1. INTRODUCTION
We have entered the age of antibiotics with the discovery of penicillin in 1929 and streptomycin in 1943 where treatment of previously considered deadly bacterial infections was easy. The time frame from 1950 to 1960 was viewed as the brilliant period of anti-microbial revelation as one portion of the anti-microbial utilized today were found in the 20th century. Since then, there has been a worldwide increase in the number of microbial infections over the previous years because of the increase of antimicrobial resistance in organisms resulting in a phenomenon called multidrug resistance. The multidrug resistance may be described as the resistance shown by various microbial strains towards known and notable antibiotics. This resistance of bacterial strains toward antibiotics has been developed several years ago right after the invention of the very first miracle drug named penicillin. Bacteria when exposed to a particular drug for a protracted period can develop survival strategies toward the antibiotic to which it was previously susceptible to. The foremost reasons behind this acquired resistance are (a) overuse of medication; (b) misuse of medicine; (c) consumption of unprescribed drugs; (d) natural selection process in microorganisms; (e) not taking the prescribed dosage of medicine; (f) and negligence toward the correct usage of antibiotics by a good range of the population. Some familiar multidrug-resistant organisms (MDROs) are methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococci, and extended-spectrum beta-lactamases-producing Gram-negative bacteria.
Multidrug resistance has become a universal threat as an outsized number of organisms including various potent pathogens have significantly acquired resistance toward valuable drugs within the market. This increasing resistance to antibiotics shown by pathogens is extremely dangerous since it limits the treatment options and has adverse effects on an individual’s health.
Organisms involved in nosocomial infections (hospital-acquired infection) are various bacteria, fungi, parasites, and other infectious agents that have now become immune to the standard antibiotics because of poor sanitation practices in hospitals and continuous usage of the identical drugs for a protracted period. The communicable disease Society of America refers to the pathogens that cause nosocomial infections as ESKAPE pathogens [1]. ESKAPE is that the abbreviation for Enterococcus species, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species. Data from the National Nosocomial Infections television system suggests that within the US the incidence of nosocomial MRSA infections is steadily increasing which accounts for greater than 60% of the medical aid unit admissions. Staphylococcus aureus has developed resistance to several drugs including second- and third-line drugs [2]. Resistance shown by these pathogens toward multiple drugs has become a burden on medical examiners and economic systems. The results of this are high mortality and morbidity rates, prolonged illness, diagnostic uncertainties, and also ends up in the dearth of trust of patients on the system. Because of these devastating effects of multidrug resistance on society it is become a serious challenge for all scientists and medical workers to guard the planet from life-threatening situations which will be caused because of these MDROs and thus bring back the trust of individuals within the system.
Since multidrug resistance has become a worldwide issue many experts are working to search out the mechanisms in bacterial cells that cause this mischievous behavior. By taking a more in-depth look at the within of those resistant organisms five basic mechanisms (Fig. 1) are described that might be to blame for their throw off antimicrobial drugs. The mechanisms are as follows: a. enzymatic inactivation of drug/alteration of the drug, b. modification of drug binding site, c. reduced intracellular drug accumulation, d. biofilm formation, and e. efflux pumps: this is often the foremost commonly used mechanism by organisms to amass resistance toward drugs.
2. EFFLUX PUMP
Efflux pumps are major determinants of drug resistance and have an incredible role in this much-pronounced phenomenon of multidrug resistance. These efflux pumps are channels that export the antimicrobial substances out of the cell via porins and thus prevent the action of the drug on the target. However, certain molecules aid in overcoming this mechanism of resistance and are referred to as efflux pump inhibitors (EPIs). Since efflux pumps are a serious reason for resistance there need to be counter-strategies to figure against the action of this pump to nullify the effect and forestall the bacterium from escaping the antibiotics. This review together with efflux pumps also focuses on EPIs as prominent molecules in inhibiting the action of those pumps.
2.1. Insight into the Pumps
Efflux pump is a key component of obstruction seen generally in all organisms but is prominently present in Gram-negative micro-organisms and comprise a significant class of resistance determinant. The qualities for this class of protein are situated on chromosomes or plasmids [2]. These pumps have a significant ability to expel harmful material just as colors, cleansers, substantial metals, natural solvents, and above all antimicrobial substances [3–6]; by doing so the organisms are fit for adjusting and getting by in biological and physiological specialties [7]. Articulation of this efflux action is dependent upon tight guidelines by nearby and worldwide transcriptional controllers that show that efflux pumps have physiological capacities particularly under pressure transformation, improvement of pathogenesis and virulence of microorganisms [8–10].
The expulsion of antimicrobials from the bacterial cell is done by explicit cell membrane proteins which behave as pumps and decline the degrees of harmful substances within the cell. Gigantic work is being performed on examining the specific structure of these proteins to eventually consider the working of efflux pumps [11]. These pumps show wide explicitness and high productivity of drug expulsion in which drug efflux is combined with a proton intrusion and in this way these pumps are named H+ antiporters [12,13]. These antiporters based on their composition, number of transmembrane spanning region, energy sources, and substrates are distributed into five families (Fig. 2), which are described below.
 | Figure 1: Mechanisms of drug resistance shown by organisms. 1. Drug (in blue capsule) inactivation. 2. Use of alternative enzyme. 3. Decreased uptake. 4. Biofilm formation. 5. Efflux pump. [Click here to view] |
 | Figure 2: Schematic representation of the main types of bacterial efflux systems. MFS: Major facilitator superfamily; SMR: Small multidrug resistance family; MATE: Multidrug and toxic compound extrusion family; ABC: ATP binding cassette superfamily; RND: Resistance nodulation family. [Click here to view] |
2.1.1. Adenosine triphosphate (ATP) binding cassette (ABC) super-family
The ATP binding cassette superfamily is ubiquitously present in bacterial organisms and is normally recognized as ABC transporters due to the fact they are involved in the transport of substrate from the interior to exterior of the cell. The first attribute member of the ABC family is of the eukaryotic foundation being P-glycoprotein and MRP-1 [13]. ABC transporters are membrane proteins that act as efflux pumps for more than a few lipids, drugs, natural products, and peptides. These transporters act as cleaning devices expelling nonpolar compounds from membrane bilayer to the exterior by means of using power derived from ATP hydrolysis [14]. ABC carriers comprise four centre areas explicitly two Nucleotide binding domains (NBDs) and two transmembrane domains (TMDs) The NBDs are located at the cytoplasmic face of the membrane and are responsible for the power era through ATP hydrolysis whilst the TMDs are accountable for determining the transporter function [15]. This 4-domain employer is enough and indispensable for purposeful ABC transporter. Studies have shown that the two TMDs form a long chamber within the membrane that opens to the outside and constitutes a drug-binding pocket of ABC pumps.
2.1.1.1. Working. ABC transporters basically utilize ATP hydrolysis to power unidirectional transport of substrate throughout the membrane. Transport begins with the binding of the substrate in transmembrane domains which drives conformational changes in NBDs. This leads to the dimerization of NBDs and extended affinity of NBDs toward ATP. Dimerization of NBDs is accountable for closure on inward-facing substrate-binding site and opening of the outward-facing binding sites [16]. Two face-off models have been described for subsequent steps. (a) The ATP swap model: this model states that the binding of nucleotide consequences in a power stroke that affords power for the transport of substrate and then ATP hydrolysis leads to resetting of NBDs to its basal configuration [17]. (b) Alternating catalytic sites model: this model says that drug extrusion is coupled to the relaxation of excessive electricity catalytic site conformation generated through the ATP-hydrolysis step. ATP hydrolysis and subsequent release of inorganic phosphate and adenosine diphosphate cause transporter to reset to unique confirmation [18]. ABC transporter considered in Escherichia coli is MacB is a phase of the tripartite efflux system MacAB. When overexpressed the gadget confers resistance against macrolides composed of lactones. Another example is the BrcA protein of Bacillus licheniformis which is responsible for bacitracin resistance. The protein simply removes the bacitracin molecule from its membrane target This BrcA protein shares homology with hydrophilic ATP binding components of ABC transporters of transport proteins [19]. Interestingly genes coding for ABC transporters in Mycobacterium tuberculosis occupy 2.5% of its genome. 30 ABC transporters were identified in M. tuberculosis which potentially participated in drug transport and for this reason are predominant elements contributing to multidrug resistance considered in M. tuberculosis [20]. Other ABC transporters include SrmB, oleB, DrrAB from Streptomyces species, lantibiotic exporters PepT and SalXY from Streptococcus species [16].
