1. INTRODUCTION
Rheumatoid arthritis (RA) is a chronic, autoimmune disease characterized by swelling and pain in the peripheral synovial joints and a gradual loss of mobility. Due to RA being a common debilitating disease around the world, the global incidence of RA is approximately 1% of the total population [1]. The development and progression of RA has not been fully uncovered yet, and therapeutic monitoring, early diagnosis, and prognostic estimation remain at the core of RA research. In a few individuals with a more serious variation of RA, there could be involvement of major organ systems such as the heart and lungs [2,3]. The normal functioning of the immune system requires it to maintain a balance between the identification and elimination of foreign antigen and developing tolerance against self-antigens. A loss of tolerance leads to the development of autoimmune diseases. This complex process is largely due to environmental factors exerting an effect on the immunology of a susceptible individual [4].
Sjögren’s syndrome (SS) is also a chronic, autoimmune disease that affects up to 3% of the population. This disease is characterized by dry mouth (xerostomia) and dry eyes (keratitis sicca) due to dysfunctional exocrine glands [5]. The prevalence of autoimmune diseases is much higher in women than in men [6]. Researchers have noticed over the years that SS and RA have various overlapping pathophysiological markers but the mechanism of the development of these diseases is still not fully understood [7]. In some patients suffering from severe RA, development of sicca symptoms is common although they are milder. This milder, overlapping SS seen in RA patients is secondary SS and causes a much worse outcome for the patients [8]. Due to the dual nature of SS and RA, many research groups in recent years have attempted identification of biomarkers unique to SS or RA or even common between them [9]. The symptoms of both diseases overlap in serious cases of RA, with SS and other autoimmune diseases developing along with RA.
Joint inflammation in RA is due to the mobilization of immune cells which lead to a release of chemokines and pro-inflammatory cytokines in the synovial area. The gene expression analysis of RA involves the study of joint synovial tissues and peripheral blood of RA patients. A large number of gene profiling studies have already been done on RA [10-13]. These have been reported by microarray platforms and have given rise to many differentially expressed genes (DEGs). These DEGs are involved in the disease by affecting the biological processes, pathways, and molecular functions. Progression of the field of medical biotechnology has led to an increase in the knowledge of molecular and cellular pathways that are involved in disease pathology [14]. Microarray has greatly aided in the present times for diagnosis and treatment [15].
The gene expression omnibus (GEO) is an immense database housing large gene expression data generated from large-scale experiments such as microarray and NGS for all types of tissue, diseases, species, and cells [16]. Using these data and combining them with various other bioinformatic tools, a comprehensive expression profile of any condition or disease can be generated without performing an actual experiment and solve the present disadvantages [17]. After the identification of pathways and their role in the pathophysiology of RA, identification of biomarkers can be carried out, used for targeted diagnosis, prognosis, and drug targeting [18]. In this study, gene microarray analysis of the whole blood of RA patients was carried out alongside a cohort of SS. This analysis is useful for evaluating genetic susceptibility in a cohort of RA and SS patients. The purpose of this work was to identify crucial genes and pathways that are involved in the pathophysiology of RA.
2. MATERIALS AND METHODS
2.1. Data Retrieval
The microarray gene expression data in the whole blood of RA and SS patients were searched using a keyword “Rheumatoid Arthritis” and “Sjogren’s Syndrome” in the GEO database [19]. The hits obtained were further screened using following criteria: (a) Only datasets derived from whole blood were included and (b) the datasets containing the whole blood transcriptomic information from both diseased and healthy cohorts. By restricting the entry type (series), study type (expression profiling by array), and tissue sources (Homo sapiens), 5386 search hits that were not related to the purpose of this study were excluded from the study. After filtering, gene expression profiles of accession no. GSE93272 for RA and GSE84844 for SS were selected [13,20]. For further analyses, platform and series matrix file(s) of both the microarray datasets (Accession No. GSE93272 and GSE84844) were retrieved from NCBI-GEO.
2.2. Identification of DEGs
GEO2R was used to retrieve the complete list of genes with their corresponding log2 fold change and P-values. DEGs were identified from these lists based on log2 fold change cutoff of 1.0 and P ≤ 0.05. Benjamini and Hochberg FDR cutoff of 0.05 was used for multiple testing corrections. Venn diagram was generated using Venny 2.1 drawing tool to identify commonly expressed genes between both groups and genes unique to RA or SS [21]. Using the DEGs, gene ontology (GO) and comparative gene expression analysis between the three groups, that is, healthy volunteers, RA, and SS patients were performed. The normalized expression values using Z scores of each group were obtained, and subsequently, heatmap of the common genes was generated using HemI tool [22].
