1. INTRODUCTION
The widespread and long-term use of chemicals including atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine) herbicide in both agriculture and non-agricultural field is still a severe concern today. These compounds have the potential to runoff and leach through the soil leading to surface and groundwater contamination [1]. Most attentively, atrazine can cause serious human health problems such as endocrine disruption, central nervous system, reproductive system, immune system, and carcinogenic disorders [2]. Atrazine inhibits photosynthesis efficiency, superfluous energy dissipation in electron transport, and destroys cellular structure which resulted in the inhibition of growth in algae [3]. Moreover, atrazine has a moderately persistent, long half-life, and high mobility in soil than some other herbicides. Due to its high toxicity, persistence, and mobility in the environment, atrazine was prohibited by the European Union in 2004 [4], but it is still one of the most extensively used herbicides against weeds today in several countries, for example, annually 23 million kg in the USA [5], 27 million kg in Brazil, 16 million kg in Argentina [6], and 3 million kg in India [1]. Therefore, for a safe and sound environment, the rapid abolition of atrazine from the contaminated site has become very crucial.
Microorganisms have tremendous potential for bioremediation and herbicide degradation due to the presence of various catalytic enzymes [7]. The presence of such characteristics, microorganisms can degrade atrazine into different metabolites that act as a source of energy for other organisms. Many strains have been reported for their abilities in atrazine mineralization including members of the genera Pseudomonas, Bacillus, Burkholderia, Arthrobacter, Enterobacter, and Norcardioides [8-11]. In addition, several fungal species belonging to the genera Fusarium, Aspergillus, Penicillium, and Pleurotus have also been isolated and studied for degradation of atrazine [2,12,13]. Therefore, microorganisms can be chosen for easy and better strategies for the rescue of atrazine polluted sites ecofriendly.
In recent years, several review papers have been published on the degradation of atrazine in different aspects such as the impact of atrazine in the aquatic environment, technologies used to reduce the toxicity of atrazine as well as advantage and disadvantages [14,15]. In 2021, a similar review was published by Abd Rani et al. [16] that focused on only bacteria while fungi and yeast are neglected. In contrast, this review is a humble attempt to accumulate all the microbes associated with atrazine degradation in a single article that has already been gathered through vigorous research. This article also presents the clear degradation pathways along with the genes and enzymes involved in atrazine-degradation. This review will help researchers to develop a cost-effective and efficient microbiological technology for the remediation of atrazine-contaminated soil.
2. DEGRADATION OF ATRAZINE
2.1. Pathways of Atrazine-degradation
Degradation pathways of atrazine occur through three major different pathways that channel into cyanuric acid metabolism [1]. The degradation pathway is generally initiated by two enzymes, that is, atrazine chlorohydrolase and atrazine monooxygenase. The first pathway is initiated by the enzyme atrazine chlorohydrolase catalyzes the hydrolytic dechlorination of atrazine and produces hydroxyatrazine (HA) which is further converted into N-Isopropylammelide by the activity of atrazine ethylaminohydrolase and finally into Cyanuric acid later on by N-isopropylammelide isopropylaminohydrolase [17].
The second and third pathways are beginning with atrazine monooxygenase activity that degrades the atrazine into Deisopropylatrazine and Deethylatrazine, respectively [18]. In the second pathway, the enzyme s-triazine hydrolase transforms the Deisopropylatrazine into deisopropylhydroxyatrazine which is then converted into 2,4-Dihydroxy-6-(N’-ethyl)amino-1,3,5-triazine by 2,4-Dihydroxy-6-(N’- ethyl)amino-1,3,5-triazine aminohydrolase. Later, ethylaminohydrolase convert it into Cyanuric acid. In the third pathway, Deethylatrazine is transformed into deisopropyldeethylatrazine by deethylatrazine monooxygenase which is then converted into cyanuric acid through several steps [18] [Figure 1].
