1. INTRODUCTION
Agriculture farming is one of the main leading economic sectors of India with 58% of the population dependency on food as a basic necessity of life. Weeds, pests, microbiological, parasite infection, and insect activity can reduce annual crop yield by up to 20% [1]. Applying pesticides to crops have long been a standard agricultural technique to prevent crop yield loss. For the past 40 years, among all the many types of pesticides, those organophosphates with a P-O-C bond in the core structure have been utilized extensively to protect various crops [2]. Furthermore, according to estimates, the insecticide group accounted for 34% of total organophosphate usage worldwide [3]. Monocrotophos was discovered to be a systemic but non-specific organophosphate that is responsible for groundwater contamination due to its high hydrophilic properties [4]. Excessive use of monocrotophos contributes to eutrophication of water bodies, disruption of nutrient element cycling in the ecosystem, and loss of biodiversity in nature [5]. Based on toxicity, Environmental protection agency classified monocrotophos into Class I toxicity level, which is a group of highly toxic compounds [6]. Normally, monocrotophos has a half-life of 17–96 days which can be expanded up to 2500 days in the case of properly stored technical grade monocrotophos, which is generally sprayed for crop cultivation [7]. Monocrotophos residues were detected in human blood at 0.79 ng/mL, which is known to cause adverse effects because it targets acetylcholine esterase [8]. Besides human toxicity, monocrotophos was also screened in Danio rerio with a remarkable increase in micronuclei [9]. In other studies, severe effects such as paralysis in Caenorhabditis elegans [10], and growth retardation on Odontella mobilensis and Coscinodiscus centralis [11] were observed. Because of these toxic effects, there is an urgent need to remove and detoxify monocrotophos from the environment with a rapid and sustainable approach.
Conventional methods for removing monocrotophos have numerous drawbacks. Toxic compounds generated during degradation or removal, bioaccumulation, and biomagnification of these pesticides, use of hazardous chemicals, use of heat, and heavy economic load are the main drawbacks of conventional methods [3]. Thus, the use of microbial or other biological methods that primarily rely on microorganisms, photodegradation, or phytodegradation is the ultimate solution to all of these drawbacks. Because microorganisms are so diverse in terms of habitat adaptation and metabolism, this enormous diversity could be explored to remove and degrade monocrotophos from the environment. Microbial removal of monocrotophos is an environmentally friendly and cost-effective method that does not require the use of acids or chemicals [1]. Furthermore, microbes can eliminate multiple pollutants at the same time. Several soil bacteria, fungi, and actinomycetes were isolated and tested for their ability to degrade monocrotophos. Monocrotophos can be utilized by microbes as the sole source of carbon and phosphorous. Bacillus subtilis [3], P. fluorescens [12], Starkeya novella [13], Pseudomonas synxantha, Salmonella enterica [6], Agrobacterium radiobacter, Clavibacter michiganense, and Arthrobacter atrocyaneus [14] were reported for monocrotophos degradation under various conditions with varying concentrations. Fungi such as Aspergillus niger, A. flavus [15], Fusarium oxysporum, Escovopsioides nivea, and Cunninghamella echinulate were also isolated and screened for monocrotophos degradation [14].
However, the use of those selected bacteria, which can degrade pesticide rapidly and are compatible in the mixed culture at the field level, is a less evaluated approach. With the provision of broad-ranged enzymes, compatible consortia could completely mineralize hazardous compounds and convert them to less toxic or non-toxic metabolites. Furthermore, an efficient consortium could reduce the degradation time that would otherwise be increased by pure inoculum. Besides this, self-sustainability under variable environmental conditions is an another advantage of consortia which might be proved helpful in the field-level study of pesticide degradation [16]. The purpose of this study was to test the potency of monocrotophos degradation in an aqueous medium as well as in the field using a microcosm with selected pure isolates and their consortium. Plant growth promotion ability was evaluated alongside cytotoxicity and phytotoxicity analysis. As a result, this study will supplement information about the concomitant ability of monocrotophos degradation and toxicity alleviation by polluted site isolated bacteria. Optimally, formulated bacterial consortium might be used for reclamation of monocrotophos and other organophosphate contaminated sites.
2. MATERIALS AND METHODS
2.1. Sample Collection, Characterization, and Isolation of Monocrotophos Degrader
The wastewater sample was collected from Amla khadi (21° 41’ 52.02”N, 72° 01’ 09.91”E), which is located near the Gujarat Industrial Developmental Corporation (GIDC) area of Ankleshwar, Gujarat, India. The sample was collected in a sterile carboy container and transported to the laboratory with the maintenance of 4°C cooling conditions. The collected sample was proceeded for the analysis of physicochemical parameters such as chemical oxygen demand (COD), biological oxygen demand (BOD), total dissolved solids (TDS), total suspended solids (TSS), and dissolved oxygen (DO) [17].
The sample enrichment was carried out by the addition of 50 mL of collected sample into 150 mL of sterile minimal salt medium (MSM) supplemented with 100 ppm of filter-sterilized monocrotophos. Flask was incubated in an incubator shaker (120 rpm) at 37 ± 1°C for 7 days. After incubation, 0.1 mL of serially diluted (up to 10−8) samples were spread on 100 ppm of monocrotophos containing MSM agar plates. All the plates were incubated at 37°C for 3–4 days. After incubation, morphologically distinct and separated colonies were preserved on the same MSM agar slant for further study.
2.2. Monocrotophos Degradation
All bacterial isolates recovered from the wastewater samples were screened for monocrotophos tolerance ability by plate assay. The MSM agar plates having different concentrations (100–5000 ppm) of monocrotophos were tested. Among the all-bacterial isolates, coded as MCPB1, MCPB2, and MCPB3 had maximum monocrotophos tolerant ability and were further used for the subsequent experiments as individual pure bacterial inoculum and as a consortium consisting of a mixture of MCPB1, MCPB2, and MCPB3.
The pesticide degradation study was carried out in sterile MSM broth containing (per L) of of 1.5 g NH4NO3, 1.5 g K2HPO4, 0.5 g KH2PO4, 0.2 g MgSO4, 0.5 g NaCl, and pH 7.0 [18]. The filter-sterilized 500 ppm monocrotophos was added separately into the MSM broth as a sole carbon source. Activated bacterial culture (A600: 1.0) was centrifuged and the collected cell pellet was washed thrice with sterile distilled water subsequently, a resuspended bacterial cell pellet (individual pure bacterial inoculum and a consortium) was used for inoculation. All flasks were incubated in shaking condition at 120 rpm, 37°C for 15 days. 5 mL of sample was withdrawn from the flask at 24 h of a regular time interval to determine monocrotophos degradation. The absorbance of the supernatant was measured at 254 nm using a UV-vis spectrophotometer and the degradation percentage was calculated using the following equation [19].
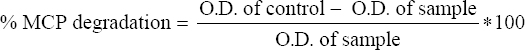
2.3. Analytical Methods for Degradation Study
Samples were prepared with thrice extraction of cell-free supernatant in ethyl acetate. The extract was passed through anhydrous sodium sulfate to remove excess water molecules. Then, the concentrated sample was redissolved in ethyl acetate and filtered using a 0.22 μm pore size syringe filter for further analysis. High performance liquid chromatography (HPLC) was performed as an analytical tool for confirmation of monocrotophos degradation. HPLC (YL 9100) was performed using the BDS HYPERSIL C18 column having a diameter of 250 × 4.6 mm and particle size 5 μL. Acetonitrile and HPLC grade water (7:3 v/v) at isocratic mode were used as carrier mobile phase. 20 μL of the sample was injected at a 1 mL/min flow rate of the mobile phase. Degradation was detected using a UV detector at a 214 nm scanning range [1].