2.3. GO and Pathway Enrichment Analysis
Database for Annotation, Visualization, and Integrated Discovery (DAVID) is a web accessible program that helps in rapid annotation and summarization for GO and pathway analysis data [23]. DAVID consists of four main modules: Annotation tool, GoCharts, KeggCharts, and DomainCharts. These modules are used to mine data accordingly and help in faster annotation. DAVID was used to generate unique ENTREZ IDs for all gene symbols to prevent duplication of data. Functional enrichment analysis of DEGs was performed to identify the associated GO terms. Protein Analysis Through Evolutionary Relationships (PANTHER) database is designed to facilitate high throughput analysis by classifying proteins according to family and sub-family, molecular function, biological processes, and pathways [24]. The website allows users to browse and query gene functions, and further to analyze large-scale experiments with relevant statistical data. For pathway analysis, DEGs were processed using Pathview web server [25]. Log2FC of gene expression is coded into the KEGG graph, rendered as colors to show low high or low expression. The Reactome database was used for in-depth analysis of the pathways [26]. Reactome is an open-source curated database of human pathways and reactions. Pathway annotations are cross-referenced to proteins, small molecules, genes, and primary text mining of an array of sources.
3. RESULTS AND DISCUSSION
3.1. Identification of DEGs
In the present study, microarray expression data of both diseased and healthy whole blood were obtained from the GEO database. In the accession no. GSE93272, the gene expression data were obtained from 45 drug-naïve RA patients and 35 healthy controls [13], whereas accession no. GSE84844 consisted of 30 primary SS and 30 healthy controls [20]. The DEGs identified were further filtered with the fold change in gene expression. Filtered DEGs of SS and RA datasets obtained after applying fold change were used to generate Venn diagram using Venny 2.1 [Figure 1]. It was observed that out of 206 genes in RA, 160 genes were unique whereas in SS, 92 genes out of 138 genes were found to be unique. Between both the datasets, only 46 genes are expressed commonly in both RA and SS [Table 1]. The expression values of the common DEGs were used to generate heatmap using HemI tool [Figure 2]. Comparative gene expression analysis between SS and RA showed up-and-down regulation of genes. Expression of three genes, that is, C-type lectin domain family 4 member D (CLEC4D), lymphocyte antigen 96 (LY96), and BCL2-related protein A1 (BCL2A1), was highly upregulated in RA. Three of the most downregulated genes in RA were BMP2-inducible kinase (BMP2K), MS4A4A, and cytochrome c oxidase subunit 7B (COX7B). The expression of COX7B gene in both RA and SS was downregulated. In case of SS, the expression of MS4A4A gene was upregulated whereas in RA, its expression was downregulated. Further analysis of these significantly upregulated genes could help us gain an insight into the progression of RA.
CLEC4D is a member of the C-type lectin domain superfamily. It is associated with diverse functions in the cell-like cell adhesion, signaling and plays a role in inflammation and immune responses. A gene expression analysis of human pancreas tissues revealed that CLEC4D was downregulated in Type 1 diabetes [27]. In osteoarthritis (OA), CLEC4D was identified as a major miRNA target that suggests its critical role in OA pathogenesis and was suggested as major influencer in OA progression [28]. This gene could be explored as an indicator for RA. The role of CLEC4D, as a C-type lectin, has been widely discussed and its activity is tightly regulated since it is a useful indicator of cell state. Various C-type lectins have been discussed because of their role in maintaining homeostasis of microbiota. They form a network of important proteins that are involved in many functions of the cell [29]. A study has also revealed the connection between disease states of tuberculosis and RA, involving many common genes and proteins [30]. The progression of RA leads to the development of tuberculosis among immune-compromised patients. LY96 is associated with toll-like receptor 4 on the lipid membrane of cells and initiates a response against lipopolysaccharides (LPSs). LY96 gene liaises with TLR4 in the innate immune response to bacterial LPS [31]. LY96 has been shown to be upregulated in patients with RA [32]. The involvement of TLR ligands’ signaling suggests a connection that results in persistent inflammation and joint destruction early on in the disease. The high upregulation of LY96 gene suggests a key role in RA progression. In the search for putative biomarkers for RA, LY96 seems to be a high return target for deeper analysis. LY96 overexpression was also reported in SS as a pro-inflammatory gene on ocular surfaces. LY96 gene is activated in the conjunctiva of SS patients finally leading to keratoconjunctivitis sicca symptoms [33]. Many cancers have also shown evidence of high LY96 expression such as thyroid carcinomas, glioblastomas, and Wilms’ tumor [34-36]. BCL2A1 encodes a member of the BCL2 protein family. This gene is involved in crucial cellular activities such as maintenance of homeostasis, programmed cell death, and tumorigenesis [37]. It is also responsible for reduction in the levels of pro-apoptotic cytochrome c to block the activation of caspase. Control of cell fate is determined by the regulated induction of apoptotic markers [38]. Out of all the roles played by BCL2A1, the function of this gene that is most studied is its role in the inflammation cascade. Interaction of BCL2A1 with pro-inflammatory cytokines such as IL-1 and TNFα. BCL2A1 overexpression has been studied to be a factor in reduction of pro-apoptotic markers to keep a check on inflammation [39]. From the irrefutable evidence gained from the study of BCL2A1, focusing on this genes’ role in severe autoimmune diseases like RA could help in further development of an outcome that could aid in alleviation of pain and inflammation that is so closely associated with these diseases. BCL2A1 activation solely rests on the checks and balances of cell death and differentiation.