2.1.2. Resistance nodulation (RND) family
The RND group of efflux system is the largest and generally noticeable among all different families. This system is generally present in Gram-negative microscopic organisms like E. coli, P. aeruginosa, Serratia marcescens, and Stenotrophomonas maltophilia. The external layer of Gram-negative microorganisms comprises an additional coverage that goes about as a boundary for a section of poisonous substrates [21]. In any case, concentrates altogether indicated that substrates like beta-lactams could pass this external layer within a moment making it evident that the external film alone can’t go about as a barrier to prevent the entry of medicine but requires another process to figure with it in collaboration and this process was revealed to be the RND efflux framework [22]. The RND efflux pumps exist in a three-way structure crossing over both external and internal membrane [23] subsequently fit for pumping out medications directly within the periplasm not permitting them to reemerge cytosol. The three-sided complex comprises three segments (a) A carrier protein (example AdeB) located within the inner layer (b) Periplasmic adornment protein or membrane fusion protein (MFP) (example AdeA) (c) Outer film protein channel (example AdeC) present within the outer film of Gram-negative bacteria [23,24]. All the three parts together are to fault for the working of RND pumps and likewise, the absence of anybody segment makes it nonuseful. The qualities encoding for MFP are found in an exceedingly single operon or could be a part of operon coding for a unique efflux pump [25]. As indicated by a survey in Acinetobacter baumanii the carrier protein AdeB catches its substrate from inside the internal layer of phospholipid bilayer film or the cytoplasm. This substrate is then shipped into the extracellular layer through MFP (AdeC) and this vehicle is intervened by means of periplasmic protein. The individuals from this group of efflux framework are proton antiporters utilizing the proton angle to control efflux, trading one proton for one drug molecule [24].
Most generally examined efflux pumps of the RND family are AcrAB-Tol arrangement of E. coli and MexAB-OprM arrangement of P. aeruginosa [26,27]. These RND efflux pumps are substrate-specific as an example the ArcAB framework in Proteus mirabilis follows up on novobiocin [24], the ArcAB-TolC arrangement of E. coli acts on a large range of substrates—beta-lactams, chloramphenicol, fluoroquinolones, macrolides, novobiocin and in a similar manner the MexAB-OprM arrangement of P. aeruginosa not just follows up on beta-lactams, chloramphenicol, fluoroquinolones, macrolides, novobiocin yet additionally on aminoglycosides, tetracycline and trimethoprim [23,24]. Different individuals from the RND framework incorporate the SdeAB-SmeC framework of S. marcescens, ArcAB arrangement of K. pneumoniae and Klebsiella oxyloca, AdeAB-AdeC arrangement of Acinetobacter baumani, and so on.
2.1.3. Small multidrug resistance (SMR) family
The SMR family is a proton-driven group of efflux framework having a little size, basic structure, and shows a double topology [28]. The individuals from this family comprise transport proteins situated within the inward layer of the cell. The polypeptide chains of SMR efflux pumps that are found in the inward film of Gram-negative microbes, are 100–140 amino acid build-ups long and contain four transmembrane helices [29,30]. The most usually contemplated SMR efflux pumps incorporate EmrE of P. aeruginosa, SsmE of S. marcescens, EmrE of E. coli, EbrAM of Bacillus subtilis. These pumps are mostly engaged with the expulsion of quaternary ammonium mixes just as antimicrobials. Aside from drug expulsion SMR relatives are likewise associated with the expulsion of guanidinium mixes by going about as guanidinium H+ antiporters, in this way sending out guanidinium particles from the cytoplasm by coupling its movement with proton motive force [28]. The SMR family can be isolated into three unique subclasses dependent on their capacity to present host protection from an expansive scope of medications [31]. The three classes are as follows: 1. Silencer of groEL mutations (silencer of groEL); 2. Paired SMR; and 3. Small multidrug pumps [30,32]. SMR proteins related to multidrug opposition are found on plasmids or associated with horizontal gene transfer. SMR family is engaged with reflux of a wide scope of substrates, for example, lipophilic medications, nicotine, polyamine, DNA intercalating colors, and other poisonous mixes also [30,33,34]. SMR efflux pump AbeS in A. baumanii shows protection from antibiotics, hydrophobic mixes, cleansers, and disinfectants [29] while the EmrE efflux arrangement of E. coli gives protection from harmful mixes like ethidium, acriflavine, tetraphenylphosphonium, benzalkonium, streptomycin, and tobramycin. Consequently, both these individuals from the SMR efflux framework are of incredible incentive for the bacterium to present protection from numerous substrates and advance endurance for a more extended timeframe. Studies have likewise demonstrated that an E. coli gene ynfA, an SMR quality relative, may be associated with the complex guidelines by which beforehand defenseless microbes become safe over the span of evolution [35].
2.1.4. Major facilitator superfamily (MFS)
The MFS is the most important family of all membrane transporter families concerned with the efflux of drugs. The contributors of this family are ubiquitously present in all domains of lifestyle such as microorganisms, archaea, and eukarya [28,36,37]. Maximum efflux pumps of this family incorporate uniporters, symporters, and antiporters that are concerned about antibiotic resistance. These pumps concerned in the extrusion of substrates are present on the plasma membrane, thereby stopping the build-up of antibiotics inside the cell [36]. The structure of MFS transporters is such that it carries transmembrane helices and structural motifs. Most of the MFS transporters encompass 400–600 amino acyl residues in duration with the central helices organized in pseudo-symmetrical helix bundles. Moreover, research has also shown that these residues play a crucial role in catalysis and the two bundles incorporate inverted topology repeats of three bundles each [38]. The structure consists of a vital pore with two domain names that switch conformations from the cytoplasmic aspect of the membrane to the periplasmic facet as a result of the gradient of Na+ or H+ ions [37]. The MFS efflux proteins may be divided into distinct clusters with either 12 or 14 transmembrane segments. The transporter category database has further grouped the MFS family into 76 subfamilies primarily based on phylogenetic analysis, substrate specificity, and mechanisms. Genes encoding for these family of transporters is ordinarily chromosomal but now and again can be located on plasmids [36]. Out of all the subfamilies family, 2 and 3 encompass drug efflux structures and own 12 and 14 transmembranes, respectively. The families are known as DHA14 and DHA12 and work as H+ antiporters to catalyze drug efflux [39].
Studies show that the MFS individuals follow a two-step mechanism for the extrusion of antimicrobials. The sooner proposed version become referred to as a rocker-switch version; however latest studies led to a replace and the model changed into named as the clamp-and-switch model [38,40,41]. The up-to-date model shows that structural modifications are concerned in the extrusion; first, the helices that line the inner pore bend and are assumed to present upward thrust to an occluded country (the clamping step). Then the domains alternatively rotate exposing the binding area to one of the alternative facets of the internal membrane (the switching step). The exit of the substrate is thru the outer membrane thru the TolC protein [40].
