1. INTRODUCTION
Gastric cancer (GC) ranks fourth among the most common cancer and ranks third in the cancer death worldwide. There were 19,293,789 cases for cancer reported in 2020, of which nearly 5.6% i.e. 1,089,103 cases were of stomach cancer. The total number of death’s in 2020 was about 9,958,133 out of which 768,793, that is, 7.7% death cases were of GC. The incidences have increased by 1.3% and mortality rate has also increased by 0.9% across the world [1]. GC is a multifactorial disease [2]. Genetic factors along with environmental condition play an important role in the cause of GC. Lifestyle changes that increase the vulnerability to GC includes smoking, high intake of salty, smoke foods, low dietary fiber, obesity, and radiation [3]. Besides, the above factors, Helicobacter pylori colonization in stomach lining is found to be the main etiological factor for GC. The gastric pathogen has the capacity of forming colonies in the mucosal membrane and evokes an immune response. This results in the generation of reactive oxygen species due to the infection which could damage the DNA. Men have two-fold higher risks than the women of developing GC. This may be due to estrogens which protect a person from developing GC. Furthermore, consumption of anti-estrogen medicines like tamoxifen might increase the incidence of a person getting GC [4]. Peritoneal metastasis (PM) occurs in around 53 to 66% of patients who has clear cut metastatic GC. CT or computed tomography scan was used most commonly as the non- invasive method to diagnose PM. Early diagnosis and detection of the GC were quite important as there would be minimal treatment required and the person could prevent unnecessary surgical treatment. However, CT detection have low sensitivity and high specificity as the clinical signs such as omental cake, parietal peritoneum thickening, and deposition of large number of ascites would appear in the later stage of cancer. Even when the patient undergoes multiple clinical tests, on an average 16.7% of peritoneal metastatic cancer are not detected. Laparoscopy should be done to patients who are potentially resectable advanced GC to detect occult PM. For the past few decades, microRNA (miRNA) has emerged in the field of cancer as biomarkers. miRNA’s, a class of non-coding RNA(~22 nucleotide long) plays a very important role in the regulation of gene expression [5,6]. miR-34a will increase the expression of genes which are related to apoptosis, miRNA-373, miRNA-10b, miRNA-520c will encourage tumor metastasis and invasion [7]. Nearly 140 miRNA were found to be up regulated (miRNA-9, miRNA-21, miRNA-17-92, miRNA-145, miRNA-224, etc.); 17 miRNA were downregulated (miRNA-449a, miRNA-223, miRNA-421, and miRNA-34) and these miRNA play an important role in GC disease diagnosis and serve as a biomarker for the early detection of GC and potential therapeutic targets [8].
2. miRNA BIOGENESIS AND MECHANISM
Various types of ribonucleic acid such as messenger RNA, miRNA, transfer RNA, and ribosomal RNA are found in eukaryotes. Besides the role of RNA in protein synthesis, miRNA’s belongs to non-coding RNA family, are involved in regulation of gene expression at the post-translational level. The biogenesis will start either co-transcriptional or post-transcription by processing RNA polymerase. The biogenesis can occur either by canonical or non-canonical pathway which are as follows [8,9].
2.1. Canonical Pathway
miRNA genes were transcribed by RNA polymerase II into initial transcript called pri-miRNA, which is about ~500 to 3000 bp long [8]. These pri-miRNA’s were further modified/transformed to hairpin shaped precursor miRNA called pre-miRNA’s (~60–70 nucleotide long) with the help of DROSHA or DGCR 8 enzyme. At this stage, the precision processing was very crucial for pre-miRNA as this would determine the target sequence in mRNA for gene silencing. The pre-miRNA was taken into the cytoplasm by exportin-5 and later on dicer (belongs to RNase III) cleaves the pre-miRNA and give rise to ~20–24 nucleotide RNA duplex, where one strand was loaded into RNAinduced silencing complex which has Argonaute [Figure 1]. This complex would muzzle/silence the target miRNA and could regulate the gene expression at post-translational level. Thus, these miRNA’s could play a key role in the animals including roles in cell apoptosis, grow, and cell proliferation [8-11].