 | Figure 1: Venn diagram showing the number of genes expressed uniquely in RA and SS datasets as well as the genes that are commonly expressed in both RA and SS datasets. [Click here to view] |
 | Table 1: Expression of common genes in rheumatoid arthritis and Sjögren’s syndrome patients. [Click here to view] |
COX7B is one of the decisive genes playing a role in oxidative phosphorylation. It acts as the terminal part of the mitochondrial electron transport chain (ETC) that eventually drives oxidative phosphorylation. Significant downregulation of COX7B gene was observed in both SS and RA genes. Downregulation of the genes of the ETC could point to further severe outcomes of the disease. However, this could also point to the fact that diseases like RA could have a better outcome with drugs that specifically target the ETC. It has been reported that downregulation of ETC genes points toward a dysfunction in the redox balance of the energy production [40]. A few studies have shown evidence of the role of COX7B gene in cardiac contraction and further in cardiomyopathy in adults as well as pediatric cases [11,41]. This gene needs to be studied in depth with the other genes of Complex IV of oxidative phosphorylation to arrive at a stronger and more conclusive role of the genes of ETC in diseases such as RA and SS.
 | Figure 2: Heatmap showing differential expression genes that are commonly expressed between RA and SS cohorts. Comparative gene expression analysis between SS and RA showed up-and-down regulation of genes. [Click here to view] |
BMP2K is a crucial gene involved in skeletal development and growth. BMP stands for bone morphogenic proteins and BMP2K acts as a putative regulatory gene involved in osteoblast differentiation. It was proposed as a novel biomarker for gallbladder cancer; since it was identified as a clathrin-coated vesicle-associated protein in the progression of gallbladder cancer [42]. Extensive mapping of this gene has not been done in autoimmune diseases but it shows immense potential in the tumor microenvironment.
The MS4A4A is the membrane spanning 4-domains A4A protein. In RA, its expression was downregulated while in SS, it is upregulated. The MS4A4A gene has been reported to present some unique expression patterns in hematopoietic and non-lymphoid tissues. It is selectively expressed by macrophage lineage cells and exists as a plasma membrane tetraspan molecule. Human MS4A4A is coexpressed with M2/M2 like molecules that are present on infiltrating macrophages from synovial tissue that shows inflammation. It has been shown that MS4A4A can act as a practical target for in-depth analysis in the search for RA biomarkers. This finding would need to be validated in with healthy controls versus RA [43]. Conversely, among genes that show differential expression in SS, MS4A4A showed the highest change in expression and has been reported to be associated with mesenchymal stem cell expression with T helper cells. A significant upregulation was reported in MS4A4A gene expression [44]. Expression of MS4A4A in mesenchymal stem cells or even with T- and B-cell differentiation can lead us to strong evidence of this gene in various altered immune system pathways in diseases.
3.2. GO Analysis
The DEGs unique to RA and SS were submitted separately to DAVID for gene function augmentation analysis to identify the most significant GO categories and pathways. The genes common to both diseases were also used to identify pathways most crucial to their function. This helped us gain more insight into the cellular localization, molecular and biological function of the proteins, as well as the pathways involved in RA and SS. After processing the gene lists on PANTHER, graphs were generated using the data obtained depicting the role of the genes in various biological processes, cellular components, molecular functions, protein class, and pathways. After processing the genes unique to RA [Figure 3], it was found that a significant number of genes were involved in catalytic activities like hydrolase activity and binding activities like nucleic acid binding. When biological processes were analyzed, the genes were mostly involved in cellular macromolecule and biosynthetic metabolic processes, cellular regulation of metabolic processes, and cellular processes like DNA-dependent transcription. Among cellular components, RA unique genes were found to be part of intracellular organelles primarily the mitochondrial nuclear lumen. Among the protein classes for RA genes, most were involved as metabolite interconversion enzymes such as oxidoreductase and hydrolase and were also involved as protein-modifying enzymes such as serine proteases. They were also involved in EGF receptor signaling pathways primarily expressed with epidermal growth factor, CCKR signaling pathways, inflammation mediated by chemokines, and cytokine signaling expressed with cyclooxygenase, phospholipase A2, and Rho proteins. In SS, unique genes were processed through PANTHER and graphs were generated from the data obtained [Figure 4]. On analyzing the molecular functions, the genes unique to SS were found to be most involved in catalytic activities such as peptidase activity and protein kinase activity. Cellular processes like protein modification by small proteins and metabolic processes like metabolism of organic macromolecules were the key biological processes that these SS genes were involved. The majority of SS genes in the cellular component were involved in part of the nuclear lumen as well as nucleotide excision repair complex. On further analysis, some key pathways that SS genes are involved in are apoptosis signaling pathway, gonadotropin-releasing hormone receptor pathway, and inflammation mediated by chemokine and cytokine signaling pathway involved with interleukin 2 and beta-arrestin proteins. The common genes between SS and RA were analyzed and revealed that most of the genes were involved in binding activities such as GTPase binding, kinase binding, phosphatase binding, and protease binding [Figure 5]. Catalytic activity involvement of these genes was limited to hydrolase and peptidase activity.