2.4. Identification of Isolates
Isolates were identified using morphological characteristics followed by molecular identification using 16s rRNA gene sequencing. DNA was isolated and extracted by the phenol/chloroform method [20]. 16s rRNA gene region was amplified using 357 forward primers and 1391 reverse primers after removing chemical contaminants and quality control of DNA. Sequencing was carried out after a reaction mixture preparation using ABI 3500xl Genetic Analyzer. The sequences were compared with the 16S rRNA gene sequences available at NCBI, GenBank, using the BLAST algorithm. Based on the sequence similarity score, first 15 sequences were aligned. Phylogenetic tree construction for evolutionary history and analysis was completed using the Neighbor-Joining (NJ) plot and Molecular Evolutionary Genetics Analysis 7 (MEGA7) tool [21,22]. The p-distance method was used for the calculation of evolutionary distance [23]. Obtained sequences were submitted to the NCBI gene bank.
2.5. Qualitative and Quantitative Estimation of Enzyme Phosphotriesterase
The selected monocrotophos degrading isolates were further screened for the production of the enzyme phosphotriesterase (esterase and phosphatase). 5 μL culture was put on the center of MSM agar plates supplemented with 500 ppm monocrotophos and 1% of tributyrin (MCP + tributyrin agar [TBA]) for the assay of esterase production. Nutrient agar plates with 1% tributyrin (NA + TBA) were also spot inoculated by isolates to check whether the secreted enzyme is inducible or constitutive. Similarly, for phosphatase production, Pikovskaya’s agar (PVK) agar plates were prepared with and without monocrotophos, to check the phosphatase production. After inoculation, plates were incubated at 37°C for 48 h [24]. The solubilization index (SI) was calculated using the following formula [25].
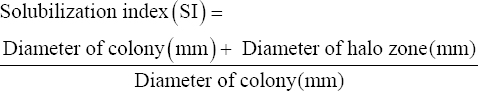
Intra and extracellular enzyme production was determined from cell pellet and supernatant. Esterase assay was performed with some modifications of Hriscu et al. (2013) [26]. Briefly, 0.5 mL cell-free supernatant and lysed pellet were mixed in a tube containing 1 mL of 50 mM tris Cl buffer (pH 8.0), 0.5 mL of 1 mmol/L para nitro phenyl acetate (p-NPA), and 2.5 mL double distilled water. The reaction was carried out at 37°C for 30 min and then the absorbance measurement was done at 400 nm. With some modifications, a phosphatase assay was performed as explained by Behera et al. (2017) [27]. Briefly, 1.0 mL of cell-free supernatant and lysed pellet were mixed in a tube containing 4 mL of 50 mM tris Cl buffer (pH = 8.0) and 0.5 mL of 1 mmol/L para nitro phenyl phosphate (p-NPP). This reaction mixture was incubated at 37°C for 30 min. After incubation, 4 mL of 0.5 M NaOH was added to stop the reaction and measured the absorbance at 410 nm.
2.6. Monocrotophos Degrading Enzyme Profile
Cell-free supernatant was subjected to 70% ammonium sulfate precipitation under cooling conditions. Precipitates were dissolved in 50 mM Tris-Cl buffer (pH 8.0) and dialyzed against the same buffer for desalting excess ammonium sulfate. After dialysis, samples were passed through the Sephadex G-100 column, already equilibrated with 50 mM Tris-Cl buffer (pH 8.0). Collected and concentrated fractions were passed through a 50 mM Tris-Cl pre-equilibrated DiEthyl Amino Ethyl (DEAE) Sepharose column. Sample elution was carried out using a NaCl gradient up to 1 M [28]. Screening of esterase assay [26], phosphatase assay [27], and estimation of protein concentration [29] was carried out at each step of purification. Purified samples were used for SDS-PAGE analysis to determine the molecular weight of organophosphate degrading enzymes. About 10% resolving gel and 5% stacking gel were prepared from monomers of acrylamide (30%) and bisacrylamide (0.8%). Samples were loaded into wells after mixing with 5X protein loading dye (Reduced). A prestained protein ladder (HiGenoMB, Himedia Pvt. Ltd.) was also loaded to compare the molecular weight of sample proteins. After successfully separating samples, the gel was stained with Coomassie brilliant blue. The destained gel was observed to determine molecular weight by comparing it with the standard ladder [30].
2.7. Identification of Degraded Metabolite
Gas chromatography–mass spectrometry (GC-MS) was performed according to the method described by Dash and Osborne (2020) [1] for the identification of metabolites generated after monocrotophos degradation. Shimadzu Nexis GC-2030 was used with a column internal diameter of 0.25 mm and length of 30 m. 1 μL of the sample was injected using helium at a 1 mL/min flow rate with a constant value. The temperature of the injector was set at 260°C. The oven temperature was kept at 60°C initially for up to 2 min and was increased up to 300°C withheld for 6 min. Results were analyzed using the National Institute of Standards Technology (NIST) library.
2.8. Toxicity Analysis
2.8.1. Cytotoxicity assay
Toxicity analysis of standard pesticide and its degraded product was performed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay on peripheral blood mononuclear cells (PBMC). PBMCs were recovered using a sterile Ficoll density gradient (HiSep™ LSM 1077 having density 1.077 + 0.0010 g/mL) and washed twice with phosphate-buffered saline. Cells were tested for viability through a 0.4% trypan blue exclusion assay. For proliferation, RPMI 1640 (HiKaryoXL™ RPMI medium consists of L-Glutamine, FBS, PHA-M, Penicillin, Streptomycin, and Sodium bicarbonate) was suspended with isolated PBMCs. 2 × 106 cells/mL were distributed in sterile and unused 96 well microtiter plates. The seeded plate was incubated in a CO2 incubator containing 5% CO2 for 24 h. After incubation, monocrotophos degraded metabolite, and standard monocrotophos were added to each well in triplicate with control. The plate was incubated for 48 h at 37°C in a CO2 incubator containing 5% CO2. MTT (5 mg/mL) was added to each well and after 4 h of incubation, 100 μL of 0.04 N HCl with isopropanol was also added. At 590 nm and 620 nm, wavelength optical density was measured and cell viability was calculated [31]. The percentage of cell viability was calculated according to the following equation [32].
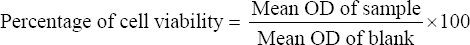
2.8.2. Field trial
Phytotoxicity screening was carried out with 100 g of active soil inoculated with each pure isolate and consortium at the density of 1 × 108 cells/mL. After the addition of 500 ppm monocrotophos, the soil was mixed properly. Surface-sterilized seeds of Cicer arietinum were seeded in each set-in triplicate. Microcosm experiment was carried out with different sets: (1) Soil + Seeds (2) Soil + Seeds + MCP (3) Soil + Seeds + isolate MCPB1 (4) Soil + Seeds + MCP + isolate MCPB1 (5) Soil + Seeds + isolate MCPB2 (6) Soil + Seeds + MCP + isolate MCPB2 (7) Soil + Seeds + isolate MCPB3 (8) Soil + Seeds + MCP + isolate MCPB3 (9) Soil + Seeds + Consortium (10) Soil + Seeds + MCP + Consortium. Random rotational cycles were controlled under greenhouse conditions to maintain a 5% moisture level. After 15 days and 30 days of incubation, screening of pesticide degradation was carried out. After germination of seeds, parameters such as root length, shoot length, dry weight of root and shoot, fresh weight of root and shoot, chlorophyll content, and Vigor index were calculated [1,33]. After a performance of a phytotoxicity assay, isolates were also screened for their plant growth-promoting ability.
2.9. Determination of Plant Growth Promoting (PGP) Activity
The indol acetic acid (IAA) production was measured using a 5 mL supernatant (with and without monocrotophos). The supernatant was mixed with 0.5 mL of orthophosphoric acid and 5 mL of Salkowaski’s reagent. The development of pink color indicates the IAA production, which was quantified at 530 nm [34]. To check siderophore production ability, Chrome Azurol S agar plate with and without monocrotophos was spot inoculated with bacterial isolates at the center and incubated for 4–5 days for pink to orange halos development around the colony. Ammonia production was estimated by inoculation of isolates in peptone water and after incubation of 96 h, positive confirmation was obtained by the development of brown color through the mixing of drops of Nessler’s reagent in the supernatant [35].
2.10. Statistical Analysis
Experiments were carried out in triplicates, and the results presented are the mean ± standard error of the three replicates.