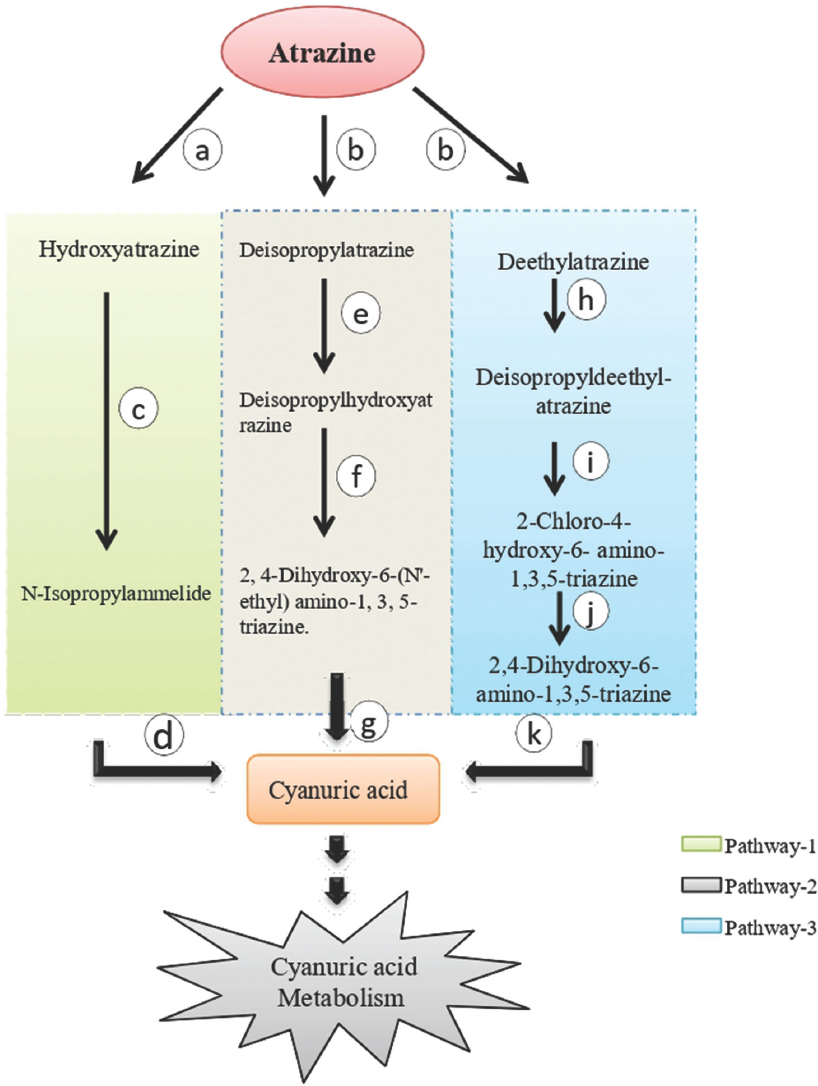 | Figure 1: Atrazine-degradation pathways. a=atrazine chlorohydrolase, b=atrazine monooxygenase, c=Hydroxydechloatrazine ethylaminohydrolase, d=N-isopropylammelide isopropylamido hydrolase, e=s-triazine hydrolase, f=2,4-Dihydroxy-6-(N’-ethyl)amino-1,3,5-triazine hydrolase, g=ethylaminohydrolase, h=deethylatrazine monooxygenase, i=s-triazine hydrolase, j=hydroxychloroatrazine ethyaminohydrolase, k=N-isopropylammelide isopropylaminohydrolase. [Click here to view] |
2.2. Atrazine-degrading Microorganisms
A large range of microorganisms involved in the degradation of atrazine leads to the production of metabolites while some other microorganisms derive their nutrients and energy by mineralizing them completely into CO2 and NH4+ [19]. Atrazine degrading microorganisms are not limited to only bacteria and fungi, but many microalgae; for example, Chlamydomonas mexicana, Chlorella sp., and Selenastrum capricornutum have also been reported by several researchers [20,21].
2.2.1. Bacteria
Bacteria are the most widely reported microorganism for atrazine elimination from polluted sites [22]. As the potential machines for bioremediation, a large variety of strains of Gram-positive and Gram-negative bacteria that degrade atrazine have been isolated and identified. Atrazine degrading bacteria produce various catalytic enzymes that break down atrazine (i.e., atrazine chlorohydrolase, allophanate amidohydrolase, HA hydrolase, N-isopropylammelide amidohydrolase, triazine hydrolase, 1-carboxybiuret amidohydrolase, and cyanuric acid amidohydrolase) and enhance the metabolic mechanisms. They decrease the degradation half-life of atrazine by the different metabolic processes including dechlorination, dealkylation, hydroxylation, and ring cleavage [23]. The atrazine-degrading strains, for example, Pseudomonas strain ADP break down atrazine into cyanuric acid through three enzymatic steps, and cyanuric acid acts as a source of nitrogen for many other bacteria [24]. Moreover, some other bacteria belonging to the genera Rhodococcus, Acinetobacter, Streptomyces, Pseudomonas, Clavibacter [25], Arthrobacter [26], Bacillus, Alcaligenes, Klebsiella, and Agrobacterium [27], transform atrazine into cyanuric acid which further metabolized and produce carbon and nitrogen source compounds. However, in the past 10 years, among the most isolated atrazine-degrading bacteria only Arthrobacter sp., Pseudomonas sp., and Bacillus sp. are reported as capable of fully degrading atrazine into carbon dioxide and ammonia [16]. In a study, two atrazine degrading bacteria such as Bacillus lichenoformis and Bacillus megaterium were isolated from soil that showed 98.6% and 99.6% degradation efficiency of atrazine after 7 days [7]. At the same time, the degradation of atrazine was faster when two strains were used in combination under the same conditions. Based on the previous data, some of the bacteria and their producing metabolites are listed in Table 1.