3.3. Pathway Enrichment Analysis
Processing the DEGs indicated that genes of two major pathways were highly expressed in RA [Figure 6a]. The arachidonic acid metabolism was affected due to the change in expression of three core enzymes, namely, leukotriene-B4-20-monooxygenease (CYP4F2 or CYP4F3) (Enzyme nomenclature: 1.14.14.94), prostaglandin endoperoxide synthase (Enzyme nomenclature: 1.14.99.1), and phospholipase A2 (Enzyme nomenclature: 3.1.1.4). Eicosanoids are autocrine and paracrine signaling molecules that modulate physiological processes including pain, fever, inflammation, blood clot formation, smooth muscle contraction and relaxation, and the release of gastric acid. The second pathway most affected by DEGs was pantothenate and coenzyme A biosynthesis. Branched chain amino acid transaminase (Enzyme nomenclature: 2.6.1.42) is affected, as shown in Figure 6b. The involvement of the arachidonic acid pathway in RA largely manifests in patients as chronic inflammation. Eicosanoids are synthesized in humans primarily from arachidonic acid (all-cis 5,8,11,14-eicosatetraenoic acid) [45] released from membrane phospholipids. Once released, arachidonic acid is acted on by prostaglandin G/H synthases (PTGS, also known as cyclooxygenases [COX]) to form prostaglandins and thromboxanes, by arachidonate lipoxygenases (ALOX) to form leukotrienes, epoxygenases (cytochrome P450s and epoxide hydrolase) to form epoxides such as 15-eicosatetraenoic acids, and omega-hydrolases (cytochrome P450s) to form hydroxyeicosatetraenoic acids [41]. Levels of free arachidonic acid in the cell are normally very low so the rate of synthesis of eicosanoids is determined primarily by the activity of phospholipase A2, which mediates phospholipid cleavage to generate free arachidonic acid. Phospholipase A2 also acts on phosphotidylethanolamine, choline plasmalogen, and phosphatides removing the fatty acid attached to the 2nd position. Pantothenate or Vitamin B5 acts as precursor of coenzyme A, one of the five coenzymes of the citric acid cycle. Dysregulation of the citric acid cycle affects the mitochondrial ETC. Alteration in the expression of tricarboxylic acid cycle has been implicated toward the progression of autoimmune diseases [46].
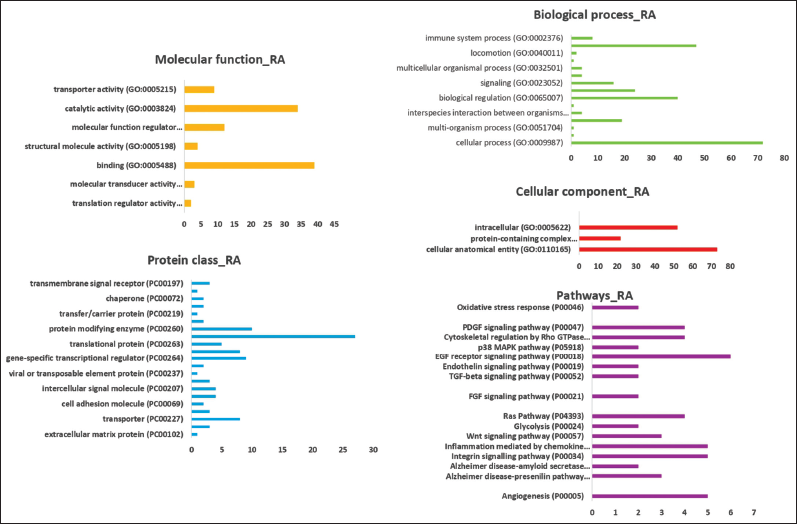 | Figure 3: Gene ontology analysis of genes that are unique to RA. Analysis consists of genes involved in molecular function, biological processes, cellular components, protein classes, and pathways. [Click here to view] |
In the present study, the genes that were found to be unique to SS were also processed through Pathview [Figure 7]. The major pathway that was found to be involved was the RIG-1-LIKE receptor signaling pathway. The genes associated with the pathway are known to play a key role in sensing viral RNA to modulate antiviral immunity. Innate immunity and inflammation are triggered on detection of self-RNA in the cytoplasm. This receptor signaling pathway is also involved in signal crosstalk between innate immune components and other factors that can impart a stronger immune response [47]. The continuous production of Type 1 interferons (IFNs) against self-antigens leads to the advancement of autoimmune disorders. It is this development that steadily progresses to severe outcomes of autoimmune diseases eventually leading to the downstream activation of pain and inflammation pathways [48]. From the diagram, it is evident that interferon regulatory factor 7 (IRF7) is upregulated. It is a critical transcription factor in the innate immune response. IRF7 is known to play a role in the transcriptional activation of virus-inducible genes. Overexpression of this gene leads to an increase in viral replication. IRF7 overexpression has a direct role to pay in IFN signaling as well. Type I IFNs typically recruit JAK1 and TYK2 proteins to transduce their signals to STAT1 and 2; in combination with IFN-regulatory factor 9 (IRF9), these proteins form the heterotrimeric complex IFN-stimulated gene factor 3 (ISGF3). Type I IFNs are composed of various genes including IFN alpha (IFNA), beta (IFNB), omega, epsilon, and kappa. The IFNA/B stimulation of the IFNA receptor complex leads to the formation of two transcriptional activator complexes: IFNA-activated factor (AAF), which is a homodimer of STAT1 and ISGF3, which comprises STAT1, STAT2, and a member of the IRF family, IRF9/P48. AAF mediates activation