1. INTRODUCTION
Phenylalanine hydroxylase (PAH) enzyme is the main factor for dopamine biosynthesis which is responsible for converting phenylalanine to tyrosine and then to other essential components like dopamine, melanin, adrenaline, and noradrenaline with the same pathway in plants, animals, and humans [1–3]. Previous studies have indicated the importance of this enzyme with satisfying levels in the body of humans and animals [4–8].
Liver and kidney are the main organs that excrete the PAH enzyme [9,10]. The deficiency of PAH enzyme leads to the accumulation of phenylalanine (Phe) amino acid in the blood, causing phenylketonuria (PKU), a disease that causes mental retardation in infants and children [11–21]. Moreover, phenylalanine deficiency in the blood leads to low dopamine levels in the body, which is the only reason for Parkinson’s disease and L- dihydroxyphenylalanine (L-DOPA)-induced dyskinesia (LID) [7,22,23]. Therefore, to sustain normal health, the PAH enzyme must be found in its active form and normal ratio in humans, animals, and plants.
For animals, normal phenylalanine level is vital due to its responsibility in the secretion of different essential hormones like prolactin [24,25]. Also, the direct influence of phenylalanine on animal behavior was cleared. For example, in different types of locusts where phenylalanine is the basic molecule in the defending cycle against predatory birds, phenylacetonitrile, a compound synthesized from phenylalanine, plays an important role by converting into hydrogen cyanide, which is toxic to predatory birds [26].
Additionally, phenylalanine is a precursor of many plant phenolic compounds, especially in faba bean [27]. Many studies have indicated that Sakha 3 faba bean genotype has the highest amount of phenylalanine [28–31]. Direct diet was applied to increase phenylalanine levels in rabbits’ blood which reflected a significant increase in PAH gene expression [32–34],
Our investigation aimed to evaluate the influence of a direct diet with fresh and dry forms of faba bean (Sakha 3 genotype) on PAH enzyme levels in rabbits. PAH genes of rabbits were PCR amplified, sequenced, and aligned with the reference gene in the GenBank.
2. MATERIALS AND METHODS
2.1. Plant Materials
Faba bean (Vicia faba L., Sakha 3 genotype) seeds were obtained from Sakha Agricultural Research Station and planted for a whole season during September 2018–March 2019 in suitable soil with recommended agriculture conditions. Plants were used as rabbit food in two forms (dry and fresh) with different quantities, as shown in Table 1.
2.2. Animal Samples and Feeding
New Zealand male rabbits (Oryctolagus cuniculus), 6 months old, were used in this study. Rabbits were grown and treated in the laboratories of the Faculty of Science, Tanta University, according to the regulations and rules of scientific ethical research with license no. IACUC-SCI-TU-0126. Rabbits were fed fresh and dry beans as a supplemented diet in different quantities for a period of 6 months, as shown in Table 1. Control rabbits were fed a normal diet without faba bean. After the feeding period, blood samples were collected from the ear vein, after sterilization with 70% ethyl alcohol. Blood samples were drawn using a medical syringe and collected in separate tubes. Rabbits were sacrificed and their livers were collected and stored for further work.
2.3. Quantification of Phenylalanine Amino Acid
Phenylalanine assay kit (Abcam, ab83376) was used according to manufacturer’s protocol for direct quantification of phenylalanine amino acid levels in rabbit blood samples. The results were recorded as an average of three sample rabbit replicates.
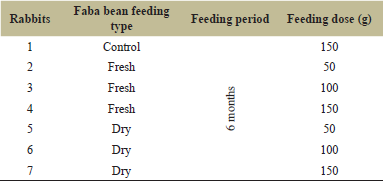 | Table 1: Faba bean feeding experiment design. [Click here to view] |
2.4. PCR Amplification of PAH Genes
PAH gene fragments of rabbits (O. cuniculus) were amplified, eluted, and sequenced. Nucleotide sequences of treated rabbit samples were aligned and compared with control as well as with reference sequence in the GenBank.
Total genomic DNAs were purified according to the manufacturer’s protocol of the GeneJET Genomic DNA purification kit (K0721/Thermo Fisher). Forward and reverse primers specific to the PAH gene (accession number 013672) were designed using the Primer-BLAST tool (www.ncbi.nlm.nih.gov/tools/primer-blast/primertool), as shown in Table 2. Then, Dream Taq PCR Master Mix (2×) was used to amplify gene fragments.