 | Figure 1: miRNA activation by canonical pathway. [Click here to view] |
2.2. Non-Canonical Pathway
As shown in Figure 2 the proteins involved in the non-canonical pathways are of different sets of proteins when compared to canonical pathways. Dicer-independent pathway and DGCR8/Drosha-independent pathway are the two types of non-canonical pathways. In DGCR8/Drosha-independent pathway, the pre-miRNA produced will resemble the dicer substrate. During splicing of introns of mRNA, mirtrons are produced which are example of such Pre-miRNA. Through exportin 1 without drosha cleavage, in cytoplasm the nascent RNAs are exported. These miRNA has a 7-methylguanosine cap that will not allow them to load into the Argonaute. Due to dicer substrate’s insufficient length in cytoplasm to complete the maturation of pre-miRNAs AGO2 is required. This also promotes loading of all pre-miRNA into AGO2-dependent and AGO2 independent slicing of 3p strand. To complete the maturation of 5p strand 3’-5’ trimming is required. Thus these miRNA’s could play a key role in the animals including roles in cell apoptosis, grow and cell proliferation [12].
3. miRNA AS PROMISING BIOMARKERS FOR CANCER
miRNA plays an important role in the metabolism, immunity etc. Dysregulation of miRNA expression is closely correlated with tumorogenesis, prognosis and progression of cancer. miRNA can be classified as tumor suppressor and oncogenic miRNA. Mutations, deletions or dysregulation of miR-15/16, miR-34, and miR-200 family can cause colon cancer, multiple myeloma, chronic lymphocytic leukemia, follicular lymphoma, lung cancer, ovarian cancer, GC, bladder cancer, pancreatic cancer, etc. Oncogenic miRNA such as miR-17/92 and miR-222/221 can cause prostate cancer, lung cancer, breast cancer, Ras-induced senescent fibroblast, glioblastoma, non-small cell lung cancer, and hepatocellular carcinoma [13].
Various findings have suggested that biomarkers such as carbohydrate antigen 19-9, carcinoembryonic antigen, carbohydrate antigen 125, and alpha-fetoprotein will improve the responsiveness while diagnosis [7]. miRNA-34a has given us the evidence that it has impact on expression of genes that are associated to apoptosis other than this miRNA-373, miRNA-10b, and miRNA-520c have resulted to encourage tumor metastasis and invasion. miRNA-17-5p/20a inhibits cell apoptosis through cyclin dependent kinase (CDK) inhibitor P21 and the transcriptional modulation of tumor suppressor protein p53INP1 and it upregulates GC cell cycle progression. MiR-93 and miR-106b will hamper the function of BCL 2L11 which is an anti-apoptotic protein by damaging TGF beta [7].
4. miRNA IN GC PROGRESSION AND METASTASIS
As shown in Figure 3 miR-150 has exhibited to influence the GC growth by selecting EGR2 which is a tumor suppressive transcription factor. In most of the cases miR-375 is downregulated but when it is over expressed it will decrease the cell viability [7,14]. MiRNA-146a is ectopically expressed in cancer tissues which inhibits the invasion and migration of the GC cells and affects the EGFR expression. MiRNA-148a and miRNA-152 are correlated with increase in tumor size when the expression are downregulated, while low expression levels of miR-204, miR-146a, or miR-142-5p are correlated with increased tumor size [7,8]. As shown in Table 1, expression of the miRNA-125a-5p is associated with an enhanced malignant potential such as tumor size and depth and poor clinical prognosis. Proto oncogene ERBB 2 is a direct target of miRNA-125a-5p which potently suppresses the proliferation of GC cells. miR-21 is downregulated in GC and it is a well-known for its expression in tumor suppressor [7]. The most explored single nucleotide polymorphism in the GC are associated with miR-421, miR-608, miR-492, miR-27a, and miR-146a genes. There are 2 miRNA’s (miR-155, miR-223) which have exhibited increase in correa’s cascade in both the regions, that is, antrum and corpus mucosa [2,9]. VGL4 suppresses proliferation of GC cell by over activation of YAP-TEAD signal and expression pattern of VGL4 and miR-222 is negatively correlated [15]. MiR-204 is negatively correlated with Bcl-2 proteins and increasing in the Bcl-2protein promises for therapeutic and preventive strategy against GC [16,17]. During progression of GC upregulation of miR-181b plays an important role and miR-181b is negatively correlated with metalloproteinase TIMP3 which inhibits the proliferation of tissues [18]. There was significant difference in the level of serum miR-20a in various types of cancer such as GC, breast cancer, nasopharyngeal cancer against non cancerous control. In additional, the level of miR-20a is directly correlated to age, lymph node metastasis, differentiated degree, and tumor stage in GC [19]. Table 1 gives us a brief idea about the list of miRNAs which is associated with cancer.