of the IRF-1 gene by binding to GAS (IFNG-activated site), whereas ISGF3 activates several IFN-inducible genes including IRF3 and IRF7, as evident from the diagram. The ISG-encoded proteins include direct effectors which inhibit viral infection through diverse mechanisms as well as factors that promote adaptive immune responses [49]. ISG15 also conjugates some viral proteins, inhibiting viral budding and release. TRAF3 acts as a scaffold for the assembly of a signaling complex composed of IKK epsilon/TBK1, leading to the activation of transcription factors IRF3/IRF7. TRAF6 is crucial for both RIG-I- and MDA5-mediated antiviral responses. The absence of TRAF6 resulted in enhanced viral replication and a significant reduction in the production of type I IFNs and IL6 after infection with RNA virus. The continuous production of type 1 IFN against self-antigens leads to the advancement of autoimmune disorders. It is the development that steadily progresses to severe outcomes of autoimmune diseases eventually leading to the downstream activation of pain and inflammation pathways.
 | Figure 4: Gene ontology analysis of genes that are unique to SS. Analysis consists of genes involved in molecular function, biological processes, cellular components, protein classes, and pathways. [Click here to view] |
 | Figure 5: Gene ontology analysis of genes that are common between RA and SS. Analysis consists of genes involved in molecular function, biological processes, cellular components, protein classes, and pathways. [Click here to view] |
 | Figure 6: (a) Pathview analysis showing the expression of DEGs of RA in various pathways. Two major pathways affected are the arachidonic acid metabolism pathway and the pantothenate and coenzyme A biosynthesis pathway. Arachidonic acid metabolism is mainly involved in the pain mechanism directly leading to dysregulation in its breakdown. (b) Pathview analysis showing the expression of DEGs of RA in various pathways. Pantothenate and coenzyme A biosynthesis pathway involvement lead to a dysregulation of the production of L-valine and L-isoleucine. [Click here to view] |
 | Figure 7: Pathview analysis showing the expression of DEGs of SS in various pathways. From the diagram, it is seen that the genes upregulated in SS are involved in viral gene expression and also play an important role in innate immunity as well as inflammation. [Click here to view] |
The common genes between RA and SS were processed and revealed four pathways most affected by the expression of these genes [Figure 7]. Glycolysis or gluconeogenesis is most affected by the enzymes involved. Phosphoglucomutase or glucose phosphomutase (Enzyme nomenclature: 5.4.2.2) activity is achieved at maximum level only in the presence of α-D-glucose-1,6-bisphosphate [Supplementary Figure 1]. The eventual dysregulation of glyceraldehyde-3-phosphate eventually leads to an imbalance in the production of bisphosphoglycerate mutase (Enzyme nomenclature: 5.4.2.4), thereby affecting the pentose phosphate pathway as well as the citric acid cycle. Oxidative phosphorylation is another pathway showing the involvement of the common genes between RA and SS. Cytochrome c reductase (Enzyme nomenclature:7.1.1.8) in conjunction with ubiquinol was upregulated. It is a part of the ETC present on the mitochondrial surface of all aerobic eukaryotes. The enzyme extrudes either two or four protons into the non-cytoplasmic compartment from the cytoplasm, depending on the physiological conditions. Cytochrome P450 4F22 (CYP4F22) [Supplementary Figure 2] is converted to 20-hydroxylate trioxilin A3 (TrXA3), an intermediary metabolite from the 12(R)-lipoxygenase pathway. This pathway is implicated in proliferative skin diseases. Upregulation of QCR8 gene which encodes the Ubiquinol-Cytochrome C Reductase Complex III Subunit 8 is a part of the mitochondrial respiratory chain. The respiratory chain contains three multi-subunit complexes that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. COX7B and COX7C are both highly differentially regulated during oxidative phosphorylation. These genes are involved in catalyzing the electron transfer from reduced cytochrome c to oxygen. It is a heteromeric complex consisting of three catalytic subunits that are encoded by mitochondrial genes and numerous structural subunits encoded by nuclear genes. COX7B has a direct involvement in cardiac muscle contraction.