Thermal cycler (CreaCon, Holland) was used with the following conditions: 40 cycles; each cycle consisted of denaturation at 95°C for 30 seconds, followed by annealing at 37°C for 1 minute and extension at 72°C for 2 minutes. There was an initial delay for 15 minutes at 95°C at the beginning of the first cycle and a 10 minutes delay at 72°C at the end of the last cycle as a postextension step. Agarose gel (1.5%) was applied to migrate amplicons via MultiSUB Mini Horizontal Electrophoresis System with Power Pro 300 Power supply (Cleaver Scientific, UK) and imaging with gel documentation system (OmniDOC, Cleaver Scientific, UK). Data analysis was carried out using TotalLab analysis software (ww.totallab.com, ver.1.0.1).
2.5. Sequencing and Alignment of PAH Genes
Amplified PCR fragments were purified with E.Z.N.A.®Gel Extraction Kit (D2500-01, Omega BIO-TEK, USA). Samples were sent to Micron-Corp., Korea, for sequencing with ABI PRISM® 3100 Genetic Analyzer. Aligned PAH sequences were analyzed on the NCBI website (http://www.ncbi.nlm.nih.gov/website) using BLAST to confirm their identity. Genetic distances and multialignments were computed by the pairwise distance method using Clustal Omega software analysis (https://www.ebi.ac.uk/Tools/msa/clustalo/). The nucleotide sequences were also compared with the highest homology sequences available in the GenBank.
2.6. Quantitative RT-qPCR Analysis of PAH Genes
Quantitative RT-qPCR was applied to detect PAH gene expression after feeding rabbits (O. cuniculus) on dry and fresh beans. Gene expression was determined using Maxima SYBR Green/Fluorescein qPCR Master Mix by Rotor-Gene Q (Qiagen, USA) using a two-step cycling protocol and glyceraldehyde-3-phosphate as a housekeeping gene. TRIzols reagent (15596026, Life Technologies, USA) was applied for total RNA purification from liver samples. Yield and quality of total RNA were determined spectrophotometrically at 260 and 260/280 nm ratios, respectively.
 | Table 2: Sequence features of specific PAH primers. [Click here to view] |
2.7. Western Blot Analysis of PAH Protein
Western blot technique was applied to evaluate the PAH protein expression. Total soluble proteins were extracted and separated via the SDS-PAGE technique using OmniPage. A vertical protein electrophoresis system with PowerPro 300 power supply (Cleaver Scientific, UK) was conducted according to Laemmli [35] and Brunelle et al. [36]. After documentation with OmniDOC (Cleaver Scientific, UK) gel documentation system, PAH protein bands were detected with their specific molecular weight. Protein bands were transferred to Amersham Hybond-N+ (GE Healthcare, USA) via electroBLOT Blotter (Cleaver Scientific, UK) and hybridization was carried out against the anti-PAH antibody (Abcam, ab178430) according to the manufacturer’s protocol. Finally, PAH bands were evaluated via the chemiPRO Western blot imaging system [37].
3. RESULTS
3.1. Levels of Phenylalanine Amino Acid in Blood
Phenylalanine content was estimated in rabbits’ blood to evaluate the effect of the faba bean-feeding diet. Data presented are the average of three replicates of rabbit samples. Generally, a positive correlation was found between faba bean dose and phenylalanine levels in the blood. As shown in Figure 1 and Table 3, rabbits fed a high dose (150 g) of dry bean showed the greatest increase in phenylalanine level (311 nmol/l), whereas rabbits fed a low dose (50 g) showed a low level of phenylalanine (120 nmol/l). Similarly, rabbits fed a high dose (150 g) of fresh faba bean had high levels of phenylalanine (98 nmol/l), whereas rabbits fed a low dose (50 g) had low levels of phenylalanine (77 nmol/).
3.2 PAH Gene Sequence
PAH gene fragments (≈1,400 bp) were amplified and cleared on agarose gel (Fig. 2). PAH genes of control and the six faba bean-fed rabbits were sequenced and aligned with the reference gene (accession number 013672).
The first 320 pb of PAH sequences representing the N-terminal of the gene was almost identical as shown in sequence alignment (Fig. 3). High genetic similarity values were detected in PAH gene sequences for both control and faba bean-fed rabbits with the reference gene sequence (Table 4). PAH gene sequence for the control sample and rabbits fed 100 g of dry faba bean reflected the highest genetic similarity with the reference sequence (99.80%). The lowest genetic similarity (97.40%) was recorded for rabbits fed 150 g of fresh faba bean.