Table 1: List of some Atrazine-degrading bacteria.
| Bacteria | Gram status | GenBank accession no. | Detected metabolites | References |
|---|---|---|---|---|
| Bacillus licheniformis ATLJ-5 | + | MH879786 | Hydroxyatrazine and N-isopropylammelide | [7] |
| Bacillus megaterium ATLJ-11 | + | MH879805 | ||
| Pseudaminobacter sp. | - | nd | Hydroxyatrazine and N-ethylammelide | [51] |
| Nocardioides sp. | + | |||
| Bacillus atrophaeus | + | MH685187 | nd | [43] |
| Paenarthrobacter sp. W11 | + | nd | nd | [52] |
| Arthrobacter sp.C2 | + | MF405158 | Hydroxyatrazine, N-isopropylammelide, cyanuric acid, and deisopropylhydroxyatrazine (DIHA) | [26] |
| Klebsiella variicola FH-1. | - | nd | nd | [53] |
| Pseudomonas spp.strains ACB and TLB | - | nd | nd | [10] |
| Variovorax sp.strain 38R | - | CP062121 | nd | [11] |
| Arthrobacter sp.strain TES | + | CP062235 | ||
| Chelatobacter sp.strain SR38 | - | CP062112 | ||
| Myriophyllum spicatum | - | nd | Hydroxyatrazine (HA), deelthylatrazine (DEA), didealkylatrazine (DDA), cyanuric acid (CYA), and biuret | [22] |
| Acetobacter sp. | - | |||
| Arthrobacter sp.strain HB-5 | + | nd | nd | [23] |
| Ensifer sp. | - | nd | N-isopropylammelide, Cyanuric acid (CA) | [54] |
| Nocardioides | + | nd | N-isopropylammelide (IPA), ammelid, biuret, and cyanuric acid | [55] |
| Arthrobacter, | + | |||
| Bradyrhizobium, | - | |||
| Burkholderia, | - | |||
| Methylobacterium | - | |||
| Mycobacterium, | + | |||
| Clostridium. | + | |||
| Rhodococcus sp. strain MB-P1 | + | FN357284 | De-ethyl de-isopropyl atrazine, De-isopropyl atrazine, De-ethyl atrazine | [56] |
| Citricoccus sp. strain TT3. | + | nd | nd | [57] |
| Klebsiella variicola Strain FH-1 | - | MH250202 | 2-hydroxyl-4-ethylamino-6-isopropylamino-1,3,5-triazine (HEIT) 2-hydroxyl-4,6-bis (ethylamino)-1,3,5-triazine (MEET), and 4,6-bis (ethylamino)-1,3,5-triazin-2 (1H)-one (AEEO) | [58] |
In the table, nd=no data obtained in the cited reference.
2.2.2. Fungi
Fungi are another main element of soil microflora involved in atrazine degradation after bacteria. They degrade atrazine at different rates and produce different metabolites through N-dealkylation of either alkylamino group [28]. The application of fungi may be the most important way to remove atrazine from contaminated soil. They are very effective in bioremediation as they can use different carbon sources for metabolism by producing different enzymes which catabolize different steps during the transformation of chemicals [29].
Among fungi, wood-degrading basidiomycetes are also a key player in atrazine degradation. The ability of chemical degradation of white-rot fungi is due to the existence of the ligninolytic system [30]. Fungi belonging to basidiomycetes and ascomycetes produce both extracellular and intracellular enzymes that biotechnologically and industrially valued molecules are responsible for herbicides and pesticide degradation [31]. The purpose of white-rot fungi in atrazine degradation may be advantageous because they can tolerate a wide range of environmental circumstances, including varying temperature, moisture, and nutrient contents. For example, Trametes versicolor belonging to basidiomycetes can actively grow and degrade atrazine in nonsterile soil under low water availability conditions [32]. A well-known white-rot fungus Phanerochaete chrysosporium has been reported to degrade a large variety of environmentally persistent chemicals.
The potential roles of mycorrhizal fungi in the degradation of atrazine have been addressed by several authors. Axenic cultures of ectomycorrhizae fungi can degrade atrazine, and degradation was increased when they were full-fledged in symbiosis with plants [33]. Besides, ericoid mycorrhizal fungi have also been reported to degrade atrazine when they are axenically cultured [34]. Moreover, arbuscular mycorrhiza fungi have remarkable potential for atrazine degradation. They enhance soil microbial activity and increase the activities of soil enzymes [28]. Glomus caledonium and G. etunicatum can accumulate in fugal hyphae or the associated roots and atrazine dissipation in the near rhizosphere and bulk soils [35]. Some of the fungi associated with atrazine degradation are listed in Table 2.
Table 2: List of some atrazine-degrading fungi.
