1. INTRODUCTION
The Tomato spotted wilt virus (TSWV) and Tomato mosaic virus (ToMV) are two of the most dangerous viral infections that threaten tomato (Solanum lycopersicum L.) crops around the world [1,2]. Genetic resistance to viral diseases by using resistance genes has been applied for 80 years to decrease crop losses. Up to now, several resistance loci have been found in different crops and wild species [3]. The TSWV is a member of the genus Tospovirus in the family Bunyaviridae. TSWV symptoms are necrotic spots, curling, bronzing, and stunting of plants [4]. The TSWV infects both monocotyledons and dicotyledons [5]. TSWV resistance sources were identified in different tomato genotypes, e.g., Solanum habrochaites and S. habrochaites var. glabratum (“PI134417” and “LA1223”) [1]. Up to now, many resistance loci to the TSWV have been defined, namely, Sw1a, Sw1b, sw2, sw3, sw4, Sw-5 (Sw-5a to Sw-5e), Sw-6, and Sw-7 [1,6–8]. Sw-5 is one of the TSWV resistance alleles which has been used to generate TSWV-resistant tomato cultivars. Sw-5, which is found on chromosome 9 of S. peruvianum, is known to give resistance to Tospoviruses such as the TSWV [9–11]. The Sw-5 protein is made up of three domains: a coiled-coil (CC) domain, a nucleotide-binding site (NBS), and a leucine-rich repeat (LRR) domain [7].
The ToMV belongs to the genus Tobamovirus, which is one of the family Virgaviridae. Three ToMV resistance alleles have been introduced into domesticated tomatoes: Tm-1, Tm-2, and Tm-22. The tomato gene Tm-1 has been discovered in S. habrochites “PI126445” mapped to chromosome 2 [12,13]. It has been known that resistance to the ToMV is due to inhibition of movement of the virus into the plant cells. The Tm-2 and Tm-22 resistance alleles conferred a higher level of resistance than the Tm-1 allele in a wild-type tomato, S. peruvianum [14,15]. Furthermore, tomato plants with Tm-2 or Tm-22 show a hypersensitive response to the ToMV [16,17]. Tm-2 and its allele Tm-22 have been found near the centromere of chromosome 9 [18]. Besides, the resistance locus Tm-22 is more durable than Tm-2 [19]. Therefore, the Tm-22 resistance locus is both economically and practically important. It is used in tomato breeding programs as a source of ToMV resistance. Tm-2 and Tm-22 are resistance (R) loci in the plant host which encode members of the CC/nucleotide-binding-ARC/LRR protein family [20]. Tm-22 and Tm-2 have seven nucleotide variations in their open reading frames, resulting in four amino acid changes at the protein level. Two of these distinctions belong to the NBS domain, whereas the other two belong to the LRR domain [21].
Here in this investigation, a set of allele-specific markers were used to identify the resistance alleles Sw-5 and Sw-5b as well as Tm-1, Tm-2, and Tm-22, responsible for resistance to the TSWV and ToMV, respectively, in 19 tomato genotypes. Therefore, the functional markers can be used as a powerful tool in tomato breeding programs for TSWV and ToMV resistance.
2. MATERIALS AND METHODS
2.1. Plant Materials
Nineteen tomato lines, including accessions and commercial cultivars, were utilized in this investigation, as mentioned in Table 1. Each tomato seed was cultivated in a pot containing peat moss:vermiculite:sand in a ratio of 1:1:1. The pots were kept in a glasshouse in 27°C light/16°C dark, a photoperiod of 16 hours light:8 hours dark cycle, and 68%–75% relative humidity [22,23].
2.2. Virus Resistance Tests
2.2.1. Source of virus isolates
The TSWV and ToMV isolates were obtained from the Virology Laboratory, Department of Agricultural Microbiology, Faculty of Agriculture, University of Ain Shams. The TSWV and ToMV were maintained on Nicotiana tabacum cv. White Burley and Datura metel L. plants, respectively (Figs. 1 and 2).
2.2.2. Virus inoculation
One-month-old tomato lines cultivated in the glasshouse were mechanically inoculated with TSWV- or ToMV-infected tomato sap according to Green [24]. Virus symptoms were recorded for 4 weeks after inoculation. These materials were evaluated in two successive seasons, 2019–20 and 2020–21, in the greenhouse.