A few efflux participants of the MFS family are MefA undercover agent of Streptococcus pyogenes that is a macrolide-efflux protein and is also observed in Streptococcus pneumoniae and Lactococcus lactis, Orf 1 Sco a transmembrane protein of Streptomyces coelicolor, TetV Msm a tetracycline-resistant determinant found in Mycobacterium smegmatis, faucet multidrug resistance efflux pump of Mycobacterium fortuitum and so on [42]. Mefa expels 14 membered macrolides which include erythromycin and oleandomycin whereas any other characterized drug efflux pump Cmr of Corynebacterium glutamicum confers resistance to tetracycline, puromycin and bleomycin. The genome of S. aureus has greater than 30 putative efflux pumps out of which the antiporter DHA-2 belonging to the MFS own family is the maximum enormous. Alongside the DHA-2 family members also export bile salts and dyes; NorA, NorB, QacA/B are a few different most studied MFS efflux individuals in S. aureus [36]. Every other efflux pump of S. aureus Tet38 has involved in resistance to tetracyclin while NorA and NorB are involved within the extrusion of fluoroquinolones. Regulators for the participants of the MFS family of efflux pump include emrAB of E. coli, expression of MdrT pump beneath negative manage of TetR like repressor or BtrA in Listeria monocytogenes, and the farAB operon in Neisseria gonorrhoeae [37,43,44]. Consequently, because of all the above-stated traits of the MFS family, they take place to grow to be a primary motive for multidrug resistance in each Gram-positive in addition to Gram-negative microorganism.
2.1.5. Multidrug and toxic compound extrusion (MATE) family
MATE is another group of the efflux framework that guides the bacterium in the expulsion of harmful mixes, xenobiotics, an enormous number of anti-infection agents, and in this manner causes the bacterium to give opposition against a wide assortment of substrates coming about in multidrug obstruction. Like the MFS family, these efflux frameworks are likewise pervasively present among all spaces of life that incorporate archaea, microscopic organisms, and eukarya. Individuals from this family trade anti-infection agents or substrates by means of transmembrane H+ or Na+ particle gradients [45,46]. MATE relatives are natural cation exporters that discharge metabolic wastes or xenobiotic cations from the microbe and are unequipped for trading substrates conveying a negative charge [47,48]. The regular substrates for the MATE group of efflux frameworks are fluoroquinolones, aminoglycoside antitoxins, and some anticancer chemotherapeutic agents [47]. The MATE family is separated into three subfamilies dependent on their amino acid arrangement similarity (a) DinF (damage inducible protein F), (b) NorM, and (c) eukarya subfamily [33,37,39]. The main MATE carrier structure that was contemplated was the NorM carrier in 1988 [49]. X-ray crystallography uncovered the presence of 12 alpha transmembrane helices with N and C terminal domains. The NorM structure showed a V shape with each arm of the V being made out of six transmembranes, isolating the carrier into two comparably collapsed areas: the N (TM1-TM6) and C space (TM7-TM12). The V-molded structure has the ability of adjusting itself into internal confronting conformation and outward-confronting conformation. The instrument of expulsion is with the end goal that when Na+ particles tie to cation free, drug bound, extracellular confronting carrier it triggers the movement of TM7 and TM8 making the medication be separated from the carrier. The cation bound, without drug carrier at that point changes to an intracellular adaptation to block another medication from the cytoplasm [48]. The most examined carrier of this family is NorM since it is associated with protection from an enormous assortment of anti-microbials, for example, ciprofloxacin, norfloxacin, kanamycin, streptomycin trimethoprim, acriflavine, and ethidium bromide (EtBr). Further investigations prompted the identification of more MATE relatives, for example, AbeM from A. baumanii; HmrM from Hemophilus influenzae; VchM, VcmB, VcmD, and VcrM from Vibrio cholerae; and VmrA from Vibrio parahaemolyticus [37]. As of late a quality coding for MATE family efflux siphon was contemplated: EmmdR from the chromosome of Enterobacter cloacae and this efflux siphon speaks to an arising and promising objective for putative efflux siphon inhibitors.
Aside from the five referenced groups of efflux siphon, specialists have discovered two new families that have a place with this framework and are named as Proteobacterial antimicrobial compound efflux (PACE) family and p-aminobenzoyl-glutamate transporter (ABgt) family [37]. The PACE group of efflux carriers are generally present in Gram-negative microorganisms, for example, Pseudomonas, Klebsiella, Enterobacter, Salmonella, and Burkholderia species and are engaged with giving obstruction toward a scope of biocides utilized as disinfectants and sterilizers, for example, chlorhexidine and acriflavin [46]. All the PACE proteins contemplated show four transmembrane helices that are coordinated into two couple bacterial transmembrane pair domains [50]. The significance of PACE family proteins is probably going to reach out in this field of medication obstruction. The ABgt group of efflux carriers is a class of anti-metabolite carrier and can be a noteworthy objective for the improvement of anti-microbial to fix microbial diseases [51].
3. EFFLUX PUMP INHIBITORS
Since efflux pumps are known to have such a noticeable job in multidrug resistance obstruction developing strategies to cancel these pumps would permit the antitoxins or antibiotics to work effectively and execute the objective. Six fundamental strategies (Fig. 3) have been proposed to sidestep the efflux activity: 1. Adjusting the structure of medication to which microscopic organisms have gotten safe or resistant; 2. Disturbance of the efflux pump gets together; 3. Down managing the outflow of efflux pumps by genetic adjustments; 4. Imploding the energy that is needed for the framework to work; 5. Use of competitive or noncompetitive inhibitor with drugs; and 6. Blocking of inner/outer membrane proteins [52]. EPIs are particles that repress the transport of drugs to the outside of cell and along these lines can collect the antibiotics inside the cell and in the end assist the medications with following up on its objective. Analysts are playing out various examinations to discover the different manners by which EPIs can work, one of them being the capacity of these atoms to rejuvenate the action of antitoxins. Peptidomimetic EPI-MC-207,110 [phenylalanyl-arginyl-naphthylamide (PaN)], an important EPI was discovered for the first time after several years of struggle with multi drug resistance. It demonstrates erythromycin and levofloxacin’s antibacterial activity against clinical isolates of P. aeruginosa that overexpress MexAB-OprM [52]. An ideal EPI should have the following characteristics:
 | Figure 3: Strategies to inhibit efflux pump. 1. Altered drug structure. 2. Suppressed expression. 3. Collapsing efflux energy.4. Application of competitive/noncompetitive drug. 5. Blocking inner/outer membrane proteins. 6. Disruption of pump assembly. [Click here to view] |
- It should not be toxic to the human body.
- It should provide synergistic effects in combination with other drugs.
- It should have a simple structure and should be effective on various targets.
3.1. Classifications of EPIs
Many molecules have shown potential as EPIs, but the mode of action is not known for most of the molecules. Therefore, these compounds are categorized based on their sources rather than their mechanism of action that include 1) natural sources, i.e., EPIs derived from plant products and microorganisms and 2) synthetic compounds. Various classes of EPIs with their detailed chemical structure are mentioned in Table 1.
3.1.1. Natural sources
The application of knowledge regarding the natural sources of the traditional physicians provided helpful information for investigating new drugs. The antibiotic potential of naturally derived substances is studied widely in different parts of the world [53]. These products have provided important bioactive molecules which are active against infectious and noninfectious diseases.
3.1.2. EPIs derived from plants
EPIs derived from medicinal plants are very promising as they provide secondary metabolites which are chemically and structurally diverse with multiple pharmacological properties. Studies have shown that medicinal plants contain phytochemicals that block the efflux pump in both Gram-positive and Gram-negative bacteria and potentially restore the activity of the antibiotics, thus allowing the molecules of antibiotics to reach inside of the bacterial cell at effective concentration [54]. These plant-derived EPIs work in synergy with antibiotics, which in combination provides active treatments for patients that are suffering from MDR-associated conditions [55,56]. Major classes of plant derived EPIs are listed below.