| Fungi | Division | GeneBank accession no. | Metabolite produced | References |
|---|---|---|---|---|
| Anthracophyllum discolor | Basidiomycota | nd | nd | [59] |
| Glomus caledonium | Glomeromycota | nd | Deethylatrazine (DEA) (2-amino-4-chloro-6-isopropylanine-striazine) and deisopropylatrazine (DIA) (2-amino-4-chloro6-ethylamino-s-triazine) | [35] |
| Trametes versicolor | Basidiomycota | nd | nd | [32] |
| Fusarium sp. CCLM_DF | Ascomycota | MT062480 | Deisopropylatrazine (DIA) and deethylatrazine (DEA) | [2] |
| Fusarium sp. CCLM_GU | MT062481 | |||
| Fusarium sp. CCLM_GW | MT062482 | |||
| Fusarium sp. CCLM_IB | MT062483 | |||
| Aspergillus niger | Ascomycota | nd | nd | [60] |
| Pleurotuso streatus INCQS 40310 | Basidiomycota | nd | Deisopropylatrazine (DIA) and deethylatrazine (DEA) | [12] |
| Pluteus cubensis SXS320, | Basidiomycotina | nd | nd | [61] |
| Gloelophyllum striatum MCA7, and Agaricales MCA17 | ||||
| Bjerkandera adusta | Basidiomycota | EF441742 | nd | [62] |
| Metarhizium robertsii | Ascomycota | nd | 2-hydroxy atrazine and desethylatrazine | [63] |
| Aspergillus niger AN 400 | Ascomycota | nd | Deethylatrazine ( DEA), deisopropilatrazine (DIA), hydroxyatrazine (HA) | [64] |
| Penicillium chrysogenum NRRL 807 | Ascomycota | nd | nd | [13] |
| Saccharomyces cerevisiae | Ascomycota | nd | Hydroxyatrazine, deethylatrazine, deisopropylatrazine | [22] |
| Aspergillus fumigatus | Ascomycota | nd | nd | [19] |
| Penicillium citrinum |
In the table, nd=no data obtained in the cited reference.
2.2.3. Yeast
Apart from bacteria and filamentous fungi, yeast also has atrazine-degrading potential. A novel yeast strain Pichia kudriavzevii strain Atz-EN-01 was isolated from atrazine-contaminated soil which showed the efficient degradation in liquid culture media and soil [36]. This strain breaks atrazine down into three intermediates such as HA, N-isopropylammelide, and cyanuric acid. Another species of Pichia has (Pichia pastoris strain X-33) also been reported as the ability to transform atrazine into hydroxylisopropylatrazine, atraton (2-methoxy-4-ethylamino-6-isopropylamino-1,3,5-s-triazine), demethylated atrazine, HA [37], Hydroxy-dehydrogenated atrazine, and 2-OH-isopropyl-IPU [38]. Moreover, a yeast species called Cryptococcus laurentii was isolated from atrazine-contaminated agricultural soil and GC-MS analysis showed several metabolites such as HA, deethylatrazine, deisopropylatrazine, and deethyldeisopropylatrazin during atrazine degradation when conducting an in vitro experiment [39]. The role of Saccharomyces cerevisiae in atrazine degradation was also reported by Zhu et al. [40]. However, in recent times, most of the research is focused on bacteria and filamentous fungi while little information is available on the role of yeast in atrazine degradation.
2.3. Genes Involved in Atrazine-degradation
Atrazine a commonly known herbicide is used as a carbon and nitrogen source by different soil microflora by breaking it into CO2 and NH4+ [41]. The degradation and utilization of atrazine by microflora are possible because of the complex catabolic pathway mediated by a diverse array of enzymes encode by a series of genes [16]. Different genes are involved in different steps throughout the degradation pathways that lead to the transformation of atrazine to its intermediate cyanuric acid [42]. To the best of our knowledge, the total number of eight genes involved in the atrazine metabolic pathway has been reported such as atzA, atzB, atzC, atzD, atzE, atzF, trzN, and trzD. The genes atzABC identified from Pseudomonas sp. strain ADP that homology to five atrazine-degrading microbial isolates which gives a piece of strong evidence for the genes are widespread. Some other bacteria such as Arthrobacter agilis and Nocardioides nanhaiensis are harbored atzA/trzN genes that code for atrazine chlorohydrolase that catalyze the dechlorination of atrazine into HA [42]. The same authors stated that atzD/trzD was involved in the conversion of cyanuric acid into biuret through ring cleavage by encoding an enzyme called cyanuric acid amidohydrolase. The other genes like atzB/atzC are associated with the dealkylation catabolic step while atzE and atzF/trzF are involved in biuret deamination and hydrolysis of allophanate, respectively. Different genes and their encoded enzymes involved in different steps in atrazine degradation are shown in Table 3.
Table 3: Microbial genes involved in atrazine-degradation.
| Gene | Enzyme encoded | Step catalyzed | References |
|---|---|---|---|
| atzA | Atrazine chlorohydrolase | Atrazine → Hydroxyatrazine (HA) | [63] |
| atzB | Hydroxydechloroatrazine ethylaminohydrolase | Hydroxyatrazine (HA) → N-isopropylammelide | [64] |
| atzC | N-isopropylammelide isopropylamido hydrolase | N-isopropylammelide → Cyanuric acid+isopropylamine | [16] |
| atzD | Cyanuric acid amidohydrolase | Cyanuric acid → Biuret | [42] |
| atzE | 1-Carboxybiuret hydrolase | Biuret → Allophanic acid | [42] |
| atzF | Allophanate hydrolase | Allophanic acid → CO2+NH4+ | [42] |
| trzD | Cyanuric acid amidohydrolase | Cyanuric acid → Biuret | [42] |
| trzN | Atrazine chlorohydrolase | Atrazine → Hydroxyatrazine (HA) | [65] |
2.4. Factors Affecting Microbial Degradation of Atrazine
Several factors influence the microbial degradation of atrazine. The microbial population is influenced by both biotic and abiotic factors in the soil and they directly or indirectly affect the rate of degradation of atrazine.