When HGMA 2 is highly expressed, it has direct connection with the tumor invasion which is contrarily regulated by the let -7 family. When miR-214regulates the hedgehog signaling pathway, it results in the contribution for GC.
 | Figure 2: miRNA functional activation by non-canonical pathway. [Click here to view] |
5. microRNA IN BIOLOGICAL FLUIDS
miRNA such as miRNA-106b, miRNA-21, miRNA-106a, and miRNA-17-5p are over expressed in the plasma of GC patients. miR-196a/b has higher specificity in diagnosing the GC when compared with carcinoembryonic antigen and carbohydrate antigen 19-9. Heterogeneity nature of the tumor makes single miRNAs in blood a non ideal technique for the GC diagnosis. That is why to improve their performance for diagnosis of GC combination of plasma miRNA’s used. Patients suffering with GC highly expressed miR-21-5p in urine compared with healthy people and after surgical resection of cancer miR-21-5p expression was significantly reduced [23].
MiRNA’s have an excellent stability and they can sustain any of the conditions like boiling, freeze-thawing, high/low pH which indicates that these miRNA’s can be used as biomarkers. miRNA-129, miRNA-21, miRNA-129-1-3p, miRNA-133a, miRNA-106a, miRNA-421a, etc., are found in the gastric juice and can be used in GC diagnosis in the early stages [Table 2]. MiR-133a, miR-421 levels were lower in the GC patients when compared to normal individuals [23].
6. miRNA IN GC DIAGNOSIS
miRNAs play a key role in the GC diagnosis. It was suggested that there are certain miRNAs which do not operate alone rather they form different clusters with other miRNA and share their functions. As shown in Table 3, miR-21 plays a very important role as it is upregulated in most types of the cancer and it plays a key role in tumorigenesis [8]. In almost 92% human GC, miR-21 is said to be over expressed hence it can be used as a promising biomarker. There are a lot of methods available for medical attention/treatment for GC but some of important therapy such as radiation therapy, surgery, and chemotherapy; however, the detection of the cancer can be done using the miRNA. When miR-181b was transfected into the GC patient it showed increase in cell migration, invasion, and cell proliferation. Hence, this can be used as a potential target for the anticancer therapeutics in GC. miR-7 plays a key role in blocking the movement of tumor cells by targeting the insulin like growth factor-1 receptor [61]. When miRNA merge with the target messenger RNA it will give rise to negative regulation of the proteins. We can use this like a secret weapon to up or down regulate the expression of tumor associated genes. When prohibiting is down regulated it will suppress miR-27a which hamper the GC cell growth and even it will support the hypothesis that miR-27a functions as oncogenes. MiR-34 has potential role in the downstream pathways of p53 protein and for some of the target genes like NOTCH, Bcl-2 and HGMA2 where it acts as a potential tumor suppressor as they are entailed in cancer stem cell renewal and survival [8].