3.3. Expression of PAH Gene
Quantitative RT-qPCR was used to evaluate the effect of feeding by fresh and dry faba bean on PAH gene expression levels. As shown in Table 5, rabbits fed 150 g of dry faba bean reflected a superior increase in gene expression level, almost threefold, compared with those fed 150 g of fresh faba bean which increased gene expression level with twofold the gene expression of control. Our findings indicated overexpression of the PAH gene as a direct influence of feeding on dry faba beans.
3.4. Activity of PAH Enzyme
Western blot analysis was used to detect the effect of the faba bean diet on PAH protein in rabbits. As shown in Figure 4, total proteins from rabbit samples were fractionated and compared with control. A specific PAH protein band between 50 and 60 kDa was detected for all samples (Fig. 4A). Rabbits fed 150 g of dry beans reflected a remarkable increase in PAH protein expression level compared with controls and rabbits fed 150 g of fresh faba bean (Fig. 4B). These results indicate a clear positive correlation between feeding on dry faba bean and overexpression of PAH enzyme.
4. DISCUSSION
In this investigation, rabbits (O. cuniculus), as a biological model, were used for studying PAH gene sequence, expression, and enzyme activity. Perfect matching dopamine biosynthesis pathway between rabbits and humans and high similarity of PAH gene sequence between rabbits and humans were reported [38,39].
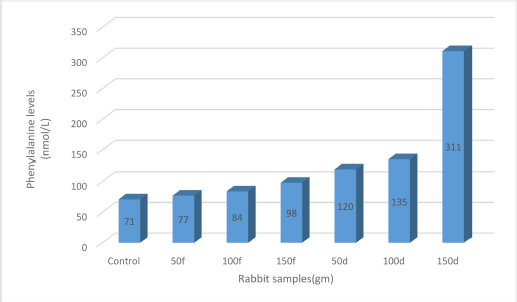 | Figure 1: Phenylalanine levels (nmol/l) for control rabbits (lane 1), fresh beans-fed rabbits (lanes 2–4), and dry beans-fed rabbits (5–7). [Click here to view] |
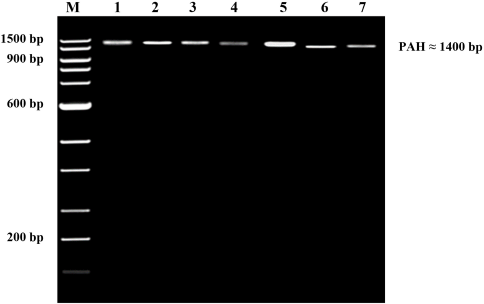 | Figure 2: PCR genomic products of PAH gene with ≈1400 bp for control rabbits (lane 1), fresh beans-fed rabbits (lanes 2–4), and dry beans-fed rabbits (lanes 5–7). [Click here to view] |
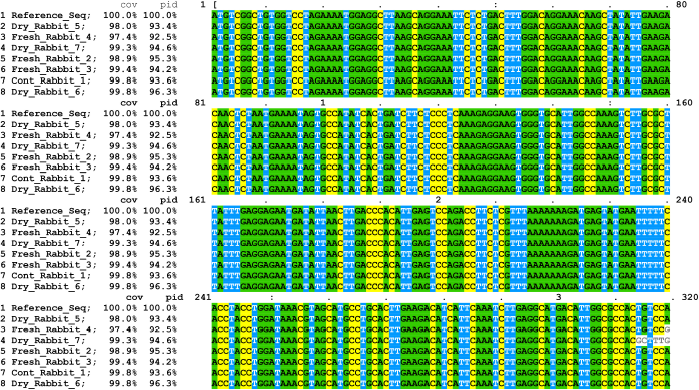 | Figure 3: Nucleotide sequence alignment of N-terminal (320 bp) PAH gene fragment amplified from control and six rabbit samples with the reference sequence (accession number 013672) in the GenBank. [Click here to view] |
Faba bean (Vicia faba L.) Sakha 3 genotype was chosen as a diet rich in phenylalanine to study the effect of different feeding conditions on PAH expression and enzyme activity. Previous studies showed that faba bean plants contain high amounts of phenylalanine and phenolic amino acids [28,40,41].