The 60S ribosomal protein L24e encoded by the gene RPL24 and 60s ribosomal protein L22 encoded by gene RPL22 are both located in the cytoplasm and involved in mRNA translation during viral infection [Supplementary Figure 3]. RPL24 dysregulation directly causes immunodeficiency 60. Immunodeficiency 60 is a dominant autosomal primary immunologic disorder characterized by inflammatory bowel disease and recurrent sinus infections. This happens due to dysregulation of the innate immune system, leading to a decrease in T-reg cells and overall weakened lymphocyte maturation. The methionine residue that initiates the downstream processing of RPL22 is post-translationally removed. The protein can bind specifically to Epstein–Barr virus-encoded RNAs 1 and 2. As shown in various genes encoding ribosomal proteins, there exist multiple pseudogenes of both RPL24 and RPL22. RPS7 (small ribosomal subunit protein ES7) encodes a protein that is a component of the 40S subunit of ribosomes. RPL24 interacts with DBA8 protein which is involved in the progression of Diamond-Blackfan anemia. This protein is phosphorylated by NEK6 and deubiquitinated by DESI2 that further lead to its stabilization. The dysregulation of the genes involved commonly between RA and SS also has a crucial role to play in cardiac muscle contraction. Cardiac muscle contraction [Supplementary Figure 4] is seen when the sodium-calcium exchanger (NCX) is underexpressed, leading to a direct outcome on the contractility of cardiac tissue. Altered expression and regulation of this gene distorts the Ca2+ homeostasis finally leading to arrhythmia and heart failure.
The expression of the arachidonic acid metabolism pathway as well as the RIG-1-like receptor signaling pathway helps us in visualizing the wide area from which the autoimmune microenvironment starts to take shape. Starting from the continuous recognition of self-antigens and mounting an attack against them, to sustenance of the downstream pain pathways that are characteristic of RA and SS progression, it would be prudent to assume that targeted biomarkers for RA and SS can be studied within these crucial pathways. Activation or blocking of central genes or even modulation of their functions could help in further validating the importance of these signaling pathways. Investigations into the activation of IFNs and into the mechanism of IFNN overproduction could help us in screening drugs against the same. Different patients of autoimmune diseases have different manifestations of the disease; hence, a detailed picture of molecular phenotypes of these genes and proteins could help in discovery of personalized biomarker targeting for each patient.
4. CONCLUSION
This study was done to identify key genes and pathways that play a role in RA and SS. In the pursuit of the identification of genes and proteins that may help in better and deeper comprehension of the diseases, some genes were found with high expression to most likely play a role in RA and SS. Through stringent screening by bioinformatics’ tools, it was understood that some genes such as CLEC4D, LY96, MS4A4A, and BCL2A1 may play a crucial role in the pathogenesis of RA and in the future could guide targeted therapy and act as a diagnostic biomarker platform. Pathways such as the arachidonic acid pathway and RIG-1-like receptor pathway were important in further analysis of the mechanism of the action of these diseases. Even in SS, some of the highly downregulated genes may play a role in the pathophysiology of the disease as most of the downregulated genes were also involved in other autoimmune diseases. These results may contribute to an improvement in our understanding of these two crucial autoimmune diseases and also provide an insight into the molecular events associated with RA. These findings need to be confirmed by other studies for further research.
5. ACKNOWLEDGMENT
KC likes to acknowledge the STRF fellowship provided by GGSIPU, New Delhi. The authors also like to acknowledge GGSIPU for infrastructural support and encouragement.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This study has not been funded by any external funding agency.
8. CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
The gene expression datasets analyzed in the present study are available in the Gene Expression Omnibus (GEO) database with accession No. GSE93272 and GSE84844.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Almutairi K, Nossent J, Preen D, Keen H, Inderjeeth C. The global prevalence of rheumatoid arthritis: A meta-analysis based on a systematic review. Rheumatol Int 2021;41:863-77. CrossRef
2. Mantel Ä, Holmqvist M, Andersson DC, Lund LH, Askling J. Association between rheumatoid arthritis and risk of ischemic and nonischemic heart failure. J Am Coll Cardiol 2017;69:1275-85. CrossRef
3. Spagnolo P, Lee JS, Sverzellati N, Rossi G, Cottin V. The lung in rheumatoid arthritis: Focus on interstitial lung disease. Arthritis Rheumatol 2018;70:1544-54. CrossRef
4. Wysocki T, Olesi?ska M, Paradowska-Gorycka A. Current understanding of an emerging role of HLA-DRB1 gene in rheumatoid arthritis from research to clinical practice. Cells 2020;9:1127. CrossRef
5. Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, Grundahl KM, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nat Genet 2013;45:1284-92. CrossRef
6. Oliver JE, Silman AJ. Why are women predisposed to autoimmune rheumatic diseases? Arthritis Res Ther 2009;11:1-9. CrossRef
7. Wang L, Wu LF, Lu X, Mo XB, Tang ZX, Lei SF, et al. Integrated analyses of gene expression profiles digs out common markers for rheumatic diseases. PLoS One 2015;10:e0137522. CrossRef
8. Hajiabbasi A, Masooleh IS, Alizadeh Y, Banikarimi AS, Parsa PG. Secondary Sjogren’s syndrome in 83 patients with rheumatoid arthritis. Acta Med Iran 2016;54:448-53.
