 was not involved in apoptosis when it was triggered with model chemotherapeutic agents [26]. Coincidentally, they concluded that there was some non-specific p53 pathway in fish. The same happened in As2O3 and sodium arsenite (NaAsO2) triggered toxicity that no increased expression occurs. Normally, p53 was activated it resulting in either growth arrest or apoptosis [Figure 6].
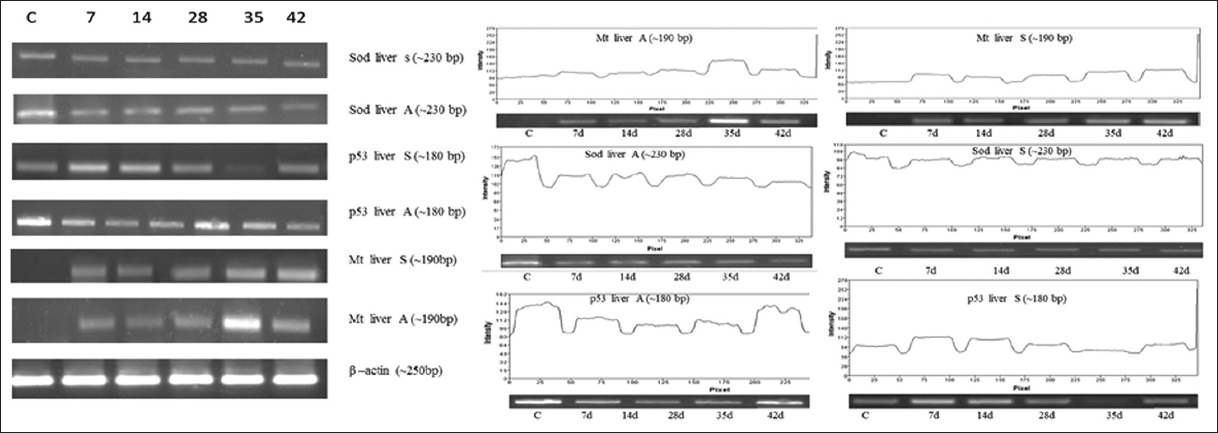 | Figure 6: Effect of As2O3 (A) and NaAsO2 (S) on liver. Analysis of Mt, p53, SOD genes expression was performed on heart by means of semi quantitative reverse transcriptase PCR. Amplification was done for 7 days to 42 days samples with respective genes (C for control). [Click here to view] |
Any aquatic organisms exposed with heavy metals such as arsenic, the inside reaction on cell apparently end up with damage to DNA, ROS production results in either upregulation or activation of p53 pathway and thus arrests G1 phase in cell division got arrested. In such belongings, the activated p53 signaling pathway permits those affected cells to do DNA editing and repairing. It was either followed by apoptosis or by usual cell division [27-29]. Even the cell was got arrested on G1 phase the damaged cells was repaired by DNA repair mechanism and allow them to survive either for certain period or permanently [30]. Conversely, the amounts of arsenic actually accumulated were not directly proportional to SODs mRNA levels in tissues such as brain, heart, and liver in fishes: the highest arsenic accumulation occurred in liver, followed by the brain and heart. Comparable conclusion of metallothionein (Mt) gene expression was observed in common carp exposed to another heavy metal like cadmium [31,32].
4. CONCLUSION
The conclusion of the current study reported that arsenic exposure to promoted oxidative stress activation and apoptosis in an endemic fish D. tambraparniei. Consequently, the expression profiling analysis of selected marker genes such as SOD, p53 and Mt indicates that impact of stress analysis by concentration and time specific manner. Stress-specific SOD excess and imbalance of oxidative stress could be considered as an early sign of cellular damage. Therefore, the results of the present study strongly suggested biomonitoring as a valuable technique for detecting heavy metal pollution in freshwater ecosystems for its direct application and analysis.
5. AUTHORS’ CONTRIBUTIONS
Dr. Selvakumar Sakthivel has initiated conception, design, analysis and interpretation of data; Dr. Anand Raj Dhanapal and Dr. Vijay Velu has continued interpretation of data and reviewing the primary draft; Mr. Venkataa Suresh Munisamy, Mr. Mohammed Parvez Nasirudeen and Mr. Varatharaji Selvaraj has continued acquisition of data and formatting; Dr. Annadurai Gurusamy has conceptualized the primary draft construction following final drafting, editing, reviewing, all correspondence, and supervision.
6. FUNDING
There is no funding to report.
7. CONFLICT OF INTEREST
The authors declare no conflicts of interest.
8. ETHICAL APPROVALS
Principles of laboratory animal care were followed and all procedures were conducted according to the guidelines established by the National Institutes of Health (Guidelines for the use of fishes, 2004), and every effort was made to minimize suffering. This study was approved by the institutional ethical committee of Manonmanaiam Sundaranar University [Ethical Clearance (EC), Biosafety and Animal Welfare Committee].
9. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
11. ACKNOWLEDGMENTS
We thank to Department of Molecular Biology, School of Biological Sciences, Madurai Kamaraj University, Madurai, Tamil Nadu, India, for provide space and animal maintaining during this work.
REFERENCES
1. DeSimone LA, Hamilton PA, Gilliom RJ. Quality of water from domestic wells in principal aquifers of the United States, 1991-2004 Overview of major findings. Water. Circular 1:1991-2004. USA;U.S. Geological Survey;2009. [CrossRef]
2. Abdul KS, Jayasinghe SS, Chandana EP, Jayasumana C, De Silva PM. Arsenic and human health effects:A review. Environ Toxicol Pharmacol 2015;40:828-46. [CrossRef]
3. Ro SH, Bae J, Jang Y, Myers JF, Chung S, Yu J,
4. Dunn AM, Hofmann OS, Waters B, Witchel E. Cloaking Malware with the Trusted Platform Module. Vol. 25. In:Proceeding. 20th USENIX Security Symposium. 2011. 395-410.