2.3. Evaluation of TSWV and ToMV Infection Under Greenhouse Conditions
Disease rating scales of 0 to 4 were used according to Hutton and Scott [25].
2.3.1. Serological assay using BIOREBA immunostrips for detection of TSWV and ToMV
All tomato plants inoculated with the TSWV or ToMV were tested for the presence of virus by AgriStrips using the virus-specific polyclonal. The TSWV and ToMV AgriStrips are a one-step assay that was developed and manufactured by BIOREBA AG, Reinach, Switzerland.
2.3.2. Isolation of DNA
Using a DNA purification kit (Bio Basic, Inc., Markham, Canada), DNA was isolated from tissues of 19 tomatogenotypes.
2.3.3. Polymerase chain reaction (PCR) amplification of resistance alleles
PCR-based markers [sequence characterized amplified regions (SCAR) and amplification-refractory mutation system (ARMS)] were carried out as mentioned below. The conditions were adjusted in 25 μl reactions containing 2.5 μl 2.5 mM dNTPs, 5 μl 5× buffer, 2.5 μl 2.5 mM MgCl2, 0.1 μl (0.5 units) Taq DNA polymerase (Promega Corp., Madison, WI), 2.5 μl of each forward and reverse primer at 10 μM, 1 μl of DNA extract, and 8.9 μl dsH2O. PCR cycles were 94°C for 4 minutes, 35 cycles of 94°C for 30 seconds, annealing temperature (Table 2) for 1 minute, and 72°C for 1.5 minutes. These cycles were followed by 72°C for 10 minutes, and then the reaction was held at 4°C. PCR reactions were performed in the thermocycler (Biometra, biomedizinische Analytik GmbH).
2.3.4. Gel electrophoresis
All the PCR products were separated on 1% agarose gel electrophoresis in a 1xTBE (Tris/Borate/EDTA buffer), stained with the RedSafe Nucleic Acid Staining Solution (1/20,000) (iNtRON Biotechnology, Inc. Kr), and were visualized with UV light.
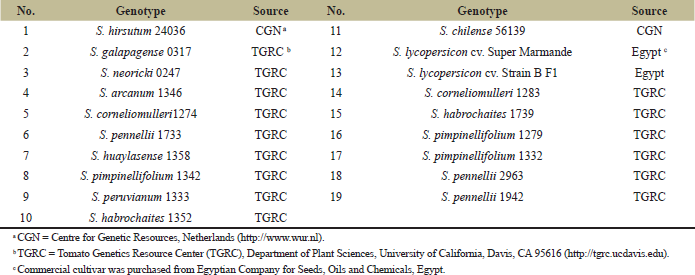 | Table 1: Tomato genotypes used in this study. [Click here to view] |
 | Figure 1: Photographs of N. tabacum cv. White Burley leaf after inoculation with TSWV isolate showing local necrotic lesions (I), compared with the healthy control (C). [Click here to view] |
 | Figure 2: N. tabaccum cv. White Burley (left) and D. metel L (right) inoculated with ToMV, appeared (severe mosaic and malformation), (necrotic local lesions) symptoms, respectively. C = healthy control; I = inoculated plants. [Click here to view] |