3. RESULTS AND DISCUSSION
3.1. Sample Collection, Characterization, and Isolation of Monocrotophos Degrader
Microorganisms are known as omnipresent and play an important role in ecological balance. Microbes found in polluted areas are well known for their ability to develop resistance as well as degrade and detoxify a wide range of pollutants and toxic compounds. One such contaminated site is present in Ankleshwar, known as Amla khadi. As it receives various types of liquid and solid wastes from different industries, that is, dyes, pharmaceuticals, agro-chemicals, and fertilizer units; it was declared a red zone by GIDC [36,37]. Pseudoxenthomonas, Rhodococcus, and Burkholderia were isolated from Amla khadi and studied for degradation of xenobiotic compounds such as phenanthrene, chrysene, and pyrene [38-40]. In this study, Amla khadi was further explored for the isolation of monocrotophos degrading microbes.
At the sample collection site, a temperature of 29°C and a pH of 7.4 were recorded. The physicochemical parameters COD, BOD, DO, TDS, and TSS were determined to be 204 mg/L, 63 mg/L, 4.40 mg/L, 892 mg/L, and 290 mg/L, respectively. COD and BOD levels were found to be higher than the acceptable limit, indicating the presence of organic, inorganic, and domestic waste material, as well as elements from industrial effluent [41,42]. Furthermore, the BOD: COD ratio was found to be 0.308, which is <0.5, indicating the presence of some chemical compounds in the collected sample [43]. Hence, based on this analysis, the collected sample was found polluted. A total of 15 different bacterial isolates were obtained after enrichment and were further screened for monocrotophos tolerance ability.
3.2. Monocrotophos Degradation
A total of 15 bacterial isolates were tested for tolerance to monocrotophos. The most potent bacterial isolates, namely, MCPB1, MCPB2, and MCPB3, were discovered to be capable of tolerating monocrotophos up to 5000 ppm. Maximum growth was observed at 500 ppm of provided monocrotophos from various concentration ranges. The spectrophotometric method was used to examine preliminary monocrotophos degradation. After 24 h of incubation in shaking conditions, 30%, 28%, 25%, and 45% degradation were observed for MCPB1, MCPB2, MCPB3, and consortium for 500 ppm of monocrotophos, respectively. After 48 and 72 h of incubation, 52%, 50%, 49%, 62%, and 75%, 73%, 70%, 86% degradation was observed, respectively. After 96 h of incubation, up to 82%, 81%, 78%, and 95% degradation was achieved by all three pure bacterial isolates and consortium.
HPLC was used to perform a confirmatory monocrotophos degradation study. Figure 1 depicts a comparison HPLC profile of undegraded and degraded monocrotophos. The observed standard monocrotophos retention time was 2.61 min. The gradual decrease of the extracted samples’ peak confirms monocrotophos degradation [Table 1]. Thus, degradation analysis revealed that consortium is more effective than pure bacterial isolates for monocrotophos degradation. Another study found that Rhodococcus phenolicus and R. ruber mediated degradation ranged from 30% to 45% after 96 h of incubation [18]. After 17 days of incubation, Sidhu et al. (2015) [6] reported 67.8%, 16.58%, and 6.67% degradation using isolated P. synxantha, B. subtilis, and S. enterica. Within 8 days, Bacillus megaterium and A. atrocyaneus could tolerate and degrade monocrotophos up to 1000 ppm [44]. After 6 days, B. subtilis KPA1 can degrade 1000 ppm of monocrotophos [3]. Serratia marcescens and Bacillus aryabhattai can degrade monocrotophos at 1200 ppm and 4500 ppm, respectively [2,1]. In our study, rapid degradation and a high tolerance of monocrotophos were observed with a selected consortium, suggesting the compatibility of bacteria.
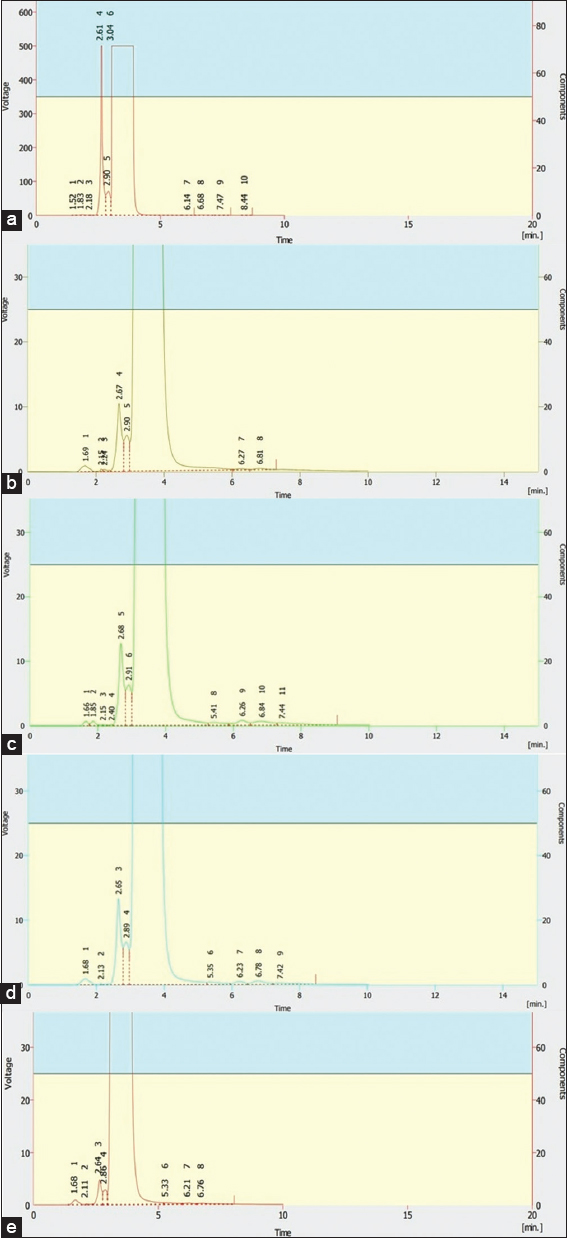 | Figure 1: Confirmation of monocrotophos (500 ppm) degradation. (a) High performance liquid chromatography chromatogram of standard undegraded monocrotophos. (b) Isolate MCPB1 treated monocrotophos in minimal salt medium (MSM). (c) Isolate MCPB2 treated monocrotophos in MSM. (d) Isolate MCPB3 treated monocrotophos in MSM. (e) Bacterial consortium treated monocrotophos in MSM. [Click here to view] |
Table 1: Confirmation of monocrotophos degradation carried out by HPLC.
| Isolated bacteria | Retention time (min) | Peak area (mV/s) | Peak height (mV) |
|---|---|---|---|
| Monocrotophos (500 ppm) | 2.61 | 30366.585 | 4999.919 |
| MCPB1 | 2.67 | 1076.204 | 104.287 |
| MCPB2 | 2.68 | 1036.651 | 127.555 |
| MCPB3 | 2.65 | 1365.613 | 133.618 |
| Consortium | 2.64 | 511.610 | 47.130 |
HPLC: High performance liquid chromatography.
3.3. Identification of Isolates
The microscopic analysis indicated that isolate MCPB1 was found to be Gram-positive, while colonials features showed orange pigment production with a convex surface and size of 1 mm diameter. MCPB2 had green diffusible pigmentation, translucent, alveolate surface, and colony size of 4 mm diameter with Gram-negative staining. MCPB3 was found to be rod-shaped Gram-negative, with a smooth colony surface of 3 mm in size and red pigmentation. 16s rRNA gene sequencing indicated that the coded MCPB1, MCPB2, and MCPB3 were identified as Gordonia terrae (1383 bp of 16s rRNA gene sequence), Pseudomonas aeruginosa (1461 bp of 16s rRNA gene sequence), and Serratia surfactantfaciens (1495 bp of 16s rRNA gene sequence) consequently. The nucleotide sequences were submitted to the NCBI gene bank with accession numbers MZ352177, MZ352166, and MZ352238.