3.1.2.1. Alkaloids. a. Reserpine is an indole alkaloid that is extracted from the roots of Rauwolfia serpentina and Rauwolfia vomitoria and is used as an antipsychotic drug. It shows efflux pump inhibitory activity against the MFS and RND efflux pump superfamily [57]. It is an effective EPI of tetracycline efflux which is exhibited by the Gram-positive B. subtilis via the BMR pump [58]. Reserpine involves direct binding to the B. subtilis membrane protein (BMR) pump that contains two phenylalanine residues (at positions 143 and 306) and one valine (at position 286). The studies show that Reserpine increases the activity of tetracycline by a fourfold decrease in maximum inhibitory concentration (MIC) in two MRSA isolates that contain Tet (k) and results in the eliminating the resistance possessed by the Nora efflux of S. aureus [59]. After the administration of Reserpine, the BMR protein pump expressed by B. subtilis increases the susceptibility to fluoroquinolones and other antibiotics [60]. Because of its nephrotoxic nature, Reserpine has not yet been used in human research with antimicrobial agents [61].
b. Piperine is another alkaloid that is isolated from black pepper (Piper nigrum) which acts as a NorA inhibitor of mutant strains of MRSA by possessing the ability to reverse ciprofloxacin resistance [62]. Piperine along with its derivative pipridine exhibit efflux pump inhibitory activity against the pathogenic bacteria including S. aureus and Mycobacteria spp [63]. According to the studies in M. tuberculosis H37Rv and few other clinical isolates, piperine inhibits an uncharacterized efflux pump Rv1258c by enhancing the activity of rifampicin [64].
3.1.2.2. Phenolic metabolites: flavonoids. a. 5’-methoxy-hydnocarpin is a flavolignan which is isolated from Berberis plant and acts as a successful EPI of the NorA pump. It helps in restoring the activity and also accumulates the berberine which is an antibiotic produced by the same species of plant. However, the clinical application is doubtful, due to its toxic nature [65]. Other plant-derived isoflavines isolated from Lupinus argenteus that comprises orobol, genistein, and biochanin A, has appeared to reduce the MIC of antibiotics, for example, repression of the MDR superfamily of efflux pump in S. aureus and M. smegmatis by use of berberine and norfloxacin [66].
b. Baicalein, a 5, 6, 7-trihydroflavone is a trihydroxy compound isolated from thyme leaves (Thymus vulgaris), is a weak antimicrobial flavone (MIC 100 mg/l). When administered along with antibiotics like ciprofloxacin, tetracycline, and β-lactam including cefmetazole, oxacillin, and ampicillin it enhances the susceptibility of clinical MRSA strain [67,68].
3.1.2.3. Polyphenols. a. Catechin gallate is a polyphenol, which is isolated from green tea extract, and has proven to reverse the resistance caused by the MRSA strains [69]. According to the EtBr inhibition assay, both epicatechin gallate and epigallocatechin gallate show weak inhibitory activity toward the NorA transporter, with epicatechin gallate being slightly more potent than epigallocatechin gallate. The studies suggest that these compounds bind two different sites with different affinities on NorA efflux pumps. Therefore, their activity as EPIs is only observed at higher concentrations [70].
b. Epigallocatechin gallate when administered along with other antibiotics such as tetracycline, erythromycin, and ciprofloxacin upgrades their potencies in TetK-overexpressing Staphylococci and in Campylobacter spp. However, because of its toxic nature, further preclinical investigations were not completed [71].
3.1.2.4. Phenolic diterpenes. a. Carnosic acid and carnosol are examples of phenolic diterpenes and are isolated from Rosemary (Rosmarinus officinalis) act as EPIs. These phenolic diterpenes help in increasing the activity of erythromycin and tetracycline against the macrolide-resistant strain of S. aureus which expresses ABC transporter Msr (A) and Tet (K) [72].
b. Geraniol is also known as monoterpenoid alcohol and is isolated from Helichrysum italicum. In several Gram-negative bacterial species, geraniol has been reported to modulate drug resistance by targeting MDR efflux pump mechanism. This compound decreases the MIC of chloramphenicol in Enterobacter aerogenes CM-64 strain which is known to overexpress the tripartite efflux pump, AceAB-TolC [73].
3.1.2.5. Heterocyclic macrocycles. a. Pheophorbide A is a natural porphyrin found in the Berberis plant and represents a very different class of inhibitors that acts against the NorA efflux pump. Pheophorbide A is a product of cholesterol metabolism and consists of four modified pyrrole subunits. This compound increases the susceptibility of S. aureus toward berberine, but due to the difficulties that were associated during the synthesis and purification there further development was affected, and also because of the unpredictable toxic nature caused reduce in research interest [74].
b. Orizabins a heterocyclic oligosaccharide sensitizes the resistance activity of norfloxacin by inducing a 4–16-fold increase in its activity against S. aureus strain (SA119B) that overexpresses NorA pump. When administrated in lower concentrations, these compounds show greater potency. Orizabins appear to be a promising inhibitor for developing an antimicrobial adjuvant as it shows weak cytotoxic and antitumor activity [75].
3.1.3. Mode of action of plant-derived EPI
Even though efflux pumps are primarily dependent on cellular energy, energy decoupling and the efflux activity possessed by these pumps represent an intriguing approach to efflux pump inhibition via proton gradient or ATPase activity [52]. As most of the energy utilized by the efflux system for their functioning is from H+ and Na+ electrochemical gradient, the EPIs principal mode of action is to target these H+/Na+ motive forces of the efflux pump mechanism or by targeting the competitive or noncompetitive inhibition with the binding substance. Another operation of the mode of action comprises (i) ionic gradient dissipation through the cell membrane; (ii) targeting the transcribing pathway of the particular gene that encrypts the efflux pumps; (iii) interfering with ATP hydrolysis, which compromises the efflux pump and increases permeability in the Gram-negative bacteria’s outer membrane; and (iv) causing conformational changes in the structure of efflux pump or by targeting the assembly of multicomponent pumps such as RND pumps.
3.1.4. Microbial-derived EPI
As we know that most of the EPIs have their origin from plant products or semi-synthetic/synthetic chemical compounds, there is a small fraction of EPIs that are derived from microbes. Microbial fermentation products that are obtained after the screening process can be used as EPIs. EA-371α and EA-371d are examples of inhibitors that are originated from the microbial source. They were first obtained from the fermentation extract of Streptomyces spp and are recognized as a potent inhibitor of the MexAB-OprM pump in P. aeruginosa. EA-371α and EA-371d fail to display selective activity against the other strains of P. aeruginosa [76]. These two compounds are important because of their ability to inhibit the tripartite efflux pump in multidrug resistant Gram-negative pathogens. Their novel structure is an opportunity for the researchers to manufacture derivatives that have increased bioavailability, reduced toxicity, and high potency that act against the efflux pump.
2-(2-Aminophenyl) indole (RP2) – Derivative of indole (RP2), an efflux inhibitor, is another compound that is originated from a soil bacterium. The studies show that it is a potential efflux pump that is used to overcome the efflux-mediated resistance possessed by the strains of S. aureus. This compound was further tested in combination with other antibiotics to check the action against the efflux pump that is overexpressed by the strains of S. aureus. It was also examined for uptake of EtBr, efflux inhibition assay which is the hallmark of EPI functionality, and cytoplasmic membrane depolarization. RP2 is an ideal microbial-derived EPI, which can be used as an effective therapeutic regimen along with other antibiotics in order to eradicate infections caused by S. aureus [77].