 | Figure 3: Different microRNA and their role in the tumor progression in Gastric cancer. [Click here to view] |
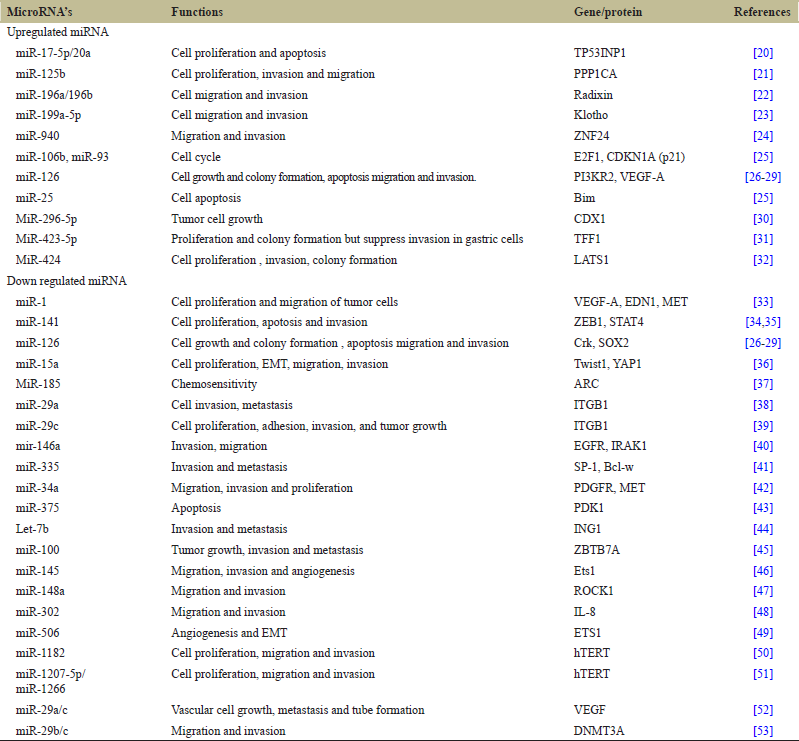 | Table 1: List of microRNA that are dysregulated in Gastric cancer. [Click here to view] |
 | Table 2: List of miRNA in biological fluids. [Click here to view] |
The expression of miR-25 was over expressed in GC patients along with lymph node metastasis. When miRNA-25 was inhibited, it repressed the proliferation, metastasis, invasion, and decreased the metastatic space of GC cells by repressing ERBB2,1 expression [85]. MiR-129 plays a key role in regulating cell proliferation by downregulating the CDK 6. miRNA-129-2 and miRNA-129-1 significantly showed lower amounts in the GC patients [85]. The microarray profiling with GC patients and normal individuals could collate the expression patterns where the researchers found that miRNA-199a-3p was notably elevated in GC patients. MiR-199a-3p was notably associated with lymph node metastasis, tumor death and stage [85]. miR-196a and miR-196b are upregulated in the gastric tumor tissues. Expression of both miRNA-196b and miRNA-196a is more efficacious than the carbohydrate antigen 19-9 or carcinoembryonic antigen as they have higher specificity and sensitivity [86]. MiR-647 plays a remarkable role in reducing metastasis tumor size as well as it is down regulated in GC patients. When miR-647 is over expressed in the cell lines it suppressed the proliferation of cells and it arrest the cell cycle at G0/G1 stage and induces cell death. Furthermore, in in vivo condition it remarkably hamper the growth of tumor. Both in vivo and in vitro studies have strongly recommended the anti-tumorigenic effects [86].
 | Table 3: Role of miRNA in the proliferation of Gastric Cancer. [Click here to view] |
7. LIMITATIONS WITH CURRENTLY AVAILABLE BIOMARKERS
Till now, there was not any single blood based biomarkers which have enough sensitivity for screening of GC. There are two biomarkers cancer antigen 72-4 (CA-72-4), carcinoembryonic antigen and cancer antigen 19-9 (CA 19-9) have been associated with GC [2]. Serological biopsy was done to test for the gastric mucosa which was a tumor based biomarker. Concentration of pepsinogen I and II correspond with AG in pre neoplastic condition in intestinal type of GC and it is commonly used in Asian countries. In Europe, the pepsinogen I and pepsinogen II boards are extended by the utilization of Gastrin -17. G-17 is created by the G-cells and stimulates the pepsinogen and hydrochloric acid (HCl) production and consequently it was essential for a similar physiological course as pepsinogen I [2].