The results of the present study indicated an increase of phenylalanine level in rabbit blood after feeding on faba bean. Our results indicated a direct correlation between increasing the amount of faba bean diet rich in phenylalanine and increasing the level of phenylalanine amino acid in rabbits’ blood, which enhances PAH gene expression and enzyme activity. This is in accordance with the study by Shebl et al. [31], who indicated the direct influence of diet rich in phenylalanine as faba bean Sakha 3 genotype on increasing phenylalanine in rabbits’ blood.
Our findings obtained by quantitative RT-qPCR reflected a high increase in PAH gene expression level in rabbits fed fresh and dry faba bean (twofold and threefold, resp.). These results are confirmed by Western blotting analysis, which indicated overexpression of PAH enzyme. Fitzpatrick [42] indicated that PAH enzyme activity in the liver responds in a cooperative fashion to phenylalanine concentrations, with low activity at basal concentrations of phenylalanine and increased activity when the concentration of phenylalanine in the blood rises. However, Lartey and Austic [43] indicated that levels of PAH mRNA were not affected by the dietary of IAA-Phe mixture to the basal diet of chicken.
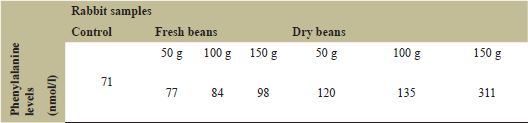 | Table 3: Phenylalanine levels in blood for control and six treated rabbits. [Click here to view] |
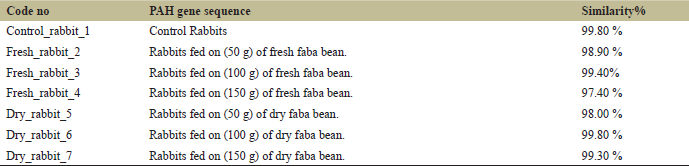 | Table 4: Sequence similarity of PAH gene from rabbit samples with Reference gene (Acc. No. 013672) from GenBank. [Click here to view] |
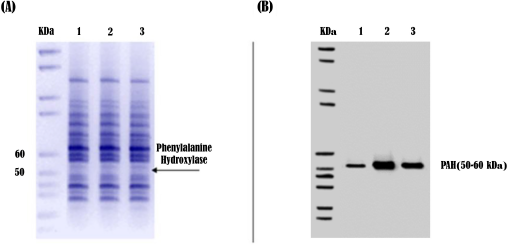 | Figure 4: Electrophoretic protein patterns (A) and PAH Western blot (B) for three rabbit samples. 1: control, 2: rabbit fed 150 g of dry beans, and 3: rabbit fed 150 g of fresh beans. [Click here to view] |
Western blotting analysis indicated a clear positive correlation between feeding rabbits on dry faba bean and overexpression of PAH enzyme. Our results are in agreement with those of Lartey and Austic [43], who reflected the positive influence of the addition of the IAA-Phe mixture on the basal diet of chicken with an increase in PAH level in blood. Our findings for using a phenylalanine-rich diet as a helpful tool to improve PAH levels are in agreement with the study by Cao et al. [44]. They indicated the effect of dietary supplementation with leucine and phenylalanine on pancreas development, PAH enzyme activity, and related gene expression in male Holstein calves. A positive correlation was detected between supplementation of phenylalanine and an increase in the number of pancreatic cells. In accordance with our results for use of Western blotting analysis to evaluate PAH activity level, Gunasekera and Hyland [45] applied this technique using the PAH 8 antibody to record and monitor the total PAH enzyme concentration level in mice. Furthermore, the applied PAH antibody in our study was supported by Silva et al. [46]. They used a monoclonal antibody raised against monkey liver PAH to detect this protein in Drosophila melanogaster.
In this investigation, PAH gene fragments were PCR amplified, sequenced, and aligned with the reference gene. High genetic similarity values were detected in PAH gene sequences for both control and faba bean-fed rabbits with the reference gene. The results indicated very little sequence variations, which have no effect on the enzyme activity in both control and faba bean-fed rabbits. Previous studies indicated that PAH genes exhibit natural mutations between different species and even among individuals of the same species. Some of these mutations are mild and do not affect the enzyme activity; however, severe mutations can occur, resulting in serious diseases such as phenylketonuria in humans [15,19,20].