9. Ibáñez-Cabellos JS, Seco-Cervera M, Osca-Verdegal R, Pallardó FV, García-Giménez JL. Epigenetic regulation in the pathogenesis of Sjögren syndrome and rheumatoid arthritis. Front Genet 2019;10:1104. CrossRef
10. Takeshita M, Suzuki K, Kondo Y, Morita R, Okuzono Y, Koga K, et al. Multi-dimensional analysis identified rheumatoid arthritis-driving pathway in human T cell. Ann Rheum Dis 2019;78:1346-56. CrossRef
11. Hao R, Du H, Guo L, Tian F, An N, Yang T, et al. Identification of dysregulated genes in rheumatoid arthritis based on bioinformatics analysis. PeerJ 2017;5:e3078. CrossRef
12. Afroz S, Giddaluru J, Vishwakarma S, Naz S, Khan AA, Khan N. A comprehensive gene expression meta-analysis identifies novel immune signatures in rheumatoid arthritis patients. Front Immunol 2017;8:74. CrossRef
13. Tasaki S, Suzuki K, Kassai Y, Takeshita M, Murota A, Kondo Y, et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat Commun 2018;9:1-12. CrossRef
14. Hasan M, Mustafa G, Iqbal J, Ashfaq M, Mahmood N. Quantitative proteomic analysis of HeLa cells in response to biocompatible Fe2C@ C nanoparticles: 16O/18O-labelling and HPLC-ESI-orbit-trap profiling approach. Toxicol Res 2018;7:84-92. CrossRef
15. Yoo SM, Choi JH, Lee SY, Yoo NC. Applications of DNA microarray in disease diagnostics. J Microbiol Biotechnol 2009;19:635-46.
16. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. CrossRef
17. Hasan M, Gulzar H, Zafar A, ul Haq A, Mustafa G, Tariq T, et al. Multiplexing surface anchored functionalized iron carbide nanoparticle: A low molecular weight proteome responsive nano-tracer. Colloids Surf B Biointerfaces 2021;203:111746. CrossRef
18. Hasan M, Yang W, Ju Y, Chu X, Wang Y, Deng Y, et al. Biocompatibility of iron carbide and detection of metals ions signaling proteomic analysis via HPLC/ESI-Orbitrap. Nano Res 2017;10:1912-23. CrossRef
19. Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol 2016;1418:93-110. CrossRef
20. Tasaki S, Suzuki K, Nishikawa A, Kassai Y, Takiguchi M, Kurisu R, et al. Multiomic disease signatures converge to cytotoxic CD8 T cells in primary Sjögren’s syndrome. Ann Rheum Dis 2017;76:1458-66. CrossRef
21. Oliveros J. An Interactive Tool for Comparing Lists with Venn’s Diagrams (2007-2015); 2018.
22. Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: A toolkit for illustrating heatmaps. PLoS One 2014;9:e111988. CrossRef
23. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 2003;4:1-11. CrossRef
24. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 2003;13:2129-41. CrossRef
25. Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013;29:1830-1. CrossRef
26. Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res 2011;39:D691-7. CrossRef
27. Yip L, Fuhlbrigge R, Alkhataybeh R, Fathman CG. Gene expression analysis of the pre-diabetic pancreas to identify pathogenic mechanisms and biomarkers of Type 1 diabetes. Front Endocrinol 2020;11:990. CrossRef
28. Li H, Bai B, Wang J, Xu Z, Yan S, Liu G. Identification of key mRNAs and microRNAs in the pathogenesis and progression of osteoarthritis using microarray analysis. Mol Med Rep 2017;16:5659-66. CrossRef
29. Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat Rev Immunol 2018;18:374-89. CrossRef
30. Badr MT, Häcker G. Gene expression profiling meta-analysis reveals novel gene signatures and pathways shared between tuberculosis and rheumatoid arthritis. PLoS One 2019;14:e0213470. CrossRef
31. Inomata M, Horie T, Into T. Effect of the antimicrobial peptide LL-37 on gene expression of chemokines and 29 toll-like receptor-associated proteins in human gingival fibroblasts under stimulation with Porphyromonas gingivalis lipopolysaccharide. Probiotics Antimicrob Proteins 2020;12:64-72. CrossRef
32. Ospelt C, Brentano F, Rengel Y, Stanczyk J, Kolling C, Tak PP, et al. Overexpression of toll?like receptors 3 and 4 in synovial tissue from patients with early rheumatoid arthritis: Toll?like receptor expression in early and longstanding arthritis. Arthritis Rheum 2008;58:3684-92. CrossRef
33. Trujillo-Vargas CM, Schaefer L, Yu Z, Pflugfelder S, Britton RA, De Paiva C. Transcriptional expression of immune genes in the Sjögren Syndrome conjunctiva. Investig Ophthalmol Visual Sci 2020;61:1379.