When compared with adjacent normal counterparts, in GC tissue 73% (44/60) of miR-421 was overexpressed and there was also no correlation with patients having poor prognosis. This shows that upregulation of miR-421 in GC could be used as diagnostic cancer at early stage. Another group of researchers found that, by real-time PCR, miR-31 was downregulated in GC tissue nevertheless the molecular mechanism has to be explicated. The clinic-pathological features such as differentiation, tumor size, stage, invasion, and metastasis were found to be correlated with the expression of miR-106a. In gastric carcinoma, the higher levels of miR-106a were detected when compared with the non-tumor tissue. Lately, five miRNA signatures with 0.85% as the receiver operating characteristic curve (miR-34, miR-20, miR-423-5p, miR-1, and miR-27a) were identified for GC as diagnostic markers, demonstrating a higher sensitivity than the conventional marker (CA19-9 or CAE). The miRNA derived from the tumors was thought to be granulated into the circulatory system. Various studies have shown that miRNA’s have a differential expression in GC patients compared to normal controls. Many studies found that conventional markers such as CA72-4, CA12-5, CEA, and CA19-9 had lower specificity and sensitivity. These markers represent a new era for circulating miRNA in cancer diagnosis. MiRNA that is deregulated could be used for GC as a diagnostic biomarker, and hopefully in plasma the deregulation of miRNA can be detected, which would aid GC in the early diagnosis. As a result, antagonizing the action of miR by designing individualized treatment for individual’s survival would be done at an early stage to improve the survival rate [87].
miRNA AS PROGNOSTIC MARKER
Most of the researchers are in search for the potential use of miRNA’s which could be used as prognostic tools other than those diagnostic applications. Nevertheless, these miRNAs must correlate with metastatic potential and clinical outcomes. In terms of cancer phenotypes differentiation profiling of miRNAs has advantage over mRNAs. A recent report on GC shows a prognostic signature which includes three protective miRNAs’, that is, miR-126, let-7a and miR-30a-5p, hazard ratio less than 1 and four risk miRNAs, that is, miR-21, miR-338, miR-223, and miR-10b, hazard ratio greater than 1 and was associated with clinical outcomes. In the patients treated with doxifluridine and S-1, low expression of miRNA-18 and miRNA-21 is associated with survival of the patient. Downregulation of miRNA-125a-3p was linked with invasion of tumor, advancement in clinical stage, metastasis, and serves as a prognostic marker for aggressive GC. Gastric cancerous growth in in vitro can be detected by abnormal expression of miRNA-125a-3p this tumor suppressive property holds the potential in clinical use. Similarly other miRNA such as miRNA-125-5p and miRNA-125a has same tumor suppressive potential in gastric and reported that, these miRNA’s are also expressed low and associated with poor prognosis [87].
8. CONCLUSION
GC ranks third among the cancer related death worldwide despite the advancement in medical field and metastasis has increased over 40% in last two decades. The significance of miRNAs in disease science has been broadly increased till date and lot of attention was given to the miRNA for GC diagnosis. Therefore, the mechanism underlying the progression and metastasis of GC is vital for cancer research. In this context, our review provides an insight into the role of miRNA in GC progression, metastasis and also with an overview of differential expression of miRNA in proliferation of GC. We need a breakthrough in some of the areas of future research to understand the origin of the circulating miRNA and their role in different stages of cancer, its prognosis, early detection, and development of effective therapeutic strategies. This review helps in the identification of miRNA targets for early diagnosis of GC, to improve the survival rate and in the development of therapeutic targets.
9. ACKNOWLEDGMENTS
The authors thank SRMIST for providing the opportunity to write this review article. Dr. Megala J for supervising the article, providing plagiarism reports before submitting to the journal.
10. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
11. FUNDING
This research did not receive any funding from in public or commercial sectors.
12. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
13. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects and hence no ethical approval was required.
14. DATA AVAILABILITY
The data generated and analyzed in this review article are included and the reference are cited in the reference section.
15. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Asia S, Asia S, Hdi H. Source: Globocan 2020; 2020. p. 1-2.
2. Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol 2018;24:3313-29. CrossRef
3. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. CrossRef
4. Choi YJ, Kim N. Gastric cancer and family history. Korean J Intern Med 2016;31:1042-53. CrossRef
5. Correa P. Gastric cancer. Overview. Gastroenterol Clin North Am 2013;42:211-7. CrossRef
6. Dong D, Tang L, Li ZY, Fang MJ, Gao JB, Shan XH, et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol 2019;30:431-8. CrossRef
7. Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol 2014;20:12007-17. CrossRef
8. Xu X, Yang X, Xing C, Zhang S, Cao J. miRNA: The nemesis of gastric cancer (review). Oncol Lett 2013;6:631-41. CrossRef
9. Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol 2014;20:5694-9. CrossRef
10. Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomed Pharmacother 2017;96:238-45. CrossRef
11. Sampath SS, Venkatabalasubramanian S, Ramalingam S. Role of MicroRNAs in the progression and metastasis of colon cancer. Endocr Metab Immune Disord Drug Targets 2020;21:35-46. CrossRef
12. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:1-12. CrossRef
13. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol Mech Dis 2014;9:287-314. CrossRef
14. Ni H, Qin H, Sun C, Liu Y, Ruan G, Guo Q, et al. MiR-375 reduces the stemness of gastric cancer cells through triggering ferroptosis. Stem Cell Res Ther 2021;12:325. CrossRef
15. Li N, Yu N, Wang J, Xi H, Lu W, Xu H, et al. miR-222/VGLL4/YAP-TEAD1 regulatory loop promotes proliferation and invasion of gastric cancer cells. Am J Cancer Res 2015;5:1158-68.
16. Sacconi A, Biagioni F, Canu V, Mori F, Di Benedetto A, Lorenzon L, et al. MiR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis 2012;3:325. CrossRef
17. Pereira AL, Magalhães L, Moreira FC, Reis-das-Mercês L, Vidal AF, Ribeiro-dos-Santos AM, et al. Epigenetic field cancerization in gastric cancer: MicroRNAs as promising biomarkers. J Cancer 2019;10:1560-9. CrossRef
18. Guo JX, Tao QS, Lou PR, Chen XC, Chen J, Yuan GB. miR-181b as a potential molecular target for anticancer therapy of gastric neoplasms. Asian Pacific J Cancer Prev 2012;13:2263-7. CrossRef
19. Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Reports 2017;6:429-34. CrossRef
20. Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, et al. MiR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer 2013;49:2010-21. CrossRef
21. Wu JG, Wang JJ, Jiang X, Lan JP, He XJ, Wang HJ, et al. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer 2015;18:729-39. CrossRef
22. Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, et al. MicroRNA-196a/-196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett 2014;351:222-31. CrossRef
23. Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Onco Targets Ther 2018;11:3891-900. CrossRef
24. Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z, et al. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget 2015;6:25418-28. CrossRef
25. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated MicroRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008;13:272-86. CrossRef
26. Liu LI, Wang WE, Zhao LI, Guo BO, Yang J, Zhao XG, et al. miR-126 inhibits growth of SGC-7901 cells by synergistically targeting the oncogenes PI3KR2 and Crk, and the tumor suppressor PLK2 2014:1257-65. CrossRef
27. Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N, et al. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget 2014;5:11873-85. CrossRef
28. Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis 2011;6:e16617. CrossRef
29. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol 2016;37:16345-55. CrossRef
30. Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li H, et al. MicroRNA-296-5p increases proliferation in gastric cancer through repression of caudal related homeobox 1. Oncogene. 2014;33(6):783-93. CrossRef