Among isolated bacteria, both P. aeruginosa and S. surfactantfaciens are members of the phylum Proteobacteria and the class Gammaproteobacteria, while G. terrae is found as a member of the phylum Actinobacteria. In phylogenetic analysis, G. terrae was found within the clade of other Gordonia lacunae strain BS2 and was compared with Mycobacterium tuberculosis H37 as an outgroup. While P. aeruginosa was classified within other P. aeruginosa clades, Acinetobacter calcoaceticus was considered as an outgroup. S. surfactantfaciens was significantly identified with a maximum score of 1784 in sequence alignments and discovered to be closest to a clade of S. marcescens strain KRED. Pseudocitrobacter anthropic was considered an outgroup. Pseudomonas and Bacillus species are the most commonly found degrader of monocrotophos, parathion, aldrin, chlorpyrifos, endosulfan, and endrin [45]. S. surfactantfaciens is a member of the genus Serratia. Another member of the same genus, S. marcescens has previously been reported to degrade other organophosphates such as parathion, fenitrothion, and chlorpyrifos in MSM as well in soil [46]. Gordonia spp. JAAS1 has also been reported to degrade chlorpyrifos into metabolite 3,5,6-trichloro-2-pyridinol within an incubation period of 72 h [47].
3.4. Enzymatic Screening
By plate assay, all three selected isolates, P. aeruginosa, S. surfactantfaciens, and G. terrae, were found to produce esterase and phosphatase (Phosphotriesterase). Table 2 shows the efficiency of monocrotophos solubilization by esterase and phosphatase production. G. terrae has the highest solubilization efficiency for esterase secretion on tributyrin supplemented NA plate and on monocrotophos + tributyrin supplemented MSM agar plate, with SI 2.26 ± 057 and 2.1 ± 0.1. As with esterase, G. terrae also has the highest solubilization efficiency for phosphatase production by plate assay on both PVK and monocrotophos supplied PVK, with SI of 1.78 ± 0.028 and 1.56 ± 0.057. Compared to all three bacterial isolates, S. surfactantfaciens has the lowest enzyme production efficiency. Furthermore, a zone of solubilization was observed around spot inoculated colonies in both types of plates; supplemented with and without monocrotophos, indicating that isolates are constitutively capable of producing extracellular phosphotriesterase. Bhadbhade et al. (2002) [44] also reported constitutive production of monocrotophos degrading phosphatase enzymes. 63.95 ± 1.52 IU/mL and 5.10 ± 1.25 IU/mL enzyme activity were found for cytosolic enzyme and membrane-bound enzyme during a study conducted by Paracoccus spp. M-1, respectively, according to Jia et al. (2006) [48].
Table 2: Solubilization index for extracellular monocrotophos degrading enzymes produced by selected bacterial isolates.
| Isolates | Esterase solubilization index | Phosphatase solubilization index | ||
|---|---|---|---|---|
| TBA+NA | TBA+MCP | PVK | PVK+MCP | |
| MCPB1 | 2.26±0.057 | 2.1±0.1 | 1.78±0.028 | 1.56±0.057 |
| MCPB2 | 1.46±0.057 | 1.36±0.057 | 1.66±0.057 | 1.47±0.046 |
| MCPB3 | 1.43±0.057 | 1.13±0.057 | 1.28±0.028 | 1.1±0.011 |
Values are means±S.E., n=3. TBA: Tributyrin agar, NA: Nutrient agar, MCP: Monocrotophos, PVK: Pikovskaya’s agar
The ability of isolates to produce enzymes and utilize substrates was also tested using a spectrophotometric assay. Extracellular esterase activities measured using p-NPA and MCP were obtained at 95.99 ± 0.89, 91.46 ± 1.13, 88.84 ± 0.55, 98.21 ± 0.07 IU/mL, and 85.38 ± 0.13, 81.27 ± 0.15, 78.69 ± 0.20, 91.95 ± 0.03 IU/mL subsequently for G. terrae, P. aeruginosa, S. surfactantfaciens and consortium after 96 h of incubation. Esterase activity increased up to 96 h after incubation. Intracellular esterase activity was found to be negligible in comparison to extracellular esterase activity. After 96 h of incubation, the maximum extracellular phosphatase activity was also obtained [Figure 2a and b]. 85.74 ± 0.60, 85.36 ± 0.10, 81.58 ± 0.41, 89.36 ± 0.11 IU/mL, and 79.94 ± 0.54, 80.46 ± 0.41, 74.88 ± 0.64, 84.32 ± 0.09 IU/mL extracellular phosphatase activities were measured using p-NPP and MCP as a substrate for G. terrae, P. aeruginosa, S. surfactantfaciens, and consortium. Similar to esterase, intracellular phosphatase activities were also found to be negligible when compared to extracellular phosphatase activities [Figure 3]. B. megaterium was found to be a higher esterase producer than A. atrocyaneus in an enzymatic screening study conducted by Bhadbhade et al. (2002) [44]. After 48 h of incubation, organophosphate hydrolase and phosphatase produced by B. aryabhattai showed 71.49 ± 0.48 IU/mL and 62.02 ± 0.78 IU/mL enzyme activities for MCP as a substrate. Using p-NPP as a substrate, 95.67 ± 0.46 IU/mL phosphatase activity was discovered [1].
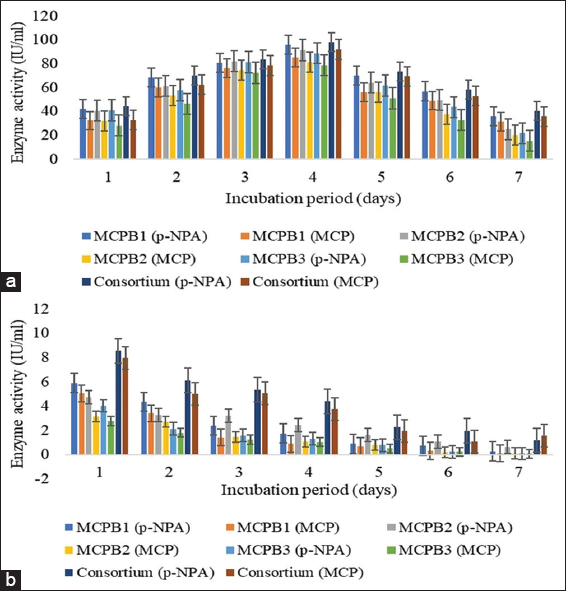 | Figure 2: Screening of esterase enzyme activity. (a) Extracellular esterase activity (IU/ml). (b) Intracellular esterase activity (IU/ml) (means ± S.E., n = 3). [Click here to view] |
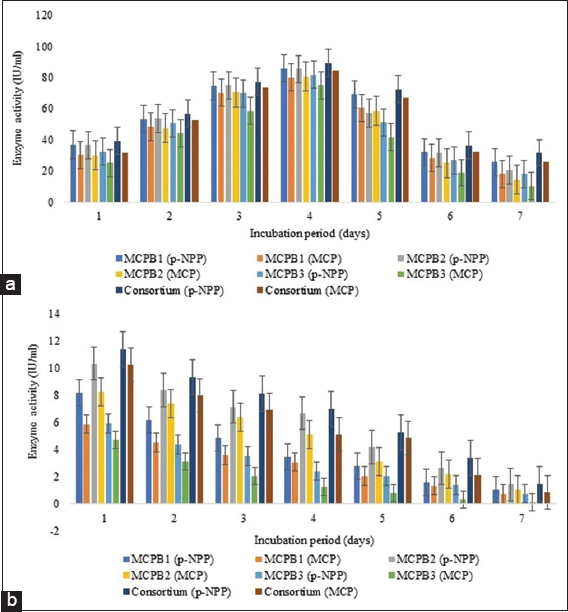 | Figure 3: Screening of phosphatase enzyme activity. (a) Extracellular phosphatase activity (IU/ml). (b) Intracellular phosphatase activity (IU/mL) (means ± S.E., n = 3). [Click here to view] |
3.5. Monocrotophos Degrading Enzyme Profile
Purification indicated that the highest purification fold was obtained for enzymes of S. surfactantfaciens with a 48.43% yield and 102.57 IU/mg specific activity. About 46.78% purification yield was observed for G. terrae and P. aeruginosa [Table 3]. Sodium dodecyl sulphate poly acrylamide gel electrophoresis of dialyzed and purified enzymes indicated bands of ~35 kDa and ~67 kDa for G. terrae ~35 kDa, ~48 kDa, and ~63 kDa for P. aeruginosa and ~35 kDa, ~45 kDa, and ~63 kDa for S. surfactantfaciens [Figure 4]. This observation was similar to the other results. Bands of 33 kDa and 67 kDa were observed during PAGE analysis for monocrotophos degrading enzyme, isolated and purified from Penicillium aculeatum and Fusarium pallidoroseum. In the same study, some metal ions Fe2+/Fe3+, Co2+/Co3+, and Hg2+ were found to have an inhibitory effect on monocrotophos degrading enzymes. Hydrolases for organophosphate parathion, two bands of 35 kDa and 43 kDa were observed from Flavobacterium spp. [28].