 | Table 2: Sequence of primers used in this study. [Click here to view] |
 | Figure 3: Phenotype of inoculated tomato genotypes three weeks after inoculation with TSWV virus. C: control, I: inoculated plants with TSWV. S. huaylasense 1358, S. habrochaites 1352 and 1739, Super Marmande, Strain B, and S. pennellii 1942 displayed mild mosaic, S. arcanum 1346 gave small leaf size, S. corneliomulleri 1283 appeared yellowing and S. pimpinellifolium 1279 showing necrotic spot. The arrow points to necrotic spots. [Click here to view] |
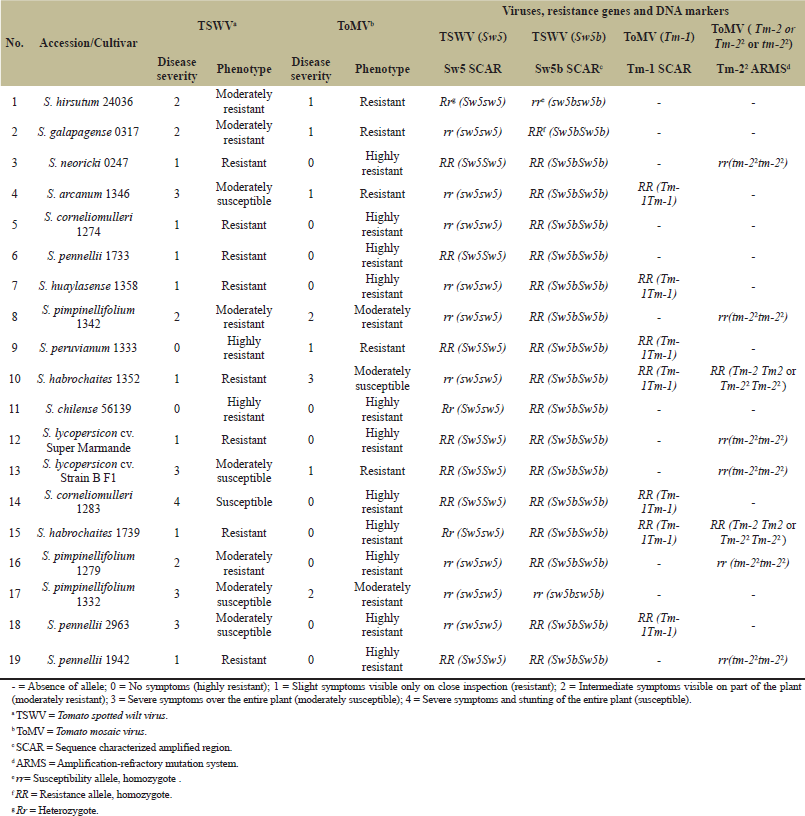 | Table 3: Tomato genotypes used to evaluate gene-based markers for resistance against TSWV and ToMV. [Click here to view] |
3. RESULTS
3.1. Characterization of ToMV and TSWV Diseases
Phenotypic characterization of 19 tomato genotypes against the TSWV and ToMV under greenhouse conditions will reflect their field performance. In fact, two tomato genotypes were categorized as highly resistant to the TSWV, eight resistant, four moderately resistant, four moderately susceptible, and one susceptible to TSWV infection (Fig. 3 and Table 3). For the ToMV, 11 lines were highly resistant; 5 genotypes were resistant, 2 moderately resistant, and 1 moderately susceptible (Fig. 4 and Table 3). The phenotype results were confirmed by TSWV and ToMV ImmunoStrip Kits. Resistance and susceptibility to viral infection were clearly distinguished by the appearance of the colored band.
 | Figure 4: Phenotype of inoculated tomato genotypes three weeks after inoculation with ToMV virus. C: control, I: inoculated plants with ToMV. S. arcanum 1346, S. habrochaites 1352, and S. pimpinellifolium 1332 showing (mild mosaic), (mild mosaic and necrotic spot) and necrotic spot, respectively. The arrows refer to necrotic spots. [Click here to view] |
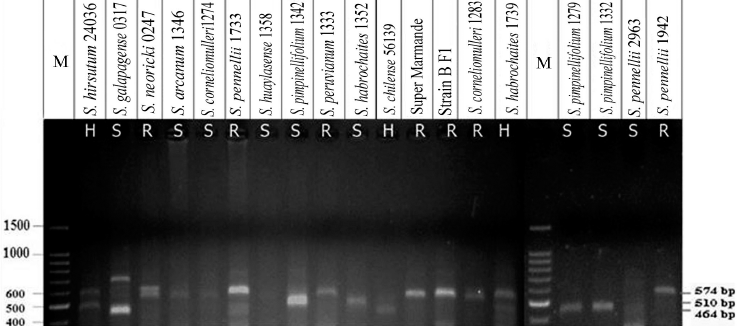 | Figure 5: PCR fragments represent primer set Sw5 SCAR amplified from 19 tomato genotypes, resolved in 1% agarose gel. Lane M: 100 bp DNA ladder; R = homozygous resistant genotypes; S = susceptible genotypes; H = heterozygote resistant genotypes. [Click here to view] |
3.2. Gene-Based Marker for Sw5 and Sw5b Resistance
Two gene-derived SCAR markers (Sw5 SCAR and Sw5b SCAR) (Table 3) were used to detect Sw5 and Sw5b resistance genes, respectively, responsible for resistance to TSWV disease.