34. Metovic J, Vignale C, Annaratone L, Osella-Abate S, Maletta F, Rapa I, et al. The oncocytic variant of poorly differentiated thyroid carcinoma shows a specific immune-related gene expression profile. J Clin Endocrinol Metab 2020;105:e4577-92. CrossRef
35. Moreno D, Sussuchi L, Leal L, Sorroche B, Santana I, Matsushita M, et al., editors. The overexpression of Immune oncology-related genes NFSF14, LY96, SLC11A1 and CTSL are associated with short survival in glioblastoma. Cancer Immunol Res 2021;9:PO005. CrossRef
36. He S, Yang L, Xiao Z, Tang K, Xu D. Identification of key carcinogenic genes in Wilms’ tumor. Genes Genet Syst 2021;96:141-9. CrossRef
37. Kønig SM, Rissler V, Terkelsen T, Lambrughi M, Papaleo E. Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level. PLoS Comput Biol 2019;15:e1007485. CrossRef
38. Berberich I, Hildeman DA. The Bcl2a1 gene cluster finally knocked out: First clues to understanding the enigmatic role of the Bcl-2 protein A1. Cell Death Differ 2017;24:572. CrossRef
39. Zheng Q, Gan G, Gao X, Luo Q, Chen F. Targeting the IDO-BCL2A1-cytochrome c pathway promotes apoptosis in oral squamous cell carcinoma. Oncotargets Ther 2021;14:1673. CrossRef
40. Derambure C, Dzangue-Tchoupou G, Berard C, Vergne N, Hiron M, D’Agostino M, et al. Pre-silencing of genes involved in the electron transport chain (ETC) pathway is associated with responsiveness to abatacept in rheumatoid arthritis. Arthritis Res Ther 2017;19:1-13. CrossRef
41. Hefti MM, Senthil K, Karlsson M, Ehinger J, Mavroudis CD, Nadkarni VM, et al. Early mitochondrial gene regulation in pediatric cardiac arrest. Circulation 2018;138 Suppl 2:A296. CrossRef
42. Song X, Li M, Wu W, Dang W, Gao Y, Bian R, et al. Regulation of BMP2K in AP2M1-mediated EGFR internalization during the development of gallbladder cancer. Signal Transduct Target Ther 2020;5:154. CrossRef
43. Seyhan AA, Gregory B, Cribbs AP, Bhalara S, Li Y, Loreth C, et al. Novel biomarkers of a peripheral blood interferon signature associated with drug-naïve early arthritis patients distinguish persistent from self-limiting disease course. Sci Rep 2020;10:8830 CrossRef
44. Hu L, Xu J, Wu T, Fan Z, Sun L, Liu Y, et al. Depletion of ID3 enhances mesenchymal stem cells therapy by targeting BMP4 in Sjögren’s syndrome. Cell Death Dis 2020;11:1-14. CrossRef
45. Scagliola A, Mainini F, Cardaci S. The tricarboxylic acid cycle at the crossroad between cancer and immunity. Antioxid Redox Signal 2020;32:834-52. CrossRef
46. Wang B, Wu L, Chen J, Dong L, Chen C, Wen Z, et al. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct Targeted Ther 2021;6:94. CrossRef
47. Ryan DG, Murphy MP, Frezza C, Prag HA, Chouchani ET, O’Neill LA, et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab 2019;1:16-33. CrossRef
48. Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity 2011;34:680-92. CrossRef
49. Kato H, Oh SW, Fujita T. RIG-I-like receptors and Type I interferonopathies. J Interferon Cytokine Res 2017;37:207-13. CrossRef
SUPPLEMENTARY FIGURES
 | Supplementary Figure 1: Pathview analysis showing the expression of common genes expressed in both RA and SS in various pathways. Involvement of the glycolysis/gluconeogenesis enzymes leads to an impairment of the pyruvate cycle and directly leads to overexpression of the pentose phosphate cycle. [Click here to view] |
 | Supplementary Figure 2: Pathview analysis showing the expression of common genes expressed in both RA and SS in various pathways. Involvement of the oxidative phosphorylation dysregulation leads to an impairment of the cellular respiration and further downstream complications leading to disease and formation of reactive oxygen species. [Click here to view] |
 | Supplementary Figure 3: Pathview analysis showing the expression of common genes expressed in both RA and SS in various pathways. Involvement of the upregulated ribosomal proteins is associated with viral translations, immunodeficiencies, and immune system dysregulation. [Click here to view] |
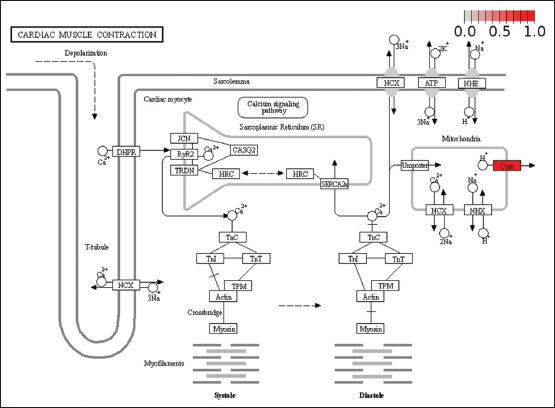 | Supplementary Figure 4: Pathview analysis showing the expression of common genes expressed in both RA and SS in various pathways. Involvement of the upregulated ribosomal proteins is associated with viral translation, immunodeficiencies, and immune system dysregulation. [Click here to view] |