Table 3: Purification of monocrotophos degrading enzymes indicating specific activity, purification fold, and yield of purification.
| Purification steps | Isolate MCPB1 | Isolate MCPB2 | Isolate MCPB3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Specific activity (IU/mg) | Yield (%) | Purification fold | Specific activity (IU/mg) | Yield (%) | Purification fold | Specific activity (IU/mg) | Yield (%) | Purification fold | |
| Crude extract | 12.41 | 100 | 1 | 12.41 | 100 | 1 | 11.2 | 100 | 1 |
| Dialysis | 22.6 | 82.32 | 1.82 | 22.6 | 82.32 | 1.82 | 19.33 | 81.19 | 1.72 |
| Sephadex G-100 purification | 86.51 | 70.79 | 6.97 | 86.51 | 70.79 | 6.97 | 90.32 | 67.24 | 8.06 |
| DEAE sepharose purification | 95.21 | 46.78 | 7.67 | 95.21 | 46.78 | 7.67 | 102.57 | 48.43 | 9.15 |
DEAE: Di-Ethyl Amino Ethyl
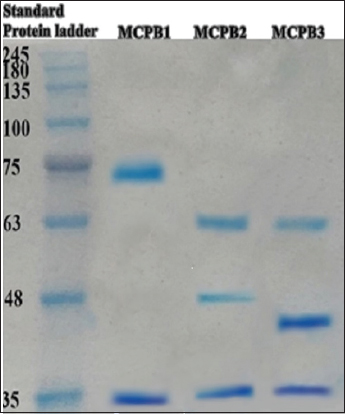 | Figure 4: Sodium dodecyl sulfate poly acrylamide gel electrophoresis analysis for determination of molecular weight (kDa) of enzymes (Phosphotriesterase) responsible for monocrotophos degradation, lane 1-standard molecular weight prestained protein ladder, lane 2-purified enzyme from isolate MCPB1, lane 3-purified enzyme from isolate MCPB2, Lane 4-purified enzyme from isolate MCPB3. [Click here to view] |
3.6. Degraded Metabolite Identification
Based on a comparison with the NIST library, degraded metabolites were identified using GC MS. For consortium treated monocrotophos, trimethyl phosphate [M=140] and acetamide, N-acetyl N-methyl (M = 115) were detected as an intermediate degraded metabolites with a retention time of 4.26 min and 5.5 min, respectively [Table 4]. Standard monocrotophos was also screened simultaneously with M = 223 with a retention time of 9.67 min [Figure 5]. In GC MS, cyclohexanone, 2-cyclohexylidene, which is used to make monocrotophos, was also detected as an intermediate metabolite. In addition to cyclohexanone, 2-cyclohexylidene, dimethyl phosphate, monomethyl phosphate, phosphoric acid, glutaraldehyde, and acetic acid, (1-methyl ethyl ester) were detected as monocrotophos degradation products [1]. Dhanya et al. (2016) [49] investigated cyclohexanone degradation using Pseudomonas with phenol production. The metabolite detected in GC MS during monocrotophos degradation in the present study is acetamide, N-acetyl N-methyl, which were also detected as an intermediate metabolite of monocrotophos degradation by Srinivasulu et al. (2017) [18]. According to Acharya et al. (2015) [3], N- methylacetoacetamide was degraded to methylamine and carbon dioxide and these products were detected during monocrotophos degradation using isolate B. subtilis KPA-1. Monomethyl phosphate, N-methylacetoacetamide, N- methyl butyramide, and ethylthiophosphoric acid were also reported as degraded metabolites of monocrotophos, studied using S. marcescens JAS16 [2]. Methyl phosphate, dimethyl phosphate, and 3-hydroxy-N-methyl butyramide were reported as intermediate products during a screening of monocrotophos contaminated soils by B. licheniformis, B. subtilis, and P. stutzeri. Saravanan et al. (2020) [24], studied monocrotophos degradation using B. subtilis isolated from Perionyx gut area, with identification of 1-[2-hydroxyphenyl]-1-butanone, 6-oxo-4-[3-fluorophenyl]-1,6-dihydro pyrimidine, 1,1-dimethyl ethyl diethyl ester phosphoric acid, 1-methyl-dimethyl ester imidazole-4,5-dicarboxylic acid, and hexahydro-4-[3-phenyl-2-propenyl]-2-thioxo-pyrimidine-4,6-dione as degraded metabolites.
Table 4: Identification of monocrotophos degraded metabolites by GC MS.
| Identified Degraded Metabolites | Retention Time (min) | M.W. of the Metabolite | Chemical Formula |
|---|---|---|---|
| Standard Monocrotophos | 9.67 | 223 | C7H14O5NP |
| Trimethyl phosphate | 4.26 | 140 | C3H9O4P |
| Acetamide, N-acetyl-N-methyl | 5.5 | 115 | C5H9NO2 |
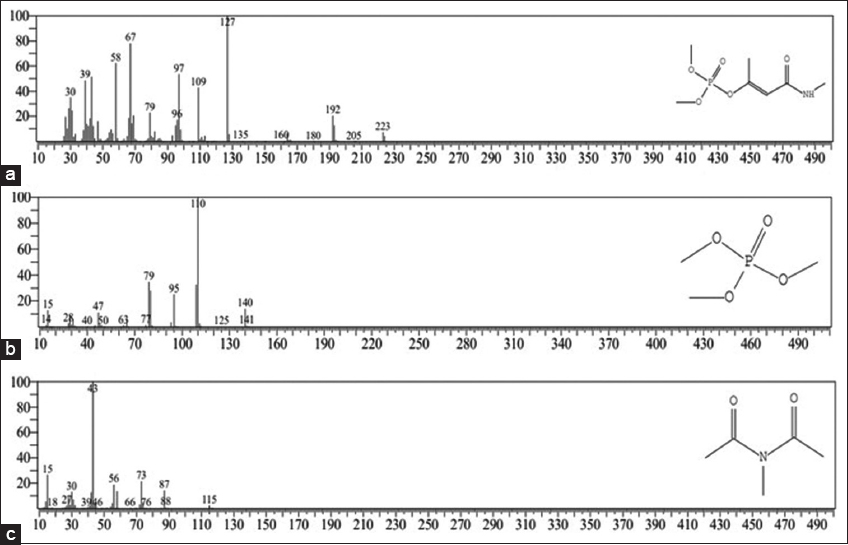 | Figure 5: Gas chromatography-mass spectrometry analysis of degraded metabolites produced by consortium treated monocrotophos (500 ppm). (a) Monocrotophos (m/z 223). (b) Trimethyl phosphate (m/z 140). (c) Acetamide N-acetyl N- methyl (m/z 115). [Click here to view] |
For the degradation of monocrotophos, different type of microbial and enzymatic metabolic reactions was reported (1) O-demethylation, (2) N-demethylation, (3) Hydroxylation at N-methyl moiety, and (4) Breakage of phosphate and crotanamide linkage [48]. In this present study, two different intermediate metabolites of monocrotophos were identified. As described earlier, phosphatase and esterase commonly known as phosphotriesterase are the organophosphate hydrolases enzymes (OPH); mainly involved in monocrotophos degradation. Based on the previous reports and detected metabolites in GC-MS, it can be assumed that monocrotophos was degraded by phosphotriesterase to intermediate trimethyl phosphate and acetamide, N-acetyl N-methyl due to C-O bond breakage. Trimethyl phosphate was later converted to phosphoric acid, while acetamide, N-acetyl N- methyl was converted to methylamine, acetic acid, and ammonia, followed by carbon dioxide [1,44]. Thus, according to this reported and presumed degradation pathway, monocrotophos degradation results in primarily less toxic end products.