3.2.1. Sw5
PCR amplification was performed with primer pair Sw5 SCAR, using genomic DNA extracted from 19 tomato lines (Fig. 5). The PCR results indicated four DNA fragments. The first group displayed a single fragment of 574 bp which was scored by nine tomato genotypes carrying the Sw-5 locus involving S. corneliomulleri 1274 and 1283, S. pennelli 1733 and 1942, S. neoricki 0247, S. peruvianum 1333, S. arcanum 1346, S. lycopersicon cv. Super Marmande, and S. lycopersicon cv. Strain B F1. The second group yielded one band of 510 bp and included two susceptible genotypes, S. pimpinellifolium 1342 and S. habrochaites 1352. The third group exhibited one band of 464 bp and consisted of the two susceptible tomato lines, S. pimpinellifolium 1279 and 1332. The fourth group gave two amplified fragments of 464 and 574 bp, e.g., S. hirsutum 24036, S. galapagese 0317, and S. habrochaites 1739. Those were heterozygous for the Sw5 locus. On the other hand, S. huaylasense 1358 and S. pennelli 2963 did not score any PCR products (Fig. 5).
3.2.2. Sw5b
PCR experiments were conducted on DNA isolated from 19 tomato lines by the primer pair Sw5b SCAR. PCR results recorded an amplicon of 541 bp in all tomato genotypes studied except S. pimpinellifolium 1332 and S. hirsutum 24036, which confer the presence of resistance gene Sw5b (Fig. 6).
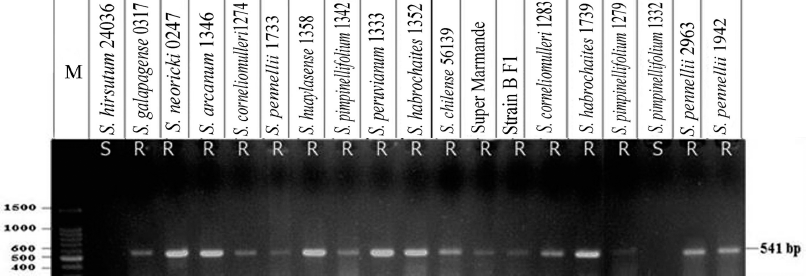 | Figure 6: PCR fragments represent primer pair Sw5b SCAR amplified from 19 tomato genotypes, resolved in 1% agarose gel. Lane M = 100 bp DNA ladder; R = homozygous resistant genotypes; S = susceptible genotypes. [Click here to view] |
 | Figure 7: PCR fragments represent primer pair Tm-1SCAR amplified from 19 tomato genotypes, resolved in 1% agarose gel. Lane M: 100 bp DNA ladder; R = homozygous resistant genotypes; S = susceptible genotypes. [Click here to view] |
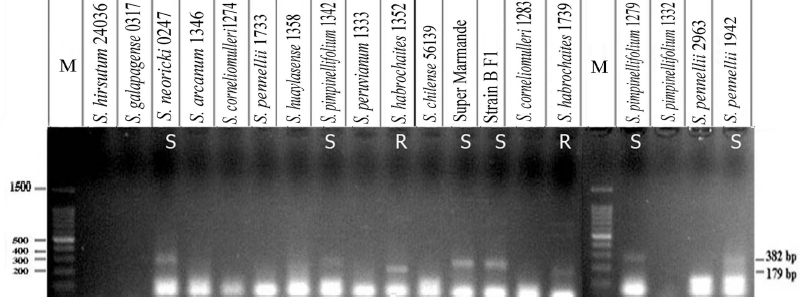 | Figure 8: PCR fragments represent primer pair Tm22 ARMS amplified from 19 tested tomato genotypes, resolved in 1% agarose gel. Lane M: 100 bp DNA ladder, R = homozygous resistant genotypes; S = susceptible genotypes. [Click here to view] |
3.3. Gene-Based Marker for Tm-1, Tm-2, Tm-22, and tm-22 Resistance Genes
Two gene-tagged markers, Tm-1 SCAR and Tm-22 ARMS related to ToMV resistance genes, were used to select tomato genotypes that have resistance genes Tm-1 and Tm-2 or Tm-22, respectively.
3.3.1. Tm-1
The primer pair Tm-1 SCAR amplified one amplicon of 1,400 bp from seven tomato lines carrying the Tm-1 resistance allele, e.g., S. arcanum 1346, S. pennellii 2963, S. corneliomulleri 1283, S. huaylasense 1358, S. habrochaites 1352 and 1739, and S. peruvianum 1333 (Fig. 7).