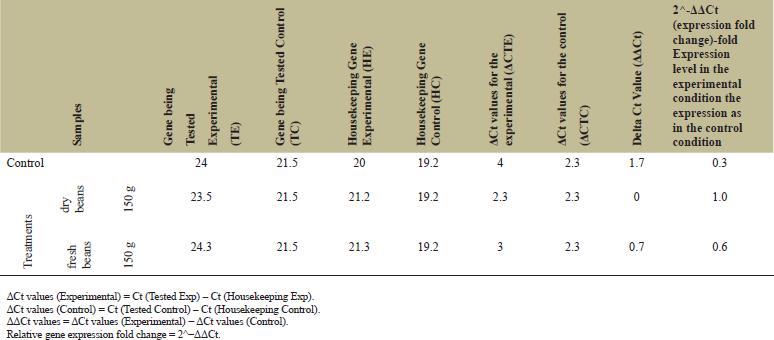 | Table 5: PAH gene expression folds in control and faba bean fed rabbits. [Click here to view] |
Our results agree with those of Mitchell et al. [12] who detected duplications and deletions in PAH gene sequences with no mutations of the enzyme activity. However, on the other hand, Güttler and Guldberg [11] and Vockley et al. [13] identified causative mutations in 686 PAH deficiency patients and indicated that the PAH mutation genotype is determinant in most patients.
Sequence analyses of mutant PAH enzymes established that the N-terminal domains are responsible for the divergent regulatory properties of the enzymes, while the C-terminal catalytic domains are responsible for substrate specificity [47]. Our results shown in Figure 3 indicated 100% sequence similarity in the first 320 pb of PAH sequences, representing the N-terminal of the gene.
5. CONCLUSION
In conclusion, our findings indicated the direct effect of fresh and dry faba bean diet on increasing phenylalanine amino acid in rabbits’ blood, which consequently increases the expression of PAH gene, thus improving the quality of life for humans.
6. DISCLOSURES
6.1. Authorship contribution
All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements.
6.2. Funding
There is no funding to report.
6.3. Conflicts of interest
The authors report no financial or any other conflicts of interest in this work.
6.4. Publisher’s note
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Teigen K, Jensen VR, Martinez A. The reaction mechanism of phenylalanine hydroxylase–a question of coordination. Pteridines 2005;16(1):27–34. CrossRef
2. Flydal MI, Martinz A. Phenylalanine hydroxylase: function, structure and regulation. JUBMB Life 2013;65(4):341–9. CrossRef
3. Simonet P, Gaget K, Parisot N, Duport G, Rey M, Febvay G, et al. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci Rep 2016;6(1):1–12. CrossRef
4. Marsden CA. Dopamine: the rewarding years. Br J Pharmacol 2006;147:S136–44. CrossRef
5. Chen S, Sun M, Zhao X, Yang Z, Liu W, Cao J, et al. Neuroprotection of hydroxysafflor yellow A in experimental cerebral ischemia/reperfusion injury via metabolic inhibition of phenylalanine and mitochondrial biogenesis. Mol Med Rep 2019;19(4):3009–20. CrossRef
6. Trivedi MN, Santani DD, Desai D, Desai U, Khemani A, Patel YD. Quantitative determination of L-dopa in seeds of Mucuna pruriens Linn. collected from different geographical sources as well as marketed formulations by high performance thin layer chromatography. Der Pharm Sinica 2015;6(7):9–20.
7. Burbulla LF, Krainc D. The role of dopamine in the pathogenesis of GBA1-linked Parkinson’s disease. Neurobiol Dis 2019;132:104545. CrossRef
8. Akram M, Daniyal M, Ali A, Zainab R, Muhammad, S, Shah A, et al. Role of phenylalanine and its metabolites in health and neurological disorders. In: Surguchov A (ed.). Synucleins—biochemistry and role in diseases. 2020. CrossRef
9. Arturo EC, Gupta K, Héroux A, Stith L, Cross PJ, Parker EJ, et al. First structure of full-length mammalian phenylalanine hydroxylase reveals the architecture of an autoinhibited tetramer. Proc Natl Acad Sci 2016;113(9):2394–9. CrossRef
10. Pecimonova M, Kluckova D, Csicsay F, Reblova K, Krahulec J, Procházkova D, et al. Structural and functional impact of seven missense variants of phenylalanine hydroxylase. Genes 2019;10(6):459. CrossRef
11. Güttler F, Guldberg P. The influence of mutations on enzyme activity and phenylalanine tolerance in phenylalanine hydroxylase deficiency. Eur J Pediatr 1996;155(1):S6–10. CrossRef
12. Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med 2011;13(8):697–707. CrossRef
13. Vockley J, Andersson HC, Antshel KM, Braverman NE, Burton BK, Frazier DM, et al. Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet Med 2014;16(2):188. CrossRef
14. Moradi P, Sarraf BS, Khamnian Z, Dolatkhah R, Hadi S, Ghafari D, et al. Distribution occurrence of phenylketonuria in the world: a systematic review and meta-analysis. Depiction Health 2016;6(4):1–12.