2.4.1. Abiotic factors
Temperature, pH, depth from the soil surface, and oxygen content of the surrounding matrix are the main abiotic factors that can influence atrazine degradation [43]. The mineralization rate of atrazine is slower in anaerobic or denitrifying conditions than in aerobic environments [44]. The water content of soil and temperature has a significant impact on atrazine degradation. The atrazine degradation is directly proportional to the temperature of its surrounding [45]. A study on the half-life of atrazine in clay soil was carried out and the results showed that the average half-life of atrazine degradation was 62 days when the water content of the soil was 20–40%. Nevertheless, when the water content of the soil was decreased by 8%, the half-life was increased up to 338 days [46]. Moreover, the half-life was increased from 44 days to 206 days while the soil temperature was decreased from 30°C to 5°C. In addition, soil layers also have a great significance in a variation of atrazine degradation rate. The occurrence of atrazine in different soil layers also influences the rate of degradation. The rate of atrazine degradation is slower on the subsurface horizon while increasing in soil depth in silty clay loam [47].
2.4.2. Biotic factors
Crops and soil management systems have a significant impact on the rate of herbicide degradation [48]. An experiment was carried out and found that in a site where corn is cultivated for several years and treated with atrazine, the adaptation of atrazine degrading microorganisms was colonized more than where alfalfa was cultivated for 4 years with no application of atrazine [49]. In addition, the role of earthworms is also not the least in atrazine degradation. Two earthworm species such as Eisenia foetida and Amynthas robust have been reported to enhance the degradation rate of atrazine [50]. However, our literature survey revealed that the effect of biotic factors on the degradation of atrazine has not been extensively studied yet.
3. CONCLUSION AND FUTURE PROSPECTS
The environment is constantly being harmed by the extended use of toxic chemicals. Several hundreds of herbicides including atrazine have been used by farmers and non-farmers to kill weeds in crop fields or resident campuses. Such type of practice has been done for many couples of the year all over the world. However, bioremediation is the soundest and default method to rehabilitate polluted sites with cost efficiency and environmental friendliness by applying microorganisms. Through the discovery of atrazine-degrading soil microorganisms, the disposing of hazardous chemicals has gained credence. In this regard, microbes can tolerate a high range of toxic chemicals and transform them into non-toxic forms by biochemical reactions, more often contaminants serve as a source of energy. Removal of persistent herbicides using microorganisms has received attention as an outstanding option. Microorganisms could be applied in different strategies. Some of the recommendations are cited below:
The gene editing and application of system biology on different microbial metabolic pathways are very important. The gene-editing tools such as clustered regularly interspaced short palindromic repeats (CRISPR-Cas), transcription activator-like effector nucleases, and zinc-finger nucleases can make it possible to design microbe with a functional gene of interest for degradation of atrazine for improved bioremediation. This will lead to optimizing the existing metabolic pathways toward the increased and efficient microbial remediation of herbicides. Genetic engineering can also open a new door for the degradation of herbicides by enhancing the capability of microorganisms. In this regard, the genetic transfer of degrading potential from one to another microbe can be a tremendous approach toward bioremediation.
Both bacteria and fungi are the main dominant degraders of atrazine. However, there are not many influential studies that have been carried out on the application of the organisms as consortia. By screening their biocompatibility, consortia can be designed that could be more effective towards bioremediation. More research on the biochemical pathways related to the catabolism of consortia could allocate for more efficient remediation and narrative applications.
Enzymes are always a major talking point in bioremediation research due to their inherent capability to degrade complex metabolites present in the pollutants. Enzyme technology could be one of the excellent techniques for the improvement of bioremediation practices. Microorganisms harbor a wide range of catalytic enzymes including chromium reductase, alkane hydroxylases, laccase, carboxylesterases, peroxidases, phytase, haloalkane dehalogenases, phosphotriesterases, and horseradish peroxidase. Using biotechnological approaches enzymes can be formulated from microorganisms for direct application in the rehabilitation of the polluted site. Moreover, with the help of enzyme engineering, the enzyme can be modified to improve its properties such as activity, stress tolerance, temperature, and pH for bioremediation.
4. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
5. FUNDING
This research did not receive any grant from funding agencies in government and non-governmental organizations.
6. CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
7. ETHICAL APPROVALS
This study does not require any ethical approval.
8. DATA AVAILABILITY
Data will be made available as per the journal policy.
9. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Mudhoo A, Garg VK. Sorption, transport and transformation of atrazine in soils, minerals and composts:A review. Pedosphere 2011;21:11-25. [CrossRef]
2. Esparza-Naranjo SB, da Silva GF, Duque-Castaño DC, Araújo WL, Peres CK, Boroski M, et al. Potential for the biodegradation of atrazine using leaf litter fungi from a subtropical protection area. Curr Microbiol 2021;78:358-68. [CrossRef]
3. Bai X, Sun C, Xie J, Song H, Zhu Q, Su Y, et al. Effects of atrazine on photosynthesis and defense response and the underlying mechanisms in Phaeodactylum tricornutum. Environ Sci Pollut Res Int 2015;22:17499-507. [CrossRef]
4. Prado B, Duwig C, Hidalgo C, Müller K, Mora L, Raymundo E, et al. Transport, sorption and degradation of atrazine in two clay soils from Mexico:Andosol and vertisol. Geoderma 2014;232:628-39. [CrossRef]
5. Malone RW, Nolan BT, Ma L, Kanwar RS, Pederson C, Heilman P. Effects of tillage and application rate on atrazine transport to subsurface drainage:Evaluation of RZWQM using a six-year field study. Agric Water Manag 2014;132:10-22. [CrossRef]
6. De Gerónimo E, Aparicio VC, Bárbaro S, Portocarrero R, Jaime S, Costa JL. Presence of pesticides in surface water from four sub-basins in Argentina. Chemosphere 2014;107:423-31. [CrossRef]
7. Zhu J, Fu L, Jin C, Meng Z, Yang N. Study on the isolation of two atrazine-degrading bacteria and the development of a microbial agent. Microorganisms 2019;7:E80. [CrossRef]
8. Manogaran M, Ahmad SA, Yasid NA, Yakasai HM, Shukor MY. Characterisation of the simultaneous molybdenum reduction and glyphosate degradation by Burkholderia vietnamiensis AQ3-12 and Burkholderia sp. AQ3-13. 3 Biotech 2018;8:117. [CrossRef]
9. Elarabi NI, Abdelhadi AA, Ahmed RH, Saleh I, Arif IA, Osman G, et al. Bacillus aryabhattai FACU:A promising bacterial strain capable of manipulate the glyphosate herbicide residues. Saudi J Biol Sci 2020;27:2207-14. [CrossRef]
10. James A, Singh DK. Atrazine detoxification by intracellular crude enzyme extracts derived from epiphytic root bacteria associated with emergent hydrophytes. J Environ Sci Health B 2021;56:577-86. [CrossRef]
11. Billet L, Devers-Lamrani M, Serre RF, Julia E, Vandecasteele C, Rouard N, et al. Complete genome sequences of four atrazine-degrading bacterial strains, Pseudomonas sp. Strain ADPe, Arthrobacter sp. Strain TES, Variovorax sp. Strain 38R, and Chelatobacter sp. strain SR38. Microbiol Resour Announc 2021;10:e01080-20. [CrossRef]
12. Lopes RD, Pereira PM, Pereira AR, Fernandes KV, Carvalho JF, França AD, et al. Atrazine, desethylatrazine (DEA) and desisopropylatrazine (DIA) degradation by Pleurotus ostreatus INCQS 40310. Biocatal Biotransformation 2020;38:415-30. [CrossRef]
13. Nicodemo SC. Potencial de Biodegradação de Atrazinapor Bacillus megaterium CCT 7935 e Penicillium chrysogenum NRRL 807 para Biorremediação de Solos Contaminados;2021. Available from:https://repositorio.ufrn.br/handle/123456789/32500 [Last accessed on 2022 Feb 10].
14. de Albuquerque FP, de Oliveira JL, Moschini-Carlos V, Fraceto LF. An overview of the potential impacts of atrazine in aquatic environments:Perspectives for tailored solutions based on nanotechnology. Sci Total Environ 2020;700:134868. [CrossRef]
15. He H, Liu Y, You S, Liu J, Xiao H, Tu Z. A review on recent treatment technology for herbicide atrazine in contaminated environment. Int J Environ Res Public Health 2019;16:E5129. [CrossRef]
16. Abd Rani NF, Kamil KA, Aris F, Yunus NM, Zakaria NA. Atrazine-degrading bacteria for bioremediation strategy:A review. Biocatal Biotransformation 2021;40:233-47. [CrossRef]
17. Hsieh HY, Lin CH, Hsu SY, Stewart GC. A Bacillus spore-based display system for bioremediation of atrazine. Appl Environ Microbiol 2020;86:e01230-20. [CrossRef]
18. Qu M, Li N, Li H, Yang T, Liu W, Yan Y, et al. Phytoextraction and biodegradation of atrazine by Myriophyllum spicatum and evaluation of bacterial communities involved in atrazine degradation in lake sediment. Chemosphere 2018;209:439-48. [CrossRef]
19. Bravim NP, Alves AF, Orlanda JF. Biodegradation of atrazine, glyphosate and pendimetaline employing fungal consortia. Res Soc Dev 2020;9:e1549119679. [CrossRef]
20. Zhao F, Li Y, Huang L, Gu Y, Zhang H, Zeng D, et al. Individual and combined toxicity of atrazine, butachlor, halosulfuron-methyl and mesotrione on the microalga Selenastrum capricornutum. Ecotoxicol Environ Saf 2018;148:969-75. [CrossRef]