3.3.2. Tm-2 or Tm-22 or tm-22
A total of 19 tomato lines were exposed to the ARMS-PCR assay. Primer set Tm-22 ARMS yielded one amplified fragment of 179 bp for the dominant allele Tm-2 or Tm-22 in two accessions, S. habrochaites 1352 and 1739. Furthermore, the same primer scored one fragment of 382 bp for recessive allele tm-22 in S. pimpinellifolium 1342, S. neoricki 0247, S. lycopersicon cv. Super Marmande, S. lycopersicon cv. Strain B F1, S. pimpinellifolium 1279, and S. pennellii 1942 (Fig. 8).
4. DISCUSSION
For the time being, several dominant and recessive resistance loci to the TSWV (Sw5b/sw5b and Sw5-2/sw5-2) and ToMV (Tm-1/tm-1, Tm-2/tm-2, and Tm-22/tm22) were identified. The majority of these genes either do not permit or prevent replication of the virus. To detect these genes, four molecular markers (three SCAR and one ARMS) were employed in this study to screen tomato genotypes that have resistance genes for marker-assisted selection (MAS) programs.
In this respect, 19 tomato lines were classified as highly resistant to the TSWV under the glasshouse conditions (S. peruvianum 1333 [30] and S. chilense 56139 [31]), eight resistant (S. neoricki 0247, S. corneliomulleri 1274 [32], S. pennellii 1733 and 1942, S. huaylasense 1358 [32], S. habrochaites 1352 and 1739 [1], and S. lycopersicon cv. Super Marmande), four moderately resistant (S. hirsutum 24036, S. galapagense 0317, and S. pimpinellifolium 1342 and 1279), and four moderately susceptible (S. arcanum 1346 [32], S. lycopersicon cv. Strain B, S. pimpinellifolium 1332, and S. pennellii 2963 [33]). These genotypes have dominant or recessive alleles with homozygous or heterozygous Sw5 or Sw5b or both. These results were confirmed by Gordillo et al. [30] who found TSWV resistance in wild tomato genotypes, and some of the resistance loci were introgressed into domesticated tomato cultivars. The Sw1a and Sw1b loci were quickly overcome by TSWV isolates. However, Sw-6 and Sw-7 confer partial resistance to a small range of TSWV isolates, but they are not well identified and not widely applied in domesticated tomato cultivars.
In the current investigation, one tomato accession, S. pimpinellifolium 1332, was found to have recessive alleles sw5 and sw5b, indicating that it is moderately susceptible to the TSWV [32]. Although line S. corneliomulleri 1283 has dominant alleles with homozygous Sw5 and Swb, it is susceptible to TSWV infection, which is attributed to some TWSV strains being able to overcome Sw resistance genes [30]. These findings were in line with those reported by Pappu et al. [34,35]; de Oliveria indicated that mutation in Sw-5 proteins identified in tomato genotypes susceptible to the TSWV does not recognize the avr protein of the virus. In addition, two amino acids (aa) exchanges in movement proteins (NSm), NSmC118Y or NSmT120N, overcome Sw-5b-mediated resistance by TSWV isolates. A single mutation in the NSs protein, T104A, overcomes Tsw-mediated resistance. Aramburu et al. [36] indicated that resistance-breaking isolates of the TSWV in Spain are able to overcome the resistance referred to by the Sw-5 locus in tomatoes.
In this article, we observed that a commercial tomato cultivar (“Super Marmande”) gave resistance to the TSWV, which carries both alleles Sw5 and Sw-5b. This result was confirmed by Shi et al. [37] who selected 14 tomato genotypes and 10 domesticated tomato genotypes for resistance against the TSWV and indicated that only three domesticated genotypes (“BHN-444,” “Sophya,” and “Talladega”) and one wild species (LA3667) carry the resistance allele Sw-5b.
