1. INTRODUCTION
Soy sauce is an essential traditional condiment with a history of more than 3,000 years in China [1]. Soy sauce not only gives a delicious flavor but also improves digestion [2]. In addition, soy sauce also has a special nutritional value, including proteins and carbohydrates, as well as being fat-free and containing a rich amount of riboflavin (vitamin B12) and other minerals (sodium, calcium, phosphorus, iron, selenium, and zinc). Every year, people produce millions of tons of soy sauce to supply the consumer market in the world. Traditionally, soy sauce is processed by fermentation and koji-making is the first step in the production of the fermentation sauce process. Koji is one of the key ingredients that were considered as starter varieties for the production of some traditional fermented foods. Aspergillus oryzae is the main fermentation agent in the production of soy sauce by microbiological methods [3]. During koji incubation, A. oryzae produces many enzymes, such as amylase, protease, invertase, maltose and etc. These enzymes break down starch and proteins into sugars and amino acids, respectively, which give the sauce a sweet taste [4]. Furthermore, A. oryzae has a specific genotype and is considered a safe organism for food production because it does not produce aflatoxin [5]. Aspergillus oryzae that has strains of the filamentous fungus and develops very quickly, forms a bunch of fibers and each strand is called mycelium [6]. Molds can only grow under fully aerobic conditions and the growth of A. oryzae is influenced by many factors, such as time, humidity, temperature and etc. [7]. Besides, the content of A. oryzae added to the koji is an important factor to study. If the percentage of molds is low, molds will not grow in enough numbers to overwhelm the bioburden. In addition, the growth of molds is inhibited by the harsh conditions of the environment that lead to the survival of microbial cells which were reduced significantly. Therefore, they have no longer beneficial effects to improve the quality of the product. If the percentage of molds is high, the product cost will increase and the quality will change in an unfavorable direction. Furthermore, the koji-making time is to obtain high quality and quantity of enzymes production [8].
Pleurotus spp. are highly nutritious foods. The protein content comes only after meat and fish, including water-soluble and essential amino acids, such as lysine and tryptophan. The composition of the main nutrients of oyster mushrooms includes carbohydrates and proteins that are the main ingredients, accounting for 70%–90% of dry weight. Oyster mushrooms are also rich in minerals (K and P) and important vitamins (B1, B2, C, etc.) [9]. Fat is very low and ranges from 1% to 2% [10]. In the past, the main ingredient used in the production of sauces was soybean. With the aim of enhancing the use of oyster mushrooms and creating a sauce that is nutritious, sensory valued, and safe for human health, oyster mushrooms are selected as raw materials for the manufacture of sauce. The research determined the effects and mutual interaction of factors (mold addition, incubation time, temperature, and pH) by using the central composite design (CCD) and response surface methodology (RSM) on enzymes production of oyster mushrooms koji.
2. MATERIALS AND METHODS
2.1. Materials
Wheat flours were roasted at 90°C for 15 minutes. Aspergillus oryzae molds powder (109 cfu/g) was provided by The Institute of Biotechnology Research and Development of Can Tho University (Vietnam), originating from the American Type Culture Collection. Pleurotus spp. mushrooms, after harvesting at the Experimental Area of An Giang University (Vietnam), were washed with clean water, chopped (0.5 × 1 cm), steamed at 90°C for 9–10 minutes, and cooled to 30°C. Each 50 g of mushrooms was mixed with 5 g of roasted wheat flour. The pH of oyster mushrooms koji was adjusted by citric acid.
2.2. Experimental Design
In order to obtain the best conditions for enzyme production (amylase and protease) from koji-making, a sequence of factorial designs was used. Using the STATGRAPHIC Centurion software (version 16.1) to optimize koji-making parameters by the RSM with the CCD 22 + star.
The first experiment was conducted with two factors, including molds addition (X1) (0.02 ÷ 0.04%) and koji-making time (X2) (24 ÷ 36 hours). The numbers of treatments were 13, including 5 replications of the central points. Each factor was surveyed with five levels, coded from −α to +α (with α = ± 1.4142). The level of encrypted variables and experimental layout is shown in Table 1.
In the second experiment, RSM and CCD were also used to investigate the effects of two factors, including incubation temperature (X3) (27 ÷ 33°C) and pH of koji medium (X4) (5.5 ÷ 6.5). Each factor was coded from −α to +α (with α = ± 1.4142). The design was 13 treatments, including 5 replications of the central points. The level of encrypted variables is shown in Table 2.
 | Table 1: Variable coding and survey levels of mold addition and koji-making time. [Click here to view] |
 | Table 2: Variable coding and survey levels of incubation temperature and pH of koji. [Click here to view] |
2.3. Analysis Methods
2.3.1. Determination of α-amylase activity
The α-amylase activity (U/g dry matter) was measured by an amylase unit expressed by the amount of enzymes that were capable of catalyzing the hydrolysis of 1 g of starch for 1 hour at 30°C and pH = 4.7 ÷ 4.8 [11]. The optical density of the solution was measured on a colorimeter with a liquid layer length of 1 cm and wavelength λ = 676 nm. The optical density difference between the control solution and the test solution is proportional to the amount of starch that has been exposed to α-amylase.