3.7. Toxicity Analysis
3.7.1. Cytotoxicity assay
The purpose of a cytotoxicity assay is to determine whether a compound has stimulatory or inhibitory effects on cells. The colorimetric method, which involves specific dye treatment and formazan production with the mitochondrial dehydrogenase enzyme, is used to determine cell death or survival. The amount of viable cells is directly related to the formation of purple-colored formazan [50]. Table 5 shows the cell viability obtained for degraded monocrotophos products and undegraded monocrotophos at 100 ppm and 500 ppm. Cell viability testing revealed that degraded products were less toxic than non-degraded monocrotophos. Another screening of the MTT assay was carried out by Al-Sarar et al. (2015) [51] on the CHO-K1 cell line for which after 24 h of incubation, an MTT50 value of 113.52 ± 10.65 was obtained for monocrotophos in serum-free media. Another genotoxicity study was carried out on lymphocytes of Rattus norvegicus through comet and micronucleus assay showed the toxic effect of monocrotophos through generation of single- and double-strand breaks in DNA and micronucleus in mono and polychromatic erythrocytes [52]. Alkahtane et al. (2018) [53] observed depolarization of mitochondria, apoptic events, and DNA strand breakage in HaCaT cells at 600 ng/mL of monocrotophos.
Table 5: Cytotoxicity assay on PBMC.
| Concentration of monocrotophos | Treatment | PBMC viability (%) |
|---|---|---|
| 0 ppm | Control | 99.48±0.49 |
| 100 ppm | Non-degraded MCP | 7.07±0.23 |
| MCPB1 degraded MCP | 86.97±0.47 | |
| MCPB2 degraded MCP | 86.62±0.72 | |
| MCPB3 degraded MCP | 84.65±0.48 | |
| Consortium degraded MCP | 91.18±0.77 | |
| 500 ppm | Non-degraded MCP | 2.53±0.20 |
| MCPB1 degraded MCP | 72.19±0.33 | |
| MCPB2 degraded MCP | 71.34±0.26 | |
| MCPB3 degraded MCP | 67.15±0.29 | |
| Consortium degraded MCP | 85.04±0.72 |
Values are means±S.E., n=3. PBMC: Peripheral blood mononuclear cell, MCP: Monocrotophos
3.7.2. Field trial
In microcosm screening, an experimental setup was carried out using different combinations with control. After 30 days of incubation, 91.8 ± 0.15% 93.05% ± 0.09, 91.88% ± 0.12, 97.52 ± 0.07% degradation was observed for MCPB1, MCPB2, MCPB3, and consortium inoculated and monocrotophos treated soil experiment set up. While active soil with only 500 ppm of monocrotophos without any isolated bacterial inoculum showed 43.36% of degradation. No more residues of monocrotophos were detected in aerial parts of the plant. This could be due to the cometabolism of monocrotophos by plant and inoculated isolates. The degradation rate in soil was found to be decelerated after 15 days of incubation. This was also supported by another monocrotophos degradation study using B. aryabhattai in soil. The degradation rate was found to be slower after 20 days of incubation. After 40 days of incubation, 99% degradation of monocrotophos was observed with a shorter half-life of monocrotophos [1]. Higher substrate availability enhances the production of extracellular phosphotriesterase enzymes, which could explain why the enzymes degrade monocrotophos so quickly in the beginning. While changes in the physiological conditions of soil such as temperature, pH, and oxygen, concentration may affect the availability of monocrotophos for isolates and thus degradation rate was observed to be slower after a certain period [15]. In another microcosm screening, carried out in soil using two isolates A. niger and A. flavus in sterilized sandy loam soil, after 30 days of incubation, 99% degradation was observed with more efficient degradation by A. niger than A. flavus using a first-order kinetic reaction [19]. Thus, a consortium of isolated bacteria was found to be more effective in microcosm and in MSM for monocrotophos degradation than a pure isolate.
3.8. PGP Characterization of Isolates
In addition to the degradation ability of monocrotophos in soil, isolates were tested for plant growth-promoting parameters. Phosphate is a limiting macronutrient that acts as a fertilizer to improve plant growth and development. Phosphate solubilizing bacteria improve plant availability of insoluble phosphate through a variety of mechanisms [54]. Siderophores are low-molecular-weight compounds secreted by microbes to scavenge iron from their surroundings. Plants use iron for basic cellular metabolic processes, regulation, growth, and metabolism. In addition to this primary function, siderophores are used to chelate copper, manganese, and cobalt [55]. The presence of monocrotophos reduces the plant growth-promoting efficiency of isolates, according to the SI value for phosphate and siderophore production [Table 6]. IAA production, which increases root surface area for efficient nutrient uptake and absorption [56], was found to range between 15 and 75 mg/mL for all three isolates. IAA production was influenced by monocrotophos in the same way that iron and phosphate solubilization were affected. MCPB3 was (S. surfactantfaciens) was detected with the maximum amount of ammonia production compared to MCPB1 and MCPB2 [Table 6].
Table 6: Screening of plant growth promoting (PGP) parameters of selected isolates.
| Isolated bacteria | Siderophore production (SI) | IAA production (mg/ml) | Ammonia production | |||
|---|---|---|---|---|---|---|
| CAS plates | CAS+MCP plates | Tryptophan | Tryptophan+MCP | Peptone water | Peptone water+MCP | |
| MCPB1 | 3.45±0.05 | 3.22±0.02 | 20.34±0.19 | 15.89±0.05 | Positive | Positive |
| MCPB2 | 2.48±0.05 | 2.33±0.03 | 71.1±0.04 | 59.61±0.24 | Positive | Positive |
| MCPB3 | ND | ND | 61.63±0.1 | 39.08±0.02 | Positive | Positive |
ND: Not detected, values are means±S.E., n=3. IAA: Indol acetic acid, CAS: Chrome Azurol S agar, MCP: Monocrotophos
Pesticide toxicity was observed in soil supplemented with only monocrotophos, which showed significant retardation in shoot, root, chlorophyll content, and Vigor index [Table 7]. In contrast, pure isolates and consortium inoculated seedlings with monocrotophos showed a significant increase in plant growth when compared to uninoculated seedlings [Figure 6]. Thus, in soil-based experiments, isolates can simultaneously perform functions of monocrotophos degradation and plant growth enhancement, as well as stress elimination caused by monocrotophos. PGP bacteria were also found to induce and accelerate monocrotophos degradation in the plant system [57]. Parallel effect of pesticide degradation and plant growth was also observed with monocrotophos degrading and PGP B. aryabhattai which was reported positive for ammonia production with IAA production ability of 161.99 ± 0.02 mg/mL, while was reported negative for iron-chelating siderophore production [1].