In our study, 11 tomato genotypes were highly resistant to the ToMV, namely, S. neoricki 0247, S. corneliomulleri 1274 and 1283 [38], S. pennellii 1733, 2963, and 1942, S. huaylasense 1358 [38], S. chilense 56139, S. lycopersicon cv. Super Marmande, S. habrochaites 1739 [12–13], and S. pimpinellifolium 1279 [38], five genotypes were resistant (S. hirsutum 24036, S. galapagense 0317, S. arcanum 1346 [38], S. peruvianum 1333 [14,15], and S. lycopersicon cv. Strain B), two accessions were moderately resistant (S. pimpinellifolium 1342 and 1332 [38]), and one line was moderately susceptible (S. habrochaites 1352). These lines have homozygous dominant or recessive alleles Tm-1 or Tm-2 or Tm-22 or tm22 separately or mixed. However, S. neoricki 0247, S. pimpinellifolium 1342 and 1279, S. lycopersicon cv. Super Marmande, S. lycopersicon cv. Strain B, and S. pennellii 1942 all have homozygous recessive allele tm22, which confers resistance to the ToMV. The resistance to the ToMV may be related to a recessive locus tm22, which is controlled by epistatic interactions. These results were synchronized with the results of Diaz-Pendon et al. [39] who showed that recessive loci have been related to plant virus resistance and have been ascribed to the plants lacking some basic factors desired for movement or replication of the virus. Consequently, it is possible that resistance to the ToMV may be polygenic and involves both dominant and recessive genes. Hashimoto et al. [40] reported that recessive resistance is caused by a mutation in a recessive allele that codes for a host component essential for virus replication. Furthermore, a lack of a negative regulator of defensive responses in the plant host, or the autoactivation of defense signaling, may confer recessive resistance. The most often used recessive resistance loci in diverse crops are eukaryotic translation initiation factor (eIF) 4E and eIF4G, which are effective against a broader range of plant viruses. Mutation in eIF4Es refers to loss of susceptibility to many viruses. It is critical to find new genetic sources for recessive resistance against plant pathogenic viruses in order to develop crops that rely on recessive resistance against a wide range of plant viruses.
In this work, some tomato accessions have no dominant or recessive loci and recorded resistance to the ToMV, e.g., S. hirsutum 24036, S. pennellii 1733, S. galapagense 0317, S. corneliomulleri 1274, S. chilense 56139, and S. pimpinellifolium 1332. This resistance may be due to the presence of new genes in the plant host which refer to resistance against the ToMV. These results agreed with Rasul’s results [38] who isolated six genes homologous to the Tm22 locus, which depending on their resistance phenotype were named ScoTm, Satm-2, Sctm-2, ShTm-2, SpiTm-2, and Sptm-3 from S. corneliomulleri LA1292, S. arcanum LA2172, S. chilense LA 2884, S. huaylasense LA1982, S. pimpinellifolium LA0722, and S. peruvianum LA0752, respectively. Ciuffo et al. [41] and Verlaan et al. [42] mentioned that several virus resistance loci (dominant or recessive) had been identified and applied in the breeding programs of different crops. The majority of these loci either do not permit or prevent replication of the virus into the plant cells.
In the present work, all studied tomato genotypes were resistant against both the TSWV and ToMV except S. arcanum 1346, S. lycopersicon cv. Strain B, S. corneliomulleri 1283, S. pimpinellifolium 1332, and S. pennellii 2963 as well as S. habrochaites 1352, which were moderately susceptible and susceptible to the TSWV and ToMV, respectively.
5. CONCLUSION
Gene-based markers screening for the detection of pathogen resistance genes in different tomato genotypes has become a valuable tool in plant viruses resistance. In this paper, we have applied DNA markers to detect resistance loci to the TSWV and ToMV in 19 tomato genotypes. In this investigation, we have identified 18 tomato genotypes bearing the dominant allele for TSWV resistance. In addition, seven lines have resistance genes to the ToMV. Therefore, the newness of this research is the identification of donor parents for producing tomato genotypes resistant to both the TSWV and ToMV in tomato breeding programs or the production of tomato lines with pyramided genes for resistance to several viruses through MAS.
6. CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest regarding the publication of this paper.
7. FUNDING
This work was funded by the National Research Centre (Project ref. 12050139), Dokki, Giza, Egypt.
8. CONSENT TO PARTICIPATE
Not applicable.
9. ETHICAL APPROVAL
Not applicable.
10. AUTHORS’ CONTRIBUTIONS
Dr. HAM carried out SCAR and ARMS markers and data analysis and interpretation, Prof. Dr. SAM performed virus resistance tests and writing of the manuscript, and MEO corrected and edited the manuscript.
11. DATA AVAILABILITY
All data generated or analyzed during this investigation already exist in this paper.
12. LOCAL AND NATIONAL REGULATIONS
All studies follow local and national regulations.