3.1.5. Fungal EPIs
EPIs that are derived from yeast are further characterized into five groups: (i) Tacrolimus (FK506) has the ability to potentiate azoles in the pathogenic strains that are known to overexpress the CDR1 and CaMDR genes [78]. (ii) Phenothiazines are known to potentiate ketoconazole in a strain that expresses Pdr5p, Snq2p, and Yor1p by showing characteristic antimicrobial activity [79]. (iii) Propafenone enhances the activity of azoles and terbinafine in the strains that do not possess Cdr1p and Cdr2p [80]. (iv) The effects of cycloheximide or cerulenin on Pdr5p-overexpressing cells are enhanced by isonitrile and enniatin and inhibiting Snq2p (a homologous of Pdr5p) [81]. Enniatins exhibit similarities with FK506 against the mutants of Pdr5p. The inhibitory action possessed by enniatin is competitive against FK506. It is reported that enniatin is more potent and also shows less toxicity than that of FK506. Nevertheless, enniatin does not effectively inhibit Snq2p (a homolog of Pdr5p), and it is known to be toxic to yeast cells [82]. (v) N-methylpiperazine (a derivative quinazolinone) and milbemycin derivatives are characterized as fungal-derived EPIs. These compounds are known to enhance the efficacy of fluconazole in Candida albicans [83,84]. While increasing the solubility of these compounds an analog was discovered which possessed improved physical properties and activity against isolates of Candida spp [85]. Further additions were made to the list of EPIs that are derived from the fungal source with the cyclic peptides: unnarmicin A and unnarmicin C. The modifications came from a catalogue of marine microbes [86] and sulfated sterols deduced from the marine sponge Topsentia sp, [87], acridone [88] derivatives, and ibuprofen [89].
3.1.6. EPIs derived from synthetic source
3.1.6.1. Peptidomimetic compounds. a. Phe-Arg-β-naphthylamide (MC-207, 110; MC-02,595; MC-04, 127), commonly known as PaβN, is a dipeptide amide which was one of the leading EPI compounds discovered through the synthetic approach. PAβN has been found useful in potentiating the activity of various different classes of antibiotics including 4-fluoroquinolones, macrolides, and chloramphenicol by inhibiting efflux pump MDR pathogens [90–92]. This compound is capable of inhibiting MexAB-OprM proteins that are overexpressed by the strains of P. aeruginosa [93]. PAβN increases the activity of levofloxacin eightfold and also increases the activity of other antibiotics against MexCD-OprJ and MexEF-OprN efflux transporters of P. aeruginosa. However, due to its cytotoxic property toward the mammalian cells, this compound along with its derivatives had limited clinical potential [94]. Therefore, PAβN is used as a research tool compound to determine the antimicrobial resistance of the newly identified EPIs.
b. Elacridar (GF120918), Biricodar (VX-710), and Timcodar (VX-853): These three synthetic compounds have the ability to potentiate the activity of drug sensitivity in a mammalian cell that expresses efflux proteins such as MRP-1 and P-gp while producing minimal in-vivo toxicity [95,96]. Elacridar is a multi-cyclic compound that targets multiple drug resistance in cancer. Elacridar (GF120918) acts as an inhibitor of ABC-transporter of MDR-1 (P-gp) and BCRP. It also increases the bioavailability of cytotoxic antitumor drugs [97,98]. Biricodar is a compound developed by Vertex Pharmaceutical and it is used as an adjunct therapy in ovarian cancer [99]. This compound is used in blocking two major multiple drug resistance mechanisms including P-glycoprotein (MDR-1) and multidrug resistance-associated protein (MRP) [100]. Timcodar (VX-853) compound belongs to the class of phenylalanine and derivatives. This compound is known to increase the potency of isoniazid and rifampicin which act as anti-tuberculosis drugs against M. tuberculosis [101].
3.1.6.2. Quinoline derivatives. Quinoline acts as an inhibitor of an antibiotic efflux pump that is found in the isolates of multiple drug-resistant bacterial pathogens such as E. aerogenes [102]. Quinoline derivative pyridoquinolones have the ability to potentiate the activity of norfloxacin against AcrAB-TolC efflux pump that is overexpressed in E. aerogenes [103]. 4-substituted thioalkyl, alkylamino, and alkoxy quinolone are other synthetic analogs that are known to enhance the activity of antibiotics such as tetracyclines, norfloxacin, and chloramphenicol in organisms like K. pneumoniae and E. aerogenes [104]. By modifying the flavone scaffold a series of derivatives have been synthesized such as 2-phenyl-4(1H)-quinolone and 2-phenyl-4-hydroxyquinoline and according to the studies these derivatives have been reported to be effective NorA EPIs in S. aureus [105].
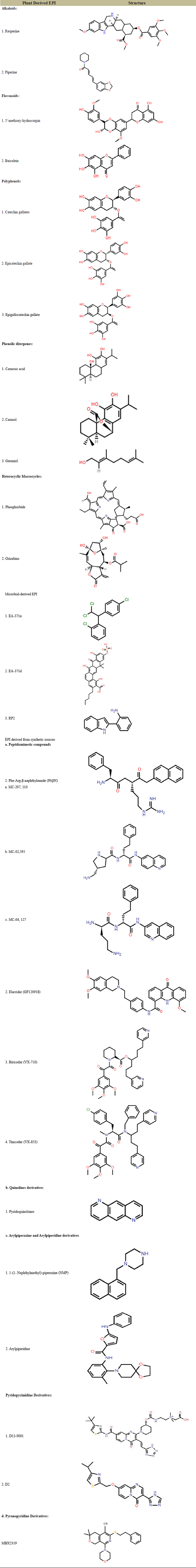 | Table 1: Various EPIs with their chemical structures. [Click here to view] |
3.1.6.3. Arylpiperazine and arylpiperidine derivatives. Arylpiperazine compound known as 1-(1-Naphthylmethyl)-piperazine (NMP) is one of the leading compounds that has shown to possesses EPI activity by inhibiting both AcrAB and AcrEF efflux pump [106,107]. These compounds have the ability to increase the susceptibility of fluoroquinolones in the clinical isolates of E. coli [108]. It was also able to show a partial reversal in drug resistance in species of Enterobacteriaceae, e.g., E. aerogenes, K. pneumoniae [109], and A. baumannii [106]. The mechanism of action of NMP is by causing interference with the functional assembly of the AcrB efflux pump and also by the movement of G-loop that plays a role in extrusion of certain substrates [110]. However, due to their relatively low potency, arylpiperazin has shown limited success as EPIs. Also, these compounds possess structural similarity with serotonin which further hindered their development [74].
According to the reports, arylpiperidine along with its derivatives such as 3-arylpipridine shows the ability to enhance the susceptibility to linezolid in E. coli [111]. Another class of analogues known as phenylpiperidines, which act as serotonin reuptake inhibitors, is known to inhibit the activity of the MDR efflux pump in S. aureus. The AcrAB-TolC pump activity in E. coli is partially affected by these compounds but does not show any effect on efflux activity on the RND efflux pump of P. aeruginosa [112].
3.1.6.4. Pyridopyrimidine derivatives. Pyridopyrimidine analogs D13-9001 and D2 were reported to show activity against the efflux pump (MexAB-OprM pump) expressed by P. aeruginosa under both in-vivo and in-vitro conditions [113,114]. D13-9001 possesses the ability to inhibit the efflux of antibiotics by binding to the specific site on the AcrB pump in E. coli and MexB pump in P. aeruginosa [115]. The compound is known to have a hydrophilic component that interacts with the substrate binding channel of AcrB and MexB pumps which helps in preventing the substrate from binding to the pump [116].
3.1.6.5. Pyranopyridine derivatives. Pyranopyridine compound MBX2319 acts as a potent inhibitor of the AcrAB pump in E. coli, but fails to show any activity in P. aeruginosa [114,117]. This compound helps in potentiating the activity of antibiotics that are substrates of the AcrAB efflux pump [117]. Polymyxin B nonapeptide when administered along with this compound shows a noticeable increase in EPI potency against P. aeruginosa. Furthermore, the crystallographic data suggest that there is an interaction between MBX2319 and hydrophobic trap [115] of the AcrB efflux pump and the pyridine ring formed a ring stacking interaction with the amino acid residues.