Table 7: Assessment of chlorophyll content and Vigor index for Microcosm experiment in the aspect of monocrotophos effect.
| Microcosum set up | Chlorophyll content (mg/mL) | Vigor index |
|---|---|---|
| Soil+seed | 0.536±0.12 | 4040 |
| Soil+MCP+seed | 0.096±0.09 | 1306.06 |
| Soil+MCPB1+seed | 0.363±0.25 | 3976 |
| Soil+MCPB1+MCP+seed | 0.186±0.18 | 3026 |
| Soil+MCPB2+seed | 0.423±0.09 | 3969 |
| Soil+MCPB2+MCP+seed | 0.256±0.20 | 3419 |
| Soil+MCPB3+seed | 0.373±0.15 | 3896 |
| Soil+MCPB3+MCP+seed | 0.18±0.18 | 3182 |
| Soil+Consortium+seed | 0.786±0.11 | 4925 |
| Soil+Consortium+MCP+seed | 0.592±0.09 | 4388 |
Values are means±S.E., n=3. MCP: Monocrotophos
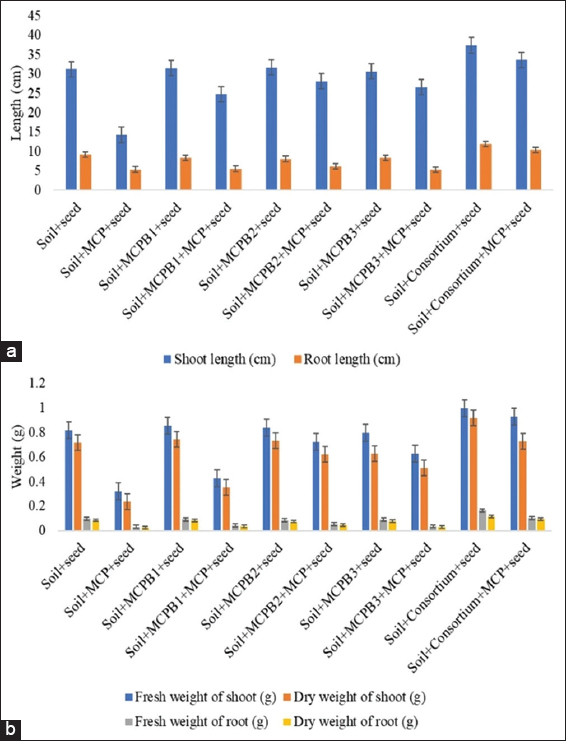 | Figure 6: Phytotoxicity assay carried out using microcosm approach. (a) Root and shoot length (cm) of Cicer arietinum with different combinations of experiment. (b) Fresh and dry weight (g) of root and shoot of C. arietinum (means ± S.E., n = 3). [Click here to view] |
4. CONCLUSION
The present study highlighted the efficacy of bacterial consortium for monocrotophos degradation. The ability of monocrotophos degradation was higher and faster with a selected consortium than that of a pure inoculum. Extracellular phosphotriesterase production aids in the breakdown of monocrotophos molecules, converting them into less toxic molecules. Furthermore, the toxicity assay represents monocrotophos bioconversion into a less hazardous end product. A group of selected bacterial isolates could also be tested for large-scale in situ monocrotophos and other organophosphate degradation.
5. ACKNOWLEDGMENT
The authors are thankful for providing the UGC-SAP-II and DST-FIST-I grants resources to initiate the experimental work. We are also sincerely thankful to Shri Ramakrishna Knowledge Foundation, Surat, Gujarat, India for their support.
6. AUTHOR CONTRIBUTIONS
Priyanka H. Jokhakar designed the study layout, performed experiments, analyzed data, and prepared the original manuscript, Pravin R. Dudhagara advised in the supervision of scientific research and proofreading for the final approval.
7. CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
8. ETHICAL APPROVAL
This study does not involve experiments on humans or on animal subjects.
9. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Dhash DM, Osborne JW. Biodegradation of monocrotophos by a plant growth-promoting Bacillus aryabhattai (VITNNDJ5) strain in artificially contaminated soil. Int J Environ Sci Technol 2020;17:1475-90. [CrossRef]
2. Abraham J, Silambarasa S. Bacterial degradation of monocrotophos and phyto-and cyto-toxicological evaluation of metabolites. Toxicol Environ Chem 2015;97:1202-16. [CrossRef]
3. Acharya KP, Shilpkar P, Shah MC, Chellapandi P. Biodegradation of insecticide monocrotophos by Bacillus subtilis KPA-1, isolated from agriculture soils. Appl Biochem Biotechnol 2015;175:1789-804. [CrossRef]
4. Singh S, Kumar V, Kanwar R, Wani AB, Gill JP, Garg VK, et al. Toxicity and detoxification of monocrotophos from ecosystem using different approaches:A review. Chemosphere 2021;275:130051. [CrossRef]
5. Buvaneswari G, Thenmozhi R, Nagasathya A, Thajuddin N. Screening of efficient monocrotophos degrading bacterial isolates from paddy field soil of Sivagangai district, Tamil Nadu, India. J Environ Sci Technol 2017;10:13-24. [CrossRef]
6. Sidhu PK, Dhanjal NIK, Cameotra SS, Sud D. Persistence and biodegradation of monocrotophos using soil microbes. Desalination Water Treat 2015;54:2293-8. [CrossRef]
7. Kaur R, Goyal D. Toxicity and degradation of the insecticide monocrotophos. Environ Chem Lett 2019;17:1299-324. [CrossRef]
8. Sharma A, Gill JP, Bedi JS, Pooni PA. Monitoring of pesticide residues in human breast milk from Punjab, India and its correlation with health associated parameters. Bull Environ Contam Toxicol 2014;93:465-71. [CrossRef]
9. D'Costa AH, Shyama SK, Kumar MK, Fernandes TM. Induction of DNA damage in the peripheral blood of zebrafish (Danio rerio) by an agricultural organophosphate pesticide, monocrotophos. Int Aquat Res 2018;10:243-51. [CrossRef]
10. Joshi AK, Nagaraju R, Rajini PS. Involvement of acetylcholinesterase inhibition in paralyzing effects of monocrotophos in Caenorhabditis elegans. J Basic Appl Zool 2018;79:33. [CrossRef]
11. Panneerselvam K, Dhandapani M, Jaikumar M. Growth inhibition effect of organophosphate pesticide, monocrotophos on marine diatoms. Indian J Geo Mar Sci 2015;44:1516-20.
12. Karunya SK, Reetha D. Biological degradation of chlorpyrifos and monocrotophos by bacterial isolates. Int J Pharm Biol Arch 2012;3:685-91.
13. Sun L, Zhu S, Yang Z, Chen Q, Liu H, Zhang J, et al. Degradation of monocrotophos by Starkeya novella YW6 isolated from paddy soil. Environ Sci Pollut Res 2016;23:3727-35. [CrossRef]
14. Bhalerao TS, Puranik PR. Microbial degradation of monocrotophos by Aspergillus oryzae. Int Biodeterioration Biodegradation 2009;63:503-8. [CrossRef]
15. Jain R, Garg V. Degradation of monocrotophos in soil, microbial versus enzymatic method. J Environ Occup Health 2015;4:44-52. [CrossRef]
16. Ma M, Zheng L, Yin X, Gao W, Han B, Li Q, et al. Reconstruction and evaluation of oil-degrading consortia isolated from sediments of hydrothermal vents in the South Mid-Atlantic Ridge. Sci Rep 2021;11:1456. [CrossRef]
17. Baird RB, Eaton AD, Clesceri LS. Standard Methods for the Examination of Water and Wastewater. 23rd ed. Washington, DC:American Public Health Association;2012.
18. Srinivasulu M, Nilanjan PC, Chakravarthi BV, Jayabaskaran C, Jaffer MG, Naga RM, et al. Biodegradation of monocrotophos by bacteria isolated from soil. Afr J Biotechnol 2017;16:408-17. [CrossRef]
19. Jain R, Garg V. Degradation of monocrotophos in sandy loam soil by Aspergillus sp. Iran J Energy Environ 2015;6:56-62. [CrossRef]
20. Wright MH, Adelskov J, Greene AC. Bacterial DNA extraction using individual enzymes and phenol/chloroform separation. J Microbiol Biol Educ 2017;18:18.2.48. [CrossRef]
21. Ziemert N, Jensen PR. Phylogenetic approaches to natural product structure prediction. Methods Enzymol 2012;517:161-82. [CrossRef]
22. Kumar S, Stecher G, Tamura K. MEGA7:Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [CrossRef]
23. Brocchieri L. Phenotypic and evolutionary distances in phylogenetic tree reconstruction. J Phylogen Evol Biol 2013;1:4. [CrossRef]
24. Saravanan P, Sridharan R, Krishnaswamy VG. Degradation of monocrotophos insecticide using Bacillus subtilis isolated from gut of perionyx sps. Austin J Environ Toxicol 2020;6:1031.