REFERENCES
1. Saidi M, Warade SD. Tomato breeding for resistance to Tomato spotted wilt virus (TSWV): an overview of conventional and molecular approaches. Czech J Genet Plant Breed 2008;44:83–92. CrossRef
2. FAOSTAT. Statistics Division of the Food and Agricultural Organization of the United Nations. 2019. Available via https://www.fao.org/faostat/en/#data/QC
3. Kang BC, Yeam I, Jahn MM. Genetics of plant virus resistance. Annu Rev Phytopathol 2005;43:581–621. CrossRef
4. German TL, Ullman DE, Moyer JW. Tospoviruses: diagnosis, molecular biology, phylogeny, and vector relationships. Annu Rev Phytopathol 1992;30:315–48. CrossRef
5. Pappu HR, Jones RAC, Jain RK. Global status of Tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res 2009;141:219–36. CrossRef
6. Finlay K. Inheritance of spotted wilt resistance in the tomato II. Five genes controlling spotted wilt resistance in four tomato types. Aust J Biol Sci 1953;6:153–63. CrossRef
7. Price DL, Memmott FD, Scott JW, Olson SM, Stevens MR. Identification of molecular markers linked to a new Tomato spotted wilt virus resistance source in tomato. Tomato Genet Coop 2007;57:35–6.
8. Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, et al. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol Breed 2001;7:151–61. CrossRef
9. Van Zijl J, Bosch S, Coetzee C. Breeding tomatoes for processing in South Africa. In: Strydom DK (ed.). International symposium on fruit & vegetables for processing 194, Acta Horticulturae, Cape Town, South Africa, pp 69–76, 1985. CrossRef
10. Stevens M, Scott S, Gergerich R. Inheritance of a gene for resistance to Tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica 1992;59:9–17. CrossRef
11. Boiteux L, Giordano LdB. Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon esculentum). Euphytica 1993;71:151–4. CrossRef
12. Pelham J. Resistance in tomato to Tobacco mosaic virus. Euphytica 1966;15:258–67. CrossRef
13. Watanabe Y, Kishibayashi N, Motoyoshi F, Okada Y. Characterization of Tm-1 gene action on replication of common isolates and a resistance-breaking isolate of TMV. Virology 1987;161:527–32. CrossRef
14. Nishiguchi M, Motoyoshi F. Resistance mechanisms of Tobacco mosaic virus strains in tomato and tobacco. In: Evered D, Harnett S (ed.). Plant resistance to viruses, John Wiley & Sons, Chichester, UK, pp 38–46, 1987. CrossRef
15. Meshi T, Motoyoshi F, Maeda T, Yoshikawa S, Watanabe H, Okada Y. Mutations in the Tobacco mosaic virus 30-kDa protein gene overcome Tm-2 resistance in tomato. Plant Cell 1989;1:515–22 CrossRef
16. Cirulli M, Alexander LJ. Influence of temperature and strain of Tobacco mosaic virus on resistance in a tomato breeding line derived from Lycopersicon peruvianum. Phytopathology 1969;59:1287–97.
17. Hall TJ. Resistance at the Tm-2 locus in the tomato to Tomato mosaic virus. Euphytica 1980;29:189–97. CrossRef
18. Young ND, Tanksley SD. RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor Appl Genet 1989;77:353–9. CrossRef
19. Fraser RSS. The genetics of resistance to plant viruses. Annu Rev Phytopathol 1990;28:179–200. CrossRef
20. Lanfermeijer FC, Dijkhuis J, Sturre MJ, de Haan P, Hille J. Cloning and characterization of the durable Tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol Biol 2003;52:1037–49. CrossRef
21. Lanfermeijer FC, Warmink J, Hille J. The products of the broken Tm-2 and the durable Tm-2(2) resistance genes from tomato differ in four amino acids. J Exp Bot 2005;56:2925–33. CrossRef
22. Dinh QD, Dechesne A, Furrer H, Taylor G, Visser RGF, Harbinson J, et al. High-altitude wild species Solanum arcanum LA385-A potential source for improvement of plant growth and photosynthetic performance at suboptimal temperatures. Front Plant Sci 2019;10:1163; doi:10.3389/fpls.2019.01163 CrossRef
23. Mahfouze SA, Mahfouze HA. Comparison between CAPS and SCAR markers for detection of Tomato yellow leaf curl virus and whitefly resistance genes in tomato genotypes. Jordan J Biol Sci 2019;12(2):123–33.