21. Sun C, Xu Y, Hu N, Ma J, Sun S, Cao W, et al. To evaluate the toxicity of atrazine on the freshwater microalgae Chlorella sp. using sensitive indices indicated by photosynthetic parameters. Chemosphere 2020;244:125514. [CrossRef]
22. Wu X, He H, Yang WL, Yu J, Yang C. Efficient removal of atrazine from aqueous solutions using magnetic Saccharomyces cerevisiae bionanomaterial. Appl Microbiol Biotechnol 2018;102:7597-610. [CrossRef]
23. Gao J, Song P, Wang G, Wang J, Zhu L, Wang J. Responses of atrazine degradation and native bacterial community in soil to Arthrobacter sp. strain HB-5. Ecotoxicol Environ Saf 2018;159:317-23. [CrossRef]
24. Delcau MA, Henry VA, Pattee ER, Peeples TL. Mode of growth and temperature dependence on expression of atrazine-degrading genes in Pseudomonas sp. strain ADP Biofilms bioRxiv 2018;1:302877. [CrossRef]
25. Popov VH, Cornish PS, Sultana K, Morris EC. Atrazine degradation in soils:The role of microbial communities, atrazine application history, and soil carbon. Soil Res 2005;43:861-71. [CrossRef]
26. Cao D, He S, Li X, Shi L, Wang F, Yu S, et al. Characterization, genome functional analysis, and detoxification of atrazine by Arthrobacter sp. C2. Chemosphere 2021;264:128514. [CrossRef]
27. Siripattanakul S, Wirojanagud W, McEvoy JM, Casey FX, Khan E. A feasibility study of immobilized and free mixed culture bioaugmentation for treating atrazine in infiltrate. J Hazard Mater 2009;168:1373-9. [CrossRef]
28. Fan X, Song F. Bioremediation of atrazine:Recent advances and promises. J Soils Sediments 2014;14:1727-37. [CrossRef]
29. Kanagaraj J, Senthilvelan T, Panda RC. Degradation of azo dyes by laccase:Biological method to reduce pollution load in dye wastewater. Clean Technol Environ Policy 2015;17:1443-56. [CrossRef]
30. Rubilar O, Tortella GR, Cuevas R, Cea M, Rodríguez-Couto S, Diez MC. Adsorptive removal of pentachlorophenol by Anthracophyllum discolor in a fixed-bed column reactor. Water Air Soil Pollut 2012;223:2463-72. [CrossRef]
31. Deshmukh R, Khardenavis AA, Purohit HJ. Diverse Metabolic capacities of fungi for bioremediation. Indian J Microbiol 2016;56:247-64. [CrossRef]
32. Bastos AC, Magan N. Trametes versicolor:Potential for atrazine bioremediation in calcareous clay soil, under low water availability conditions. Int Biodeterior Biodegradation 2009;63:389-94. [CrossRef]
33. Meharg AA, Cairney JW. Ectomycorrhizas extending the capabilities of rhizosphere remediation?Soil Biol Biochem 2000;32:1475-84. [CrossRef]
34. Donnelly PK, Entry JA, Crawford DL. Degradation of atrazine and 2,4-dichlorophenoxyacetic acid by mycorrhizal fungi at three nitrogen concentrations in vitro. Appl Environ Microbiol 1993;59:2642-7. [CrossRef]
35. Huang H, Zhang S, Shan XQ, Chen BD, Zhu YG, Bell JN. Effect of arbuscular mycorrhizal fungus (Glomus caledonium) on the accumulation and metabolism of atrazine in maize (Zea mays L.) and atrazine dissipation in soil. Environ Pollut 2007;146:452-7. [CrossRef]
36. Abigail EA, Salam JA, Das N. Atrazine degradation in liquid culture and soil by a novel yeast Pichia kudriavzevii strain Atz-EN-01 and its potential application for bioremediation. J Appl Pharm Sci 2013;3:35.
37. Huang MT, Lu YC, Zhang S, Luo F, Yang H. Rice (Oryza sativa) laccases involved in modification and detoxification of herbicides atrazine and isoproturon residues in plants. J Agric Food Chem 2016;64:6397-406. [CrossRef]
38. Lu YC, Luo F, Pu ZJ, Zhang S, Huang MT, Yang H. Enhanced detoxification and degradation of herbicide atrazine by a group of O-methyltransferases in rice. Chemosphere 2016;165:487-96. [CrossRef]
39. Evy AA, Lakshmi V, Nilanjana D. Biodegradation of atrazine by Cryptococcus laurentii isolated from contaminated agricultural soil. J Microbiol Biotechnol Res 2012;2:450-7.
40. Zhu C, Yang WL, He H, Yang C, Yu J, Wu X, et al. Preparation, performances and mechanisms of magnetic Saccharomyces cerevisiae bionanocomposites for atrazine removal. Chemosphere 2018;200:380-7. [CrossRef]
41. Shapir N, Sadowsky MJ, Wackett LP. Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J Bacteriol 2005;187:3731-8. [CrossRef]
42. Espín Y, Aranzulla G, Álvarez-OrtíM, Gómez-Alday JJ. Microbial community and atrazine-degrading genetic potential in deep zones of a hypersaline lake-aquifer system. Appl Sci 2020;10:7111. [CrossRef]
43. Zhu J, Fu L, Meng Z, Jin C. Characteristics of an atrazine degrading bacterium and the construction of a microbial agent for effective atrazine degradation. Water Environ J 2021;35:7-17. [CrossRef]
44. Sims GK, Kanissery RG. Factors controlling herbicide transformation under anaerobic conditions. Environ Res J 2012;6:355-73.