5. Crigna AT, Uhlig S, Elvers-Hornung S, Klüter H, Bieback K. Human adipose tissue-derived stromal cells suppress human, but not murine lymphocyte proliferation, via Indoleamine 2,3-dioxygenase activity. Cells 2020;9:2419. [CrossRef]
6. Montalbano G, Olivotto I, GermanàA, Randazzo B. Evaluation of the hair cell regeneration and claudin b and phoenix gene expression during exposure to low concentrations of cadmium and zinc in early developing zebrafish larvae. Comp Biochem Physiol C Toxicol Pharmacol 2021;248:109116. [CrossRef]
7. Ighodaro OM, Akinloye OA. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX):Their fundamental role in the entire antioxidant defence grid. Alexandria J Med 2018;54:287-93. [CrossRef]
8. Yadav G, Srivastava PK, Singh VP, Prasad SM. Light intensity alters the extent of arsenic toxicity in
9. Ganapathy S, Li P, Fagman J, Yu T, Lafontant J, Zhang G,
10. Webster TM, Bury N, van Aerle R, Santos EM. Global transcriptome profiling reveals molecular mechanisms of metal tolerance in a chronically exposed wild population of brown trout. Environ Sci Technol 2013;47:8869-77. [CrossRef]
11. Ferrario D, Gribaldo L, Hartung T. Arsenic exposure and immunotoxicity:A review including the possible influence of age and sex. Curr Environ Health Rep 2016;3:1-12. [CrossRef]
12. Kumari B, Kumar V, Sinha AK, Ahsan J, Ghosh AK, Wang H,
13. Silas EG. New fishes from the Western Ghats, with notes on
14. Kannan K, Johnson JA, Malleshappa H. Growth and condition of an endangered fish Dawkinsia tambraparniei (Cypriniforms:Cyprinidae) from southern Western Ghats, India. Aqua Int J Ichthyol. 2013;19:61-6.
15. Sakthivel S, Dhanapal AR, Paulraj LP, Gurusamy A, Venkidasamy SB, Thiruvengadam M,
16. Das B, Sahoo S, Mallick B. HIWI2 induces G2/M cell cycle arrest and apoptosis in human fibrosarcoma via the ROS/DNA damage/p53 axis. Life Sci 2022;293:120353. [CrossRef]
17. Kim JH, Wang SY, Kim IC, Ki JS, Raisuddin S, Lee JS,
18. Samuel MS, Datta S, Khandge RS, Selvarajan E. A state of the art review on characterization of heavy metal binding metallothioneins proteins and their widespread applications. Sci Total Environ 2021;775:145829. [CrossRef]
19. Liu Y, Chen Q, Li Y, Bi L, Jin L, Peng R. Toxic effects of cadmium on fish. Toxics 2022;10:622. [CrossRef]
20. Chakraborty SB. Non-essential Heavy Metals as Endocrine Disruptors:Evaluating Impact on Reproduction in Teleosts. Vol. 74. Proceedings of the Zoological Society;2021. 417-31. [CrossRef]
21. Chiarelli R, Martino C, Agnello M, Bosco L, Roccheri MC. Autophagy as a defense strategy against stress:Focus on
22. Bayen S. Occurrence, bioavailability and toxic effects of trace metals and organic contaminants in mangrove ecosystems:A review. Environ Int 2012;48:84-101. [CrossRef]
23. Chowdhury MJ, Baldisserotto B, Wood CM. Tissue-specific cadmium and metallothionein levels in rainbow trout chronically acclimated to waterborne or dietary cadmium. Arch Environ Contam Toxicol 2005;48:381-90. [CrossRef]
24. Filipovi?V, Raspor B. Metallothionein and metal levels in cytosol of liver, kidney and brain in relation to growth parameters of
25. AnvariFar H, Amirkolaie AK, Miandare HK, Ouraji H, Jalali MA, ÜçüncüS?. Apoptosis in fish:Environmental factors and programmed cell death. Cell Tissue Res 2017;368:425-39. [CrossRef]
26. Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development 1995;121:1065-76. [CrossRef]
27. Hale AJ, Smith CA, Sutherland LC, Stoneman VE, Longthorne VL, Culhane AC,
28. Jagoda SV, Dixon KM. Protective effects of 1,25 dihydroxyvitamin D3 and its analogs on ultraviolet radiation-induced oxidative stress:A review. Redox Rep 2020;25:11-6. [CrossRef]
29. Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression?Cell Death Differ 2018;25:104-13. [CrossRef]
30. Blackman S. A matter of life and cell death. Scientist 2005;19:22-3.
31. Hermesz E, Ábrahám M, Nemcsók J. Tissue-specific expression of two metallothionein genes in common carp during cadmium exposure and temperature shock. Comp Biochem Physiol C Toxicol Pharmacol 2001;128:457-65. [CrossRef]
32. Da Silva Alves DC, Healy B, Pinto LA, Cadaval TR Jr., Breslin CB. Recent developments in Chitosan-based adsorbents for the removal of pollutants from aqueous environments. Molecules 2021;26:594. https://doi.org/10.3390/molecules26030594 PMid:33498661 PMCid:PMC7866017