4. CHALLENGES
Many pathogenic microorganisms have become resistant over a period of time due to the different mechanisms, one of them being the efflux pump. EPIs play a major role in tackling antimicrobial resistance caused by the efflux pump mechanisms. Even though EPIs have been used as a part of laboratory experimentations for decades, many challenges are faced in order to develop successful commercial EPIs. One of the major hurdles in developing an EPI is its economic value. As EPIs are a new chemical entity (NCE), the major pharmaceutical companies tend to stay away from these compounds as they understand the problems associated with NCE. Due to this, the idea of potentiating and modifying the resistant antibiotics using EPIs was trumped [52]. EPIs that are derived from natural sources have a complex and bulky structure which makes it difficult in synthesizing them. Although syntheses of synthetic EPIs are relatively easier, these compounds have poor solubility, toxicity, and problems with cell permeability. Another challenge in developing EPIs is the lack of preclinical and clinical data. In order to develop EPIs at a commercial level, information on model organisms and patient data is required which is limited. Therefore, more work is required to be carried out at the preclinical and clinical levels for the successful development of EPIs [90]. EPIs are essentially given in combination with antibiotics in order to potentiate them. This gives rise to another challenge that is the compatibility of both partners. For successful combination therapy, the pharmacokinetics of both EPI and antibiotic must complement each other [52].
5. FUTURE PROSPECTIVE
The use of EPIs as therapeutic agents confronts several difficulties, although this should in no way detract from their relevance and benefits. EPIs are a glimmer of hope in a time when there is antibacterial resistance, as they rejuvenate the action of already accessible antibiotics. Because EPIs eliminate the need to develop new antibiotics, this method saves a great deal of the time, effort, and money involved in coming up with a new antibiotic.
Antibiotic resistance can be reversed using EPIs as therapeutic agents. Since the present economic conditions favor the large-scale manufacturing of previously optimized and stored antibiotics, it assumes a significant importance. In the realm of EPIs, X-ray crystallography and computational approaches could have a significant impact (EPIs). Long-term molecular dynamic simulations of already developed EPIs and various bacterial efflux pumps could reveal the binding locations in efflux pump morphology that are currently unknown. It may be able to generate therapeutically relevant EPIs utilizing the rational drug design approach, which could be a powerful weapon in the fight against antimicrobial resistance.
6. CONCLUSION
Due to increasing resistance shown by a wide range of organisms against antibiotics, multidrug resistance has become a major issue for the society. As discussed above, along with many other strategies, efflux pumps play a vital role in aiding the organisms to acquire resistance toward routine antibiotics. If the overuse and misuse of drugs continues, being a major cause of MDR, there is no harm in saying that the next “pandemic” hitting the world would be an “antimicrobial resistance pandemic.” However, molecules like the EPIs would provide a ray of hope in such a situation.
EPIs play a major role in inhibiting the escape of drugs. However, choosing the source of EPIs sometimes becomes challenging as the naturally derived EPIs have a complex structure and are larger molecules, while the synthetic EPIs have problems like toxicity and sometimes their synthesis becomes difficult.
Even though the use of EPIs as therapeutic agent faces a lot of challenges, its power cannot be suppressed. The most remarkable property of an EPI is that it can revive the activity of a drug to which already the bacterial cell has acquired resistance and therefore synthesizing a new drug is not essential. The use of EPIs with other drugs has shown to provide synergistic effects.
Thus, to conclude, we can say that in this new era of MDR, the use of EPIs could be a promising approach to decrease the resistance for antibiotics shown by pathogens. Speaking more specifically, EPIs in the coming future can be used to block the “escape of ESKAPE pathogens,” thus acting as “saviors of life!”
7. ACKNOWLEDGMENT
The authors express their sincere gratitude toward Principal, Abeda Inamdar Senior College Pune, for providing the necessary infrastructure conducive for their review work.
8. CONFLICT OF INTEREST
There is no conflict of interest.
9. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
10. FUNDING
There is no funding to report.
11. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
12. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
13. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
14. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 2016;4(1):14. CrossRef
2. Kumar M. Multidrug-resistant Staphylococcus aureus, India, 2013–2015. Emerg Infect Dis 2016;22:1666–7. CrossRef
3. Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 2015;28(2):337–418. CrossRef
4. Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun 2014;453(2):254–67. CrossRef
5. Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria: a study. J Pathogens 2016;2016:4065603. CrossRef
6. Lehtinen S, Blanquart F, Lipsitch M, Fraser C, Maela Pneumococcal Collaboration. On the evolutionary ecology of multidrug resistance in bacteria. PLoS Pathogens 2019;15(5):e1007763. CrossRef
7. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res Int 2016;2016 doi:10.1155/2016/2475067. CrossRef
8. Nguyen QH, Contamin L, Nguyen TV, Bañuls AL. Insights into the processes that drive the evolution of drug resistance in Mycobacterium tuberculosis. Evol Appl 2018;11(9):1498–511. CrossRef
9. Sikri N, Dalal S, Taneja R. Efflux pumps: an overview. Int J Pharm Sci Res 2018;9(3):854–61.
10. Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013;4(3):223–9. CrossRef
11. Borges-Walmsley MI, McKeegan KS, Walmsley AR. Structure and function of efflux pumps that confer resistance to drugs. Biochem J 2003;376(2):313–38. CrossRef
12. Bolla JR. Bacterial multidrug efflux pumps: structure, function and regulation. Iowa State University Ames, Ames, IA.
13. Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev 2007;71(3):463–76. CrossRef
14. Choi CH. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int 2005;5(1):1–3. CrossRef
15. Greene NP, Kaplan E, Crow A, Koronakis V. Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front Microbiol 201828;9:950. CrossRef
16. Zgurskaya HI. Multicomponent drug efflux complexes: architecture and mechanism of assembly. Future Microbiol 2009;4(7):919–32. CrossRef
17. Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol 2004;11(10):918–26. CrossRef
18. Senior AE. Catalytic mechanism of P-glycoprotein. Acta Physiol Scand Supplement 1998;643:213–8.]
19. Podlesek Z, Comino A, Herzog-Velikonja B, Žgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol 1995;16(5):969–76. CrossRef
20. Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol Rev 2000;24(4):449–67. CrossRef
21. Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 2009;1794(5):769–81. CrossRef
22. Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 2015;6:587. CrossRef
23. Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol 2011;77:1. CrossRef
24. Wieczorek P, Sacha P, Hauschild T, Zórawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacter baumannii—the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol 2008;46(3):257–67. CrossRef
25. Alvarez-Ortega C, Olivares J, Martínez JL. RND multidrug efflux pumps: what are they good for? Front Microbiol 2013;4:7. CrossRef
26. De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, et al. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2001;45(6):1761–70. CrossRef
27. Nishino K, Nikaido E, Yamaguchi A. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J Bacteriol 2007;189(24):9066–75. CrossRef
28. Kermani AA, Macdonald CB, Gundepudi R, Stockbridge RB. Guanidinium export is the primal function of SMR family transporters. Proc Natl Acad Sci 2018;115(12):3060–5. CrossRef
29. Srinivasan VB, Rajamohan G, Gebreyes WA. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob Agents Chemother 2009;53(12):5312–6. CrossRef
30. Bay DC, Turner RJ. Diversity and evolution of the small multidrug resistance protein family. BMC Evol Biol 2009;9(1):1–27. CrossRef
31. Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta 2008;1778(9):1814–38. CrossRef
32. Edwards RA, Turner RJ. Alpha-periodicity analysis of small multidrug resistance (SMR) efflux transporters. Biochem Cell Biol 1998;76(5):791–7. CrossRef
33. Ganas P, Mihasan M, Igloi GL, Brandsch R. A two-component small multidrug resistance pump functions as a metabolic valve during nicotine catabolism by Arthrobacter nicotinovorans. Microbiology 2007;153(5):1546–55. CrossRef
34. Paulsen IT, Skurray RA, Tam R, Saier Jr MH, Turner RJ, Weiner JH, et al. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol 1996;19(6):1167–75. CrossRef
35. Sarkar SK, Bhattacharyya A, Mandal SM. YnfA, a SMR family efflux pump is abundant in Escherichia coli isolates from urinary infection. Indian J Med Microbiol 2015;33(1):139–42. CrossRef
36. Lekshmi M, Ammini P, Jones Adjei LM, Shrestha U, Kumar S, Varela MF. Modulation of antimicrobial efflux pumps of the major facilitator superfamily in Staphylococcus aureus. AIMS Microbiol 2018;4(1):1. CrossRef
37. Kumar S, Floyd JT, He G, Varela MF. Bacterial antimicrobial efflux pumps of the MFS and MATE transporter families: a review. Recent Res Dev Antimicrob Agents Chemother 2013;7:1–21.