45. Andleeb S, Jiang Z, Ur Rehman K, Olajide EK, Ying Z. Influence of soil pH and temperature on atrazine bioremediation. J Northeast Agric Univ (Engl Ed) 2016;23:12-9. [CrossRef]
46. Smith AE, Walker A. Prediction of the persistence of the triazine herbicides atrazine, cyanazine, and metribuzin in Regina heavy clay. Can J Soil Sci 1989;69:587-95. [CrossRef]
47. Lavy TL, Roeth FW, Fenster CR. Degradation of 2,4-0 and atrazine at three soil depths in the field. J Environ Qual 1973;2:132-7. [CrossRef]
48. Bokszczanin K?, Wrona D, Przyby?ko S. Influence of an alternative soil management system to herbicide use on tree vigor, yield, and quality of apple fruit. Agronomy 2021;11:58. [CrossRef]
49. Stolpe NB, Shea PI. Alachlor and atrazine degradation in a Nebraska soil and underlying sediment. Soil Sci 1995;160:359-370. [CrossRef]
50. Lin Z, Zhen Z, Chen C, Li Y, Luo C, Zhong L, et al. Rhizospheric effects on atrazine speciation and degradation in laterite soils of Pennisetum alopecuroides (L.) Spreng. Environ Sci Pollut Res Int 2018;25:12407-18. [CrossRef]
51. Topp E. A comparison of three atrazine-degrading bacteria for soil bioremediation. Biol Fertil Soils 2001;33:529-34. [CrossRef]
52. Chen S, Li Y, Fan Z, Liu F, Liu H, Wang L, et al. Soil bacterial community dynamics following bioaugmentation with Paenarthrobacter sp. W11 in atrazine-contaminated soil. Chemosphere 2021;282:130976. [CrossRef]
53. Zhang J, Wu X, Zhang X, Pan H, Shearer JE, Zhang H, et al. Zn2+-dependent enhancement of Atrazine biodegradation by Klebsiella variicola FH-1. J Hazard Mater 2021;411:125112. [CrossRef]
54. Ma L, Chen S, Yuan J, Yang P, Liu Y, Stewart K. Rapid biodegradation of atrazine by Ensifer sp. strain and its degradation genes. Int Biodeterior Biodegradation 2017;116:133-40. [CrossRef]
55. Fang H, Lian J, Wang H, Cai L, Yu Y. Exploring bacterial community structure and function associated with atrazine biodegradation in repeatedly treated soils. J Hazard Mater 2015;286:457-65. [CrossRef]
56. Fazlurrahman, Batra M, Pandey J, Suri CR, Jain RK. Isolation and characterization of an atrazine-degrading Rhodococcus sp. strain MB-P1 from contaminated soil. Lett Appl Microbiol 2009;49:721-9. [CrossRef]
57. Yang X, Wei H, Zhu C, Geng B. Biodegradation of atrazine by the novel Citricoccus sp. strain TT3. Ecotoxicol Environ Saf 2018;147:144-50. [CrossRef]
58. Zhang J, Liang S, Wang X, Lu Z, Sun P, Zhang H, et al. Biodegradation of atrazine by the novel Klebsiella variicola strain FH-1. Biomed Res Int 2019;2019:4756579. [CrossRef]
59. Elgueta S, Santos C, Lima N, Diez MC. Immobilization of the white-rot fungus Anthracophyllum discolor to degrade the herbicide atrazine. AMB Express 2016;6:104. [CrossRef]
60. Herrera-Gallardo BE, Guzmán-Gil R, Colín-Luna JA, García-Martínez JC, León-Santiesteban HH, González-Brambila OM, et al. Atrazine biodegradation in soil by Aspergillus niger. Can J Chem Eng 2021;99:932-46. [CrossRef]
61. Henn C, Monteiro DA, Boscolo M, da Silva R, Gomes E. Biodegradation of atrazine and ligninolytic enzyme production by basidiomycete strains. BMC Microbiol 2020;20:266. [CrossRef]
62. Dhiman N, Jasrotia T, Sharma P, Negi S, Chaudhary S, Kumar R, et al. Immobilization interaction between xenobiotic and Bjerkandera adusta for the biodegradation of atrazine. Chemosphere 2020;257:127060. [CrossRef]
63. Szewczyk R, Ró?alska S, Mironenka J, Bernat P. Atrazine biodegradation by mycoinsecticide Metarhizium robertsii:Insights into its amino acids and lipids profile. J Environ Manage 2020;262:110304. [CrossRef]
64. Marinho G, Barbosa BC, Rodrigues K, Aquino M, Pereira L. Potential of the filamentous fungus Aspergillus niger AN 400 to degrade Atrazine in wastewaters. Biocatal Agric Biotechnol 2017;9:162-7. [CrossRef]
65. Udikovi?-Koli?N, Scott C, Martin-Laurent F. Evolution of atrazine-degrading capabilities in the environment. Appl Microbiol Biotechnol 2012;96:1175-89. [CrossRef]