15. Vieira Neto E, Laranjeira, F, Quelhas D, Ribeiro I, Seabra A, Mineiro N, et al. Mutation analysis of the PAH gene in phenylketonuria patients from Rio de Janeiro, Southeast Brazil. Mol Genet Genomic Med 2018;6(4):575–91. CrossRef
16. Tolve M, Artiola C, Pasquali A, Giovanniello T, D’Amici S, Angeloni A, et al. Molecular analysis of PKU-associated PAH mutations: a fast and simple genotyping test. Methods Protoc 2018;1(3):30. CrossRef
17. Erlandsen H, Patch MG, Gamez A, Straub M, Stevens RC. Structural studies on phenylalanine hydroxylase and implications toward understanding and treating phenylketonuria. Pediatrics 2003;112(Supplement 4):1557–65. CrossRef
18. Kabbash IA, Attalla AO, Atlam SAE. Perception of importance of premarital counseling among medical students of Tanta University, Egypt. Egypt J Commun Med. 2019;37(2):66–75. CrossRef
19. Trefz KF, Muntau AC, Kohlscheen KM, Altevers J, Jacob C, Braun S, et al. Clinical burden of illness in patients with phenylketonuria (PKU) and associated comorbidities-a retrospective study of German health insurance claims data. Orphanet J Rare Dis 2019;14(1):1–16. CrossRef
20. Shebl G, Sayed Ahmed H, Kato A, Dawoud H, Hamza M, Haider A. Detection of sequence mutations in phenylalanine hydroxylase (PAH) gene isolated from Egyptian Phenylketonuria (PKU) patients. Egypt J Exp Biol 2019;15(2):295–301. CrossRef
21. Evans S, Adam S, Adams S, Allen H, Ashmore C, Bailey S, et al. Uniformity of food protein interpretation amongst dietitians for patients with phenylketonuria (PKU): 2020 UK National Consensus Statements. Nutrients 2020;12(8):2205. CrossRef
22. Porras G, Berthet A, Dehay B, Li Q, Ladepeche L, Normand E, et al. PSD-95 expression controls L-DOPA dyskinesia through dopamine D1 receptor trafficking. J Clin Invest 2012;122(11):3977–89. CrossRef
23. Gu CP, Xie YL, Liao YJ, Wu CF, Wang SF, Zhou YL, Jia SJ. Investigation of the pharmaceutical care in one elderly Parkinson’s disease patient with psychotic symptoms. Drug Saf Case Rep 2018;5(1):14. CrossRef
24. Ahmadzadeh A, Barnes MA, Gwazdauskas FC, Akers RM. Dopamine antagonist alters serum cortisol and prolactin secretion in lactating Holstein cows. J Dairy Sci 2006;89(6):2051–5. CrossRef
25. Feng J, Kang C, Wang C, Ding L, Zhu W, Hang S. L-phenylalanine increased gut hormone secretion through calcium-sensing receptor in the porcine duodenum. Animals 2019;9(8):476. CrossRef
26. Wei J, Shao W, Cao M, Ge J, Yang P, Chen L, et al. Phenylacetonitrile in locusts facilitates an antipredator defense by acting as an olfactory aposematic signal and cyanide precursor. Sci Adv 2019;5(1):5495. CrossRef
27. Qian Y, Lynch JH, Guo L, Rhodes D, Morgan JA, Dudareva N. Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat Commun 2019;10(1):1–15. CrossRef
28. Hendawey MH, Younes AMA. Biochemical evaluation of some faba bean cultivars under rainfed conditions at El-Sheikh Zuwayid. Ann Agric Sci 2013;58(2):183–93. CrossRef
29. Soares AR, Marchiosi R, Siqueira-Soares RDC, Barbosa de Lima R, Dantas dos Santos W, Ferrarese-Filho O. The role of L-DOPA in plants. Plant Signal Behav 2014;9(4):e28275. CrossRef
30. Hu J, Kwon SJ, Park JJ, Landry E, Mattinson DS, Gang DR. LC-MS determination of L-DOPA concentration in the leaf and flower tissues of six faba bean (Vicia faba L.) lines with common and rare flower colors. Funct Food Health Dis 2015;5(7):243–50. CrossRef
31. Shebl G, Sayed Ahmed H, Kato A, Haider A. ISSR markers of some faba bean (Vicia faba L.) genotypes in relation to phenylalanine accumulation. Egypt J Exp Biol (Botany). 2018;14(1):73–82. CrossRef
32. François L, Thierry G. Recent research advances in rabbit nutrition, General secretary of the World Rabbit Science Association/Rabbit Research Laboratory, National Institute of Agricultural Research, Castanet-Tolosan, France, vol. 27, pp 1–16, 2000.