31. Liu J, Wang X, Yang X, Liu Y, Shi Y, Ren J, et al. miRNA423-5p regulates cell proliferation and invasion by targeting trefoil factor 1 in gastric cancer cells. Cancer Lett 2014;347:98-104. CrossRef
32. Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 2017;16:151. CrossRef
33. Xie M, Alwyn D, Ting D, Xing GX. MicroRNA-1 acts as a tumor suppressor microRNA by inhibiting angiogenesis-related growth factors in human gastric cancer. Gastric Cancer 2018;21:41-54. CrossRef
34. Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem 2015;36:1440-52. CrossRef
35. Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 Induced by Helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem 2014;33:1003-12. CrossRef
36. Wang T, Hou J, Li Z, Zheng Z, Wei J, Song D, et al. miR-15a-3p and miR-16-1-3p negatively regulate twist1 to repress gastric cancer cell invasion and metastasis. Int J Biol Sci 2017;13:122-34. CrossRef
37. Li Q, Wang J, He Y, Feng C, Zhang X, Sheng J, et al. MicroRNA-185 regulates chemotherapeutic sensitivity in gastric cancer by targeting apoptosis repressor with caspase recruitment domain 2014;5:e1197. CrossRef
38. He B, Xiao Y, Tang B, Wu Y, Hu C, Xie R, et al. hTERT mediates gastric cancer metastasis partially through the indirect targeting of ITGB1 by microRNA-29a. Sci Rep 2016;6:21955. CrossRef
39. Han T, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015;64:203-14. CrossRef
40. Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res 2011;17:4277-84. CrossRef
41. Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1 2012;31:1398-407. CrossRef
42. Peng Y, Guo J, Liu Y, Wu X. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep 2014;34:e00112. CrossRef
43. Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3. Cancer Res 2010;70:2339-49. CrossRef
44. Han X, Chen Y, Yao N, Liu H, Wang Z. MicroRNA let-7b suppresses human gastric cancer malignancy by targeting ING1. Cancer Gene Ther 2015;22:122-9. CrossRef
45. Shi D, Wang Y, Xing A, Gao J, Zhang H, Guo X, et al. FC/EBPα-induced miR-100 expression suppresses tumor metastasis and growth by targeting ZBTB7A in gastric cancer. Cancer Lett 2015;369:376-85. CrossRef
46. Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X, et al. Mirna-145 targets v-ets erythroblastosis virus e26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol Cancer Res 2013;11:182-93. CrossRef
47. Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res 2011;17:7574-83. CrossRef
48. Chen L, Min L, Wang X, Zhao J, Chen H, Qin J, et al. Loss of RACK1 promotes metastasis of gastric cancer by inducing a miR-302c/IL8 signaling loop. Cancer Res 2015;75:3832-41. CrossRef
49. Li Z, Liu Z, Dong S, Zhang J, Tan J, Wang Y, et al. MIR-506 inhibits epithelial-to-mesenchymal transition and angiogenesis in gastric cancer. Am J Pathol 2015;185:2412-20. CrossRef
50. Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, et al. MiR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett 2015;360:151-9. CrossRef
51. Chen L, Lü MH, Zhang D, Hao NB, Fan YH, Wu YY, et al. MiR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis 2014;5:e1034. CrossRef
52. Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, et al. Potential diagnostic, prognostic and therapeutic targets of micrornas in human gastric cancer. Int J Mol Sci 2016;17:945. CrossRef
53. Cui H, Wang L, Gong P, Zhao C, Zhang S, Zhang K, et al. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS One 2015;10:e0123926. CrossRef
54. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174-9. CrossRef
55. Huang S, Wang J, Li J, Luo Q, Zhao M, Zheng L, et al. Serum microRNA expression profile as adiagnostic panel for gastric cancer. Jpn J Clin Oncol 2016;46:811-8. CrossRef
56. Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 2011;47:784-91. CrossRef
57. Oze I, Shimada S, Nagasaki H, Akiyama Y, Watanabe M, Yatabe Y, et al. Plasma microRNA-103, microRNA-107, and microRNA-194 levels are not biomarkers for human diffuse gastric cancer. J Cancer Res Clin Oncol 2017;143:551-4. CrossRef
58. Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of MIR-21 and MIR-331. Br J Cancer 2017;117:266-73. CrossRef
59. Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun 2017;493:1322-8. CrossRef
60. Kim SY, Jeon TY, Choi CI, Kim DH, Kim DH, Kim GH, et al. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagnostics 2013;15:661-9. CrossRef
61. Ren H, Shen Y, Yang J, Li C, Sun P, Yang H. Molecular mechanism of MiR-7 inhibiting emt and invasion and metastasis of gastric cancer cells by regulating IGF1R expression. Acta Med Mediterr 2021;37:3087-92.