38. Madej MG. Function, structure, and evolution of the major facilitator superfamily: the LacY manifesto. Adv Biol 2014;2014; doi:10.1155/2014/523591 CrossRef
39. Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol Mol Biol Rev 1998;62(1):1–34. CrossRef
40. Pasqua M, Grossi M, Zennaro A, Fanelli G, Micheli G, Barras F, et al. The varied role of efflux pumps of the MFS family in the interplay of bacteria with animal and plant cells. Microorganisms 2019;7(9):285. CrossRef
41. Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol 2008;62:289–305. CrossRef
42. Saier Jr MH, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol 1999;1(2):257–79.
43. Lomovskaya O, Lewis KI, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 1995;177(9):2328–34. CrossRef
44. Lee EH, Shafer WM. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol 1999;33(4):839–45. CrossRef
45. Zakrzewska S, Mehdipour AR, Malviya VN, Nonaka T, Koepke J, Muenke C, et al. Inward-facing conformation of a multidrug resistance MATE family transporter. Proc Natl Acad Sci 2019;116(25):12275–84. CrossRef
46. Radchenko M, Symersky J, Nie R, Lu M. Structural basis for the blockade of MATE multidrug efflux pumps. Nat Commun 2015;6(1):1. CrossRef
47. Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 2006;27(11):587–93. CrossRef
48. Lu M. Structures of multidrug and toxic compound extrusion transporters and their mechanistic implications. Channels 2016;10(2):88–100. CrossRef
49. Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol 1999;31(1):394–5. CrossRef
50. Hassan KA, Liu Q, Elbourne LD, Ahmad I, Sharples D, Naidu V, et al. Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res Microbiol 2018;169(7–8):450–4. CrossRef
51. Delmar JA, Yu EW. The A bg T family: a novel class of antimetabolite transporters. Protein Sci 2016;25(2):322–37. CrossRef
52. Sharma A, Gupta VK, Pathania R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J Med Res 2019;149(2):129. CrossRef
53. Iman Islamieh D, Afshar D, Yousefi M, Esmaeili D. Efflux pump inhibitors derived from natural sources as novel antibacterial agents against Pseudomonas aeruginosa: a review. Int J Med Rev 2018;5(3):94–105. CrossRef
54. Seukep AJ, Kuete V, Nahar L, Sarker SD, Guo M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J Pharm Anal 2020;10(4):277–90. CrossRef
55. Mahmood YH, Jamshidi S, Mark Sutton J, Rahman MK. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr Med Chem 2016;23(10):1062–81. CrossRef
56. Cowan MM. Plant products as antimicrobial agents. Clin Microbiol Rev 1999;12(4):564–82. CrossRef
57. Stavri M, Piddock LJ, Gibbons S. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother 2007;59(6):1247–60. CrossRef
58. Neyfakh AA, Bidnenko VE, Chen LB. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci 1991;88(11):4781–5. CrossRef
59. Gibbons S, Udo EE. The effect of reserpine, a modulator of multidrug efflux pumps, on the in vitro activity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) possessing the tet (K) determinant. Phytotherapy 2000;14:139–40. CrossRef
60. Neyfakh AA, Borsch CM, Kaatz GW. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother 1993;37(1):128–9. CrossRef
61. Pfeifer HJ, Greenblatt DK, Koch-Wester J. Clinical toxicity of reserpine in hospitalized patients: a report from the Boston Collaborative Drug Surveillance Program. Am J Med Sci 1976;271(3):269–76. CrossRef
62. Khan IA, Mirza ZM, Kumar A, Verma V, Qazi GN. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob Agents Chemother 2006;50(2):810–2. CrossRef
63. Kumar A, Khan IA, Koul S, Koul JL, Taneja SC, Ali I, et al. Novel structural analogues of piperine as inhibitors of the NorA efflux pump of Staphylococcus aureus. J Antimicrob Chemother 2008;61(6):1270–6. CrossRef
64. Sharma S, Kumar M, Sharma S, Nargotra A, Koul S, Khan IA. Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J Antimicrob Chemother 2010;65(8):1694–701. CrossRef
65. Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci 200015;97(4):1433–7. CrossRef
66. Morel C, Stermitz FR, Tegos G, Lewis K. Isoflavones as potentiators of antibacterial activity. J Agric Food Chem 2003;51(19):5677–9. CrossRef
67. Chan BC, Ip M, Lau CB, Lui SL, Jolivalt C, Ganem-Elbaz C, et al. Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J Ethnopharmacol 2011;137(1):767–73. CrossRef
68. Fujita M, Shiota S, Kuroda T, Hatano T, Yoshida T, Mizushima T, et al. Remarkable synergies between baicalein and tetracycline, and baicalein and β-lactams against methicillin-resistant Staphylococcus aureus. Microbiol Immunol 2005;49(4):391–6. CrossRef
69. Stapleton PD, Shah S, Anderson JC, Hara Y, Hamilton-Miller JM, Taylor PW. Modulation of β-lactam resistance in Staphylococcus aureus by catechins and gallates. Int J Antimicrob Agents 2004;23(5):462–7. CrossRef
70. Gibbons S, Moser E, Kaatz GW. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med 2004;70(12):1240–2. CrossRef
71. Roccaro AS, Blanco AR, Giuliano F, Rusciano D, Enea V. Epigallocatechin-gallate enhances the activity of tetracycline in Staphylococci by inhibiting its efflux from bacterial cells. Antimicrob Agents Chemother 2004;48(6):1968–73. CrossRef
72. Oluwatuyi M, Kaatz GW, Gibbons S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004;65(24):3249–54. CrossRef
73. Lorenzi V, Muselli A, Bernardini AF, Berti L, Pagès JM, Amaral L, et al. Geraniol restores antibiotic activities against multidrug-resistant isolates from Gram-negative species. Antimicrob Agents Chemother 2009;53(5):2209–11. CrossRef
74. Zechini B, Versace I. Inhibitors of multidrug resistant efflux systems in bacteria. Recent patents on anti-infective drug discovery. Recent Pat Antiinfect Drug Discov 2009;4(1):37–50. CrossRef
75. Pagès JM, Amaral L. Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim Biophys Acta 2009;1794(5):826–33. CrossRef
76. Lee MD, Galazzo JL, Staley AL, Lee JC, Warren MS, Fuernkranz H, et al. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 2001;56(1–2):81–5. CrossRef
77. Tambat R, Jangra M, Mahey N, Chandal N, Kaur M, Chaudhary S, et al. Microbe-derived indole metabolite demonstrates potent multidrug efflux pump inhibition in Staphylococcus aureus. Front Microbiol 2019;10:2153. CrossRef
78. Maesaki S, Marichal P, Hossain MA, Sanglard D, Vanden Bossche H, Kohno S. Synergic effects of tactolimus and azole antifungal agents against azole-resistant Candida albican strains. J Antimicrob Chemother 1998;42(6):747–53. CrossRef
79. Kolaczkowski M, Kolaczkowska A, Motohashi N, Michalak K. New high-throughput screening assay to reveal similarities and differences in inhibitory sensitivities of multidrug ATP-binding cassette transporters. Antimicrob Agents Chemother 2009;53(4):1516–27. CrossRef
80. Schuetzer-Muehlbauer M, Willinger B, Egner R, Ecker G, Kuchler K. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int J Antimicrob Agents 2003;22(3):291–300. CrossRef