25. Paul D, Sinha SN. Phosphate solubilization potential and phosphatase activity of some bacterial strains isolated from thermal power plant effluent exposed water of river Ganga. CIBTech J Microbiol 2013;2:1-7.
26. Hriscu M, Chi?L, To?a M, Irimie FD. pH-Profiling of thermoactive lipases and esterases:Caveats and further notes. Eur J Lipid Sci Technol 2013;115:571-5. [CrossRef]
27. Behera BC, Yadav H, Singh SK, Mishra RR, Sethi BK, Dutta SK, et al. Phosphate solubilization and acid phosphatase activity of Serratia sp. Isolated from mangrove soil of Mahanadi river Delta, Odisha, India. J Genet Eng Biotechnol 2017;15:169-78. [CrossRef]
28. Jain R, Garg V, Dangwal K, Lily MK. Comparative purification and characterization of two distinct extracellular monocrotophos hydrolases secreted by Penicillium aculeatum and Fusarium pallidoroseum isolated from agricultural fields. Biosci Biotechnol Biochem 2013;77:961-5. [CrossRef]
29. Ranjini HS, Udupa EG, Kamath SU, Setty M, Hadapad B. A specific absorbance to estimate a protein by Lowry's method. Adv Sci Lett 2017;23:1889-91. [CrossRef]
30. Chong NF, Hussain H, Hamdin NE, Wee DHS, Nisar M, Yan WJ, et al. Higher resolution protein band visualisation via improvement of colloidal CBB-G staining by gel fixation. Chem Biol Technol Agric 2022;9:32. [CrossRef]
31. Nikbakht M, Pakbin B, Brujeni GN. Evaluation of a new lymphocyte proliferation assay based on cyclic voltammetry;An alternative method. Sci Rep 2019;9:4503. [CrossRef]
32. Kamiloglu S, Sari G, Ozdal T, Capanoglu E. Guidelines for cell viability assays. Food Front 2020;1:332-49. [CrossRef]
33. Amare T. Allelopathic effect of aqueous extracts of Parthenium (Parthenium hysterophorus L.) parts on seed germination and seedling growth of Maize(Zea mays L.). J Agric Crops 2018;4:157-63.
34. Rodrigues AA, Forzani MV, Soares RDS, Sibov ST, Vieira JD. Isolation and selection of plant growth-promoting bacteria associated with sugarcane. Pesqui Agropecuaria Trop 2016;46:149-58. [CrossRef]
35. Patel K, Jinal HN, Amaresan N. Inoculation of cucumber (Cucumis sativus L.) seedlings with salt-tolerant plant growth promoting bacteria improves nutrient uptake, plant attributes and physiological profiles. J Plant Growth Regul 2021;40:1728-40. [CrossRef]
36. Shirke KD, Pawar NJ. Enrichment of arsenic in the quaternary sediments from ankleshwar industrial area, Gujarat, India:An anthropogenic influence. Environ Monit Assess 2015;187:593. [CrossRef]
37. Chinchmalatpure AR, Camus D, Vaibhav R, Vibhute S, Kumar S, Prasad I, et al. Assessing groundwater contamination due to heavy metals and their spatial distribution in an industrial area of Western India. J Soil Salinity Water Qual 2018;10:178-85.
38. Patel V, Cheturvedula S, Madamwar D. Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J Hazard Mater 2012;201:43-51. [CrossRef]
39. Pathak H, Kantharia D, Malpani A, Madamwar D. Naphthalene degradation by Pseudomonas sp. HOB1:In vitro studies and assessment of naphthalene degradation efficiency in simulated microcosms. J Hazard Mater 2009;166:1466-73. [CrossRef]
40. Vaidya S, Jain K, Madamwar D. Metabolism of pyrene through phthalic acid pathway by enriched bacterial consortium composed of Pseudomonas, Burkholderia, and Rhodococcus (PBR). 3 Biotech 2017;7:29. [CrossRef]
41. Popa P, Timofti M, Voiculescu M, Dragan S, Trif C, Georgescu LP. Study of physico-chemical characteristics of wastewater in an urban agglomeration in Romania. ScientificWorldJournal 2012;549028. [CrossRef]
42. Agoro MA, Okoh OO, Adefisoye MA, Okoh AI. Physicochemical properties of wastewater in three typical South African sewage works. Pol J Environ Stud 2018;27:491-9. [CrossRef]
43. Messrouk H, Mahammed M, Touil Y, Amrane A. Physico-chemical characterization of industrial effluents from the town of Ouargla (South East Algeria). Energy Procedia 2014;50:255-62. [CrossRef]
44. Bhadbhade BJ, Sarnaik SS, Kanekar PP. Biomineralization of an organophosphorus pesticide, monocrotophos, by soil bacteria. J Appl Microbiol 2002;93:224-34. [CrossRef]
45. Huang Y, Xiao L, Li F, Xiao M, Lin D, Long X, et al. Microbial degradation of pesticide residues and an emphasis on the degradation of cypermethrin and 3-phenoxy benzoic acid:A review. Molecules 2018;23:2313. [CrossRef]
46. Cycon M, Zmijowska A, Wojcik M, Piotrowska-Seget Z. Biodegradation and bioremediation potential of diazinon-degrading Serratia marcescens to remove other organophosphorus pesticides from soils. J Environ Manage 2013;117:7-16. [CrossRef]
47. Abraham J, Shanker A, Silambarasan S. Role of Gordonia sp JAAS 1 in biodegradation of chlorpyrifos and its hydrolysing metabolite 3, 5, 6-trichloro-2-pyridinol. Lett Appl Microbiol 2013;57:510-6. [CrossRef]
48. Jia KZ, Cui ZL, He J, Guo P, Li SP. Isolation and characterization of a denitrifying monocrotophos-degrading Paracoccus sp. M-1. FEMS Microbiol Lett 2006;263:155-62. [CrossRef]
49. Dhanya V, Usharani MV, Jayadev P. Biodegradation of cyclohexanol and cyclohexanone using mixed culture of Pseudomonas in activated sludge process. J Environ Res Dev 2016;11:324.
50. Senthilraja P, Kathiresan K. In vitro cytotoxicity MTT assay in vero, HepG2 and MCF-7 cell lines study of marine yeast. J Appl Pharm Sci 2015;5:80-4. [CrossRef]
51. Al-Sarar AS, Bayoumi AE, Hussein HI, Abobakr Y. Cytotoxic effects of acephate, ethoprophos, and monocrotophos in CHO-K1 cells. CyTA-J Food 2015;13:427-33. [CrossRef]
52. Mishra V, Sharma S, Khatri S, Srivastava N. Evaluation of genotoxicity of monocrotophos and quinalphos in rats and protective effects of melatonin. Integr Pharmacol Toxicol Genotoxicol 2015;1:33-42. [CrossRef]
53. AlKahtane AA, Alarifi S, Al-Qahtani AA, Al-Yahya H, Ali D, Alomar SY, et al. Investigation of in vitro genotoxic effects of monocrotophos on human skin keratinocyte cells. Feb Fresenius Environ Bull 2018;27:2883-90.
54. Maldonado S, Rodriguez A, Avila B, Morales P, Gonzalez MP, Araya Angel JP, et al. Enhanced crop productivity and sustainability by using native phosphate solubilizing rhizobacteria in the agriculture of arid zones. Front Sustain Food Syst 2020;4:263. [CrossRef]
55. Ahmed E, Holmstrom SJ. Siderophores in environmental research:Roles and applications. Microbial Biotechnol 2014;7:196-208. [CrossRef]
56. Shreya D, Jinal HN, Kartik VP, Amaresan N. Amelioration effect of chromium-tolerant bacteria on growth, physiological properties and chromium mobilization in chickpea (Cicer arietinum) under chromium stress. Arch Microbiol 2020;202:887-94. [CrossRef]
57. Feng F, Ge J, Li Y, He S, Zhong J, Liu X, et al. Enhanced degradation of chlorpyrifos in rice (Oryza sativa L.) by five strains of endophytic bacteria and their plant growth promotional ability. Chemosphere 2017;184:505-13. [CrossRef]