62. Kheir TB, Futoma-Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer 2011;10:29. CrossRef
63. Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Investig 2008;88:1358-66. CrossRef
64. Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep 2012;27:1019-26. CrossRef
65. Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, et al. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res 2009;28:1-9. CrossRef
66. Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res 2009;37:1672-81. CrossRef
67. Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 2010;20:784-93. CrossRef
68. Xu Y, Deng Y, Yan X, Zhou T. Targeting miR-375 in gastric cancer. Expert Opin Ther Targets 2011;15:961-72. CrossRef
69. Wan HY, Guo LM, Liu T, Liu M, Li X, Tang H. Regulation of the transcription factor NF-κB1 by microRNA-9 in human gastric adenocarcinoma. Mol Cancer 2010;9:1-10. CrossRef
70. Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T, et al. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol 2009;44:556-61. CrossRef
71. Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL, Zhao ZH, et al. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-β signalling pathway. Nucleic Acids Res 2012;40:9286-97. CrossRef
72. Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumor Biol 2012;33:1589-97. CrossRef
73. Chen L, Yang Q, Kong WQ, Liu T, Liu M, Li X, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life 2012;64:628-35. CrossRef
74. Xia J, Wu Z, Yu C, He W, Zheng H, He Y, et al. MiR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. J Pathol 2012;227:470-80. CrossRef
75. Li C, Nie H, Wang M, Su L, Li J, Yu B, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett 2012;320:189-97. CrossRef
76. Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, et al. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J 2012;279:1252-60. CrossRef
77. Li J, Guo Y, Liang X, Sun M, Wang G, De W, et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol 2012;138:763-74. CrossRef
78. Zheng B, Liang L, Huang S, Zha R, Liu L, Jia D, et al. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene 2012;31:4509-16. CrossRef
79. Zhang Z, Liu S, Shi R, Zhao G. MiR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet 2011;204:486-91. CrossRef
80. Wang M, Li C, Nie H, Lv X, Qu Y, Yu B, et al. Down-regulated miR-625 suppresses invasion and metastasis of gastric cancer by targeting ILK. FEBS Lett 2012;586:2382-8. CrossRef
81. Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL 3 2012;323:41-7. CrossRef
82. Liu Z, Zhu J, Cao H, Ren HUI, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol 2012;40:1553-60.
83. Gao P, Xing A, Zhou G, Zhang T, Zhang J, Gao C, et al. The molecular mechanism of microRNA-145 to suppress invasion--metastasis cascade in gastric cancer. Oncogene 2013;32:491-501. CrossRef
84. Wang J, Zhang J. MicroRNA-610 inhibits the migration and invasion of gastric cancer cells by suppressing the expression of vasodilator-stimulated phosphoprotein. Eur J Cancer 2012;48:1904-13. CrossRef
85. Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol 2015;21:9838-52. CrossRef
86. Sawaki K, Kanda M, Kodera Y. Review of recent efforts to discover biomarkers for early detection, monitoring, prognosis, and prediction of treatment responses of patients with gastric cancer. Expert Rev Gastroenterol Hepatol 2018;12:657-70. CrossRef
87. Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 2014;20:10432-9. CrossRef