81. Yamamoto S, Hiraga K, Abiko A, Hamanaka N, Oda K. A new function of isonitrile as an inhibitor of the Pdr5p multidrug ABC transporter in Saccharomyces cerevisiae. Biochem Biophys Res Commun 2005;330(2):622–8. CrossRef
82. Niimi K, Harding DR, Parshot R, King A, Lun DJ, Decottignies A, et al. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a D-octapeptide derivative. Antimicrob Agents Chemother 2004;48(4):1256–71. CrossRef
83. Lemoine RC, Glinka TW, Watkins WJ, Cho A, Yang J, Iqbal N, et al. Quinazolinone-based fungal efflux pump inhibitors. Part 1: Discovery of an (N-methylpiperazine)-containing derivative with activity in clinically relevant Candida spp. Bioorg Med Chem Lett 2004;14(20):5127–31. CrossRef
84. Watkins WJ, Lemoine RC, Chong L, Cho A, Renau TE, Kuo B, et al. Quinazolinone fungal efflux pump inhibitors. Part 2: In vitro structure–activity relationships of (N-methyl-piperazinyl)-containing derivatives. Bioorg Med Chem Lett 2004;14(20):5133–7. CrossRef
85. Watkins WJ, Chong L, Cho A, Hilgenkamp R, Ludwikow M, Garizi N, et al. Quinazolinone fungal efflux pump inhibitors. Part 3: (N-methyl) piperazine variants and pharmacokinetic optimization. Bioorg Med Chem Lett 2007;17(10):2802–6. CrossRef
86. Tanabe K, Lamping E, Adachi K, Takano Y, Kawabata K, Shizuri Y, et al. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem Biophys Res Commun 2007;364(4):990–5. CrossRef
87. DiGirolamo JA, Li XC, Jacob MR, Clark AM, Ferreira D. Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp. J Nat Prod 2009;72(8):1524–8. CrossRef
88. Singh P, Kaur J, Yadav B, Komath SS. Targeting efflux pumps—in vitro investigations with acridone derivatives and identification of a lead molecule for MDR modulation. Bioorg Med Chem 2010;18(12):4212–23. CrossRef
89. Pina-Vaz C, Rodrigues AG, Costa-de-Oliveira S, Ricardo E, Mårdh PA. Potent synergic effect between ibuprofen and azoles on Candida resulting from blockade of efflux pumps as determined by FUN-1 staining and flow cytometry. J Antimicrob Chemother 2005;56(4):678–85. CrossRef
90. Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem Pharmacol 2006;71(7):910–8. CrossRef
91. Renau TE, Léger R, Hecker S, Lee V, Ohta T. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem (Print) 1999;42(24):4928–31. CrossRef
92. Lynch AS. Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view. Biochem Pharmacol 2006;71(7):949–56. CrossRef
93. Tohidpour A, Peerayeh SN, Mehrabadi JF, Yazdi HR. Determination of the efflux pump-mediated resistance prevalence in Pseudomonas aeruginosa, using an efflux pump inhibitor. Curr Microbiol 2009;59(3):352–5. CrossRef
94. Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee MA, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 2001;45(1):105–16. CrossRef
95. Mullin S, Mani N, Grossman TH. Inhibition of antibiotic efflux in bacteria by the novel multidrug resistance inhibitors biricodar (VX-710) and timcodar (VX-853). Antimicrob Agents Chemother 2004;48(11):4171–6. CrossRef
96. Minderman H, O’Loughlin KL, Pendyala L, Baer MR. VX-710 (biricodar) increases drug retention and enhances chemosensitivity in resistant cells overexpressing P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Clin Cancer Res 2004;10(5):1826–34. CrossRef
97. National Center for Biotechnology Information. PubChem Compound Summary for CID 119373, Elacridar. 2021. Available via https://pubchem.ncbi.nlm.nih.gov/compound/Elacridar (Accessed 21 January 2021).
98. Elacridar. Available via https://go.drugbank.com/drugs/DB04881
99. Seiden MV, Swenerton KD, Matulonis U, Campos S, Rose P, Batist G, et al. A phase II study of the MDR inhibitor biricodar (INCEL, VX-710) and paclitaxel in women with advanced ovarian cancer refractory to paclitaxel therapy. Gynecol Oncol 2002;86(3):302–10. CrossRef
100. Biricodar. Available via https://go.drugbank.com/drugs/DB04851
101. Grossman TH, Shoen CM, Jones SM, Jones PL, Cynamon MH, Locher CP. The efflux pump inhibitor timcodar improves the potency of antimycobacterial agents. Antimicrob Agents Chemother 2015;59(3):1534–41. CrossRef
102. Mahamoud A, Chevalier J, Davin-Regli A, Barbe J. Quinoline derivatives as promising inhibitors of antibiotic efflux pump in multidrug resistant Enterobacter aerogenes isolates. Curr Drug Targets 2006;7(7):843–7. CrossRef
103. Chevalier J, Atifi S, Eyraud A, Mahamoud A, Barbe J, Pagès JM. New pyridoquinoline derivatives as potential inhibitors of the fluoroquinolone efflux pump in resistant Enterobacter aerogenes Strains. J Med Chem 2001;44(23):4023–6. CrossRef
104. Pradel E, Pagès JM. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother 2002;46(8):2640–3. CrossRef
105. Sabatini S, Gosetto F, Manfroni G, Tabarrini O, Kaatz GW, Patel D, et al. Evolution from a natural flavones nucleus to obtain 2-(4-Propoxyphenyl) quinoline derivatives as potent inhibitors of the S. aureus NorA efflux pump. J Med Chem 2011;54(16):5722–36. CrossRef
106. Pannek S, Higgins PG, Steinke P, Jonas D, Akova M, Bohnert JA, et al. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J Antimicrob Chemother 2006;57(5):970–4. CrossRef
107. Kern WV, Steinke P, Schumacher A, Schuster S, Baum HV, Bohnert JA. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother 2006;57(2):339–43. CrossRef
108. Schumacher A, Steinke P, Bohnert JA, Akova M, Jonas D, Kern WV. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J Antimicrob Chemother 2006 ;57(2):344–8. CrossRef
109. Bina XR, Philippart JA, Bina JE. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J Antimicrob Chemother 2009;63(1):103–8. CrossRef
110. Eicher T, Cha HJ, Seeger MA, Brandstätter L, El-Delik J, Bohnert JA, et al Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci 2012;109(15):5687–92. CrossRef
111. Thorarensen A, Presley-Bodnar AL, Marotti KR, Boyle TP, Heckaman CL, Bohanon MJ, et al. 3-Arylpiperidines as potentiators of existing antibacterial agents. Bioorg Med Chem Lett 2001;11(14):1903–6. CrossRef
112. Kaatz GW, Moudgal VV, Seo SM, Hansen JB, Kristiansen JE. Phenylpiperidine selective serotonin reuptake inhibitors interfere with multidrug efflux pump activity in Staphylococcus aureus. Int J Antimicrob Agents 2003;22(3):254–61. CrossRef
113. Yoshida KI, Nakayama K, Ohtsuka M, Kuru N, Yokomizo Y, Sakamoto A, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem 2007;15(22):7087–97. CrossRef
114. Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother 2014;58(10):6224–34. CrossRef
115. Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013;500(7460):102–6. CrossRef
116. Opperman TJ, Nguyen ST. Recent advances toward a molecular mechanism of efflux pump inhibition. Front Microbiol 2015;6:421. CrossRef
117. Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D’Souza S, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother 2014;58(2):722–33. CrossRef