24. Green SK. Guidelines for diagnostic work in plant virology. 2nd edition, Asian Vegetable Research and Development Center, Taipei, Taiwan, p 63, 1991.
25. Hutton SF, Scott JW. Ty-6, a major begomovirus resistance gene located on chromosome 10. Rep Tomato Genet Coop 2014;64:14–8.
26. Dianese E´C, de Fonseca MEN, Goldbach R, Kormelink R, Inoue-Nagata AK, Resende RO, et al. Development of a locus-specific, co-dominant SCAR marker for assisted-selection of the Sw-5 (Tospovirus resistance) gene cluster in a wide range of tomato accessions. Mol Breed 2010;25:133–42. CrossRef
27. Shi A, Vierling R, Grazzini R, Chen P, Caton H, Panthee D. Identification of molecular markers for gene of Sw-5 Tomato spotted wilt virus resistance. Am J Biotechnol Mol Sci 2011a;1:8–16. CrossRef
28. Ohmori T, Murata M, Motoyoshi F. Molecular characterization of RAPD and SCAR markers linked to the Tm-1 locus in tomato. Theor Appl Genet 1996;92:151–6. CrossRef
29. Arens P, Mansilla C, Deinum D, Cavellini L, Moretti A, Rolland S, et al. Development and evaluation of robust molecular markers linked to disease resistance in tomato for distinctness, uniformity and stability testing. Theor Appl Genet 2010;120(3):655–64. CrossRef
30. Gordillo LF, Stevens MR, Millard MA, Geary B. Screening two Lycopersicon peruvianum collections for resistance to Tomato spotted wilt virus. Plant Dis 2008;92:694–704. CrossRef
31. Stevens MR, Scott SJ, Gergerich RC. Evaluation of seven Lycopersicon species for resistance to Tomato spotted wilt virus (TVSW). Euphytica 1994;80:79–84. CrossRef
32. Li J, Huang H, Zhu M, Huang S, Zhang W, Dinesh-Kumar SP, et al. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol Plant 2019;12:248–62. CrossRef
33. Stevens MR, Lamb EM, Rhoads DD. Mapping the Sw5 locus for Tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor Appl Genet 1995;90:451–6. CrossRef
34. Pappu HR, Anna E Whitfield, de Oliveira AS. Tomato spotted wilt virus (Tospoviridae). Encycl Virol 2008;133–8. CrossRef
35. de Oliveira AS, Boiteux LS, Kormelink R, Resende RO. The Sw-5 gene cluster: tomato breeding and research toward Orthotospovirus disease control. Front Plant Sci 2018;9:1055; doi:10.3389/fpls.2018.01055. CrossRef
36. Aramburu J, Galipienso L, Soler S, López C. Characterization of Tomato spotted wilt virus isolates that overcome the Sw-5 resistance gene in tomato and fitness assays. Phytopathol Mediterr 2010;49:342–51.
37. Shi A, Vierling R, Grazzini R, Chen P, Caton H, Panthee D. Molecular markers for Tm-2 alleles of Tomato mosaic virus resistance in tomato. Am J Plant Sci. 2011b;2:180–9. CrossRef
38. Rasul I. TMV resistance in different Solanum species. In: Lanfermeijer FC, Hille J (ed.). Characterization of the Tm-2² locus of tomato and its durability, University of Groningen, Groningen, Netherlands, pp 107–28, 2012.
39. Diaz-Pendon J, Truniger V, Nieto C, Garcia-Mas J, Bendahmane A, Aranda M. Advances in understanding recessive resistance to plant viruses. Mol Plant Pathol 2004;5:223–33. CrossRef
40. Hashimoto M, Neriya Y, Yamaji Y, Namba S. Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Front Microbiol 2016;7:1695; doi:10.3389/fmicb.2016.01695 CrossRef
41. Ciuffo MC, Finetti-Sialer MM, Gallitelli D, Turina M. First report in Italy of a resistance breaking strain of Tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. N Dis Rep 2005;10:48. CrossRef
42. Verlaan MG, Szinay D, Hutton SF, de Jong H, Kormelink R. Chromosomal rearrangements between tomato and Solanum chilense hamper mapping and breeding of the TYLCV resistance gene Ty-1. Plant J 2011;68:1093–103. CrossRef