33. Carabaño R, Villamide MJ, García J, Nicodemus N, Lorente A, Chamorro S, et al. New concepts and objectives for protein-amino acid nutrition in rabbits: a review. World Rabbit Sci 2009;17(1):1–14. CrossRef
34. Esteves PJ, Abrantes J, Baldauf HM, BenMohamed L, Chen Y, Christensen N,González-Gallego J, et al. The wide utility of rabbits as models of human diseases. Exp Mol Med 2018;12568:(1–10). CrossRef
35. Mu?ller-Scho?ttle F, Bogusz A, Gro?tzinger J, Herrler A, Krusche CA, Beier-Hellwig K, et al. Full-length complementary DNA and the derived amino acid sequence of horse uteroglobin. Biol Reprod 2002;66(6):1723–8. CrossRef
36. Rotbalt F, Brien DPO, Brien FJ, Goodall AH, Tuddenham GD. Purification of human factor VIII: C and its characterization by Western blotting using monoclonal antibodies. Biochemistry 1985;24(16):4294–300. CrossRef
37. Laemmli UK. 1970 Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227(5259):680–5. CrossRef
38. Brunelle JL, Green R. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods in Enzymol 2014;541:151–9. CrossRef
39. Ramesh A, Archibong AE, Hood DB, Guo Z, Loganathan BG. Global environmental distribution and human health effects of polycyclic aromatic hydrocarbons. In: Global Contamination Trends of Persistent Organic Chemicals, First Edition, Chapter 5, CRC Press, Taylor & Francis Group, Boca Raton, FL, Academic Publisher, USA, pp 97–126, 2011;
40. Saad AM. Growth behavior and productivity of faba bean (Vicia vulgaris, L.) as affected by various promoting foliar applications. Middle East J Appl Sci 2015;5(3):804–11.
41. St?tzel H, Hanafy MS. Impact of chitosan on shoot regeneration from faba bean embryo axes through its effect on phenolic compounds and endogenous hormones. Plant Arch 2020;20(1):2269–79.
42. Fitzpatrick PF. Allosteric regulation of phenylalanine hydroxylase. Arch Biochem Biophys 2012;519:194–201. CrossRef
43. Lartey FM, Austic RE. Phenylalanine hydroxylase activity and expression in chicks subjected to phenylalanine imbalance or phenylalanine toxicity. Poult Sci 2009;88(4):774–83. CrossRef
44. Cao YC, Yang XJ, Guo L, Zheng C, Wang DD, Cai CJ, et al. Effects of dietary leucine and phenylalanine on pancreas development, enzyme activity, and relative gene expression in milk-fed Holstein dairy calves. J Dairy Sci 2018;101(5):4235–44. CrossRef
45. Gunasekera RS, Hyland K. In vivo regulation of phenylalanine hydroxylase in the genetic mutant hph-1 mouse model. Mol Genet Metab 2009;98(3):264–72. CrossRef
46. Silva FJ, Bel Y, Botella LM, Cotton RGH, Ferré J. Immunological detection of phenylalanine hydroxylase protein in Drosophila melanogaster. Biochem J 1992;287(1):85–9. CrossRef
47. Daubner SC, Hillas PJ, Fitzpatrick PF. Characterization of chimeric pterin-dependent hydroxylases: contributions of the regulatory domains of tyrosine and phenylalanine hydroxylase to substrate specificity. Biochemistry 1997;36(39):11574–82. CrossRef