2.3.2. Determination of protease activity
This method was based on casein hydrolysis by enzymes in a research preparation solution [12]. Amino acids formed in hydrolysis reaction were measured by the Folin reagent. The unit of protease activity was the amount of an enzyme that converts an amount of sodium caseinate to a nonprecipitated form by tricloroacetic acid equivalent to 1 μmol of tyrosine at 30°C for 1 minute.
2.3.3. Determination of moisture
The moisture content was measured according to the standard method, as described by [13]. The sample was dried at 105°C to constant weight. The moisture W (%) was calculated using Equation (1):
where m1 is the initial mass of sample (g) and m2 is the final mass of sample (g).
2.4. Data Analysis Methods
Data were collected and processed by STAGRAPHICS Centurion 16.1 software for the analysis of variance and least significant difference test to conclude the difference between the averages of experiments at 5% confidence (p = 0.05) and Microsoft Excel software for calculating and graphing.
The appropriateness of the predicted model was assessed through the correlation coefficient R2. The equation optimizes the response surface of general form experiments according to Equation (2):
where Y is object function, βo is constant, βi is the linear coefficient, βii is the square coefficient, βij is the interaction coefficient, and Xi and Xj are survey variables.
3. RESULTS AND DISCUSSION
3.1. The Effects of Mold Addition and Koji-Making Time on Amylase and Protease Activities
The effects of molds and koji-making time on the presence of mycelium are shown in Figures 1 and 2. The results showed that the mycelium of 0.016% and 0.02% of A. oryzae developed less than mold with 0.03 ÷ 0.044% after 30 hours of incubation (Fig. 1).
Moreover, when the time of mold culture was during the period of less than 24 hours, there was no presence of mycelium. At 24 hours of koji-making, the mycelium began to grow scattered. The white mycelium covered a fairly thick layer on the medium at 30 and 36 hours of incubation. At 38.5 hours of the process, mycelium gradually turned to a yellow areca flower and appeared to have spores (Fig. 2).
The cycle of A. oryzae growth on bran could be divided into periods: the period of growth and germination of the sporangium (10 ÷ 11 hours of the first), the period of the rapid development of the fibrous system (4 ÷ 18 hours), and the strong enzyme generation period (lasting 10 ÷ 20 hours) [14]. The growth of molds also showed a contrasting relationship with the moisture of the medium [15]. Aspergillus oryzae used water on the surface of the medium to grow and create mycelium, forming spores [16]. The result in Figure 3 showed that the moisture content of the medium decreased with the increasing incubation time. In the early stage of the incubation process (less than 24 hours), the molds had not adapted to the medium, so the growth rate was quite slow and the moisture content of koji was more than 70%. In the period of 24–36 hours, the moisture of koji was 61.85% ÷ 70.39%. This result was also consistent with many research studies [17,18] where suitable moisture and time for A. oryzae to grow and produce enzymes were about 60 ÷ 68% and 30 ÷ 42 hours.
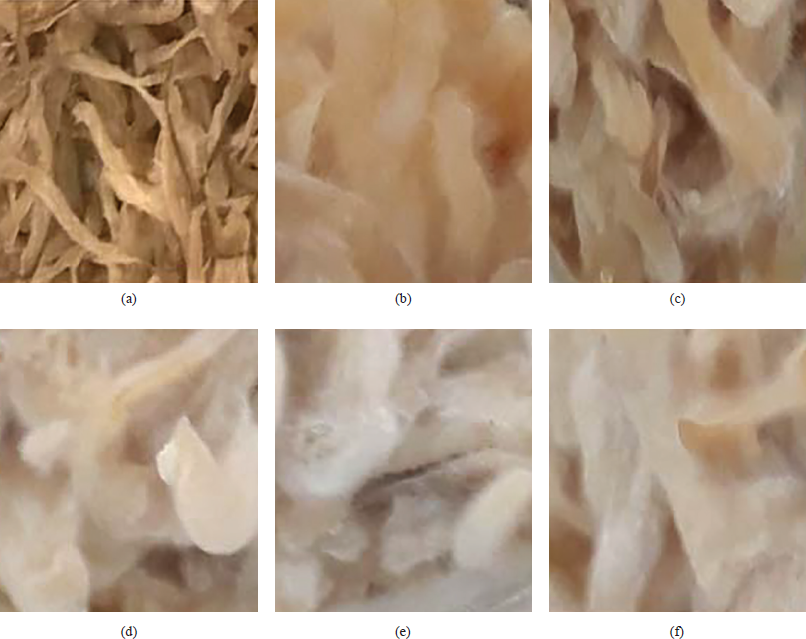 | Figure 1: Effect of mold addition on the presence of mycelium (a) initially, (b) at 0.016%, (c) at 0.02%, (d) at 0.03%, (e) at 0.04%, and (f) at 0.044%. [Click here to view] |
 | Figure 2: Koji-making time in the presence of mycelium (a) initially, (b) at 21.5 hours, (c) at 24 hours, (d) at 30 hours, (e) at 36 hours, and (f) at 38.5 hours. [Click here to view] |
In addition, the ability to produce more or less enzymes of molds means the development of mycelium [17]. The response surface models showing the effect of the mold addition and koji-making time on the activities of amylase and protease were built up (Fig. 4). The results showed that, as the addition of molds and incubation time increased, the activities of enzyme production increased to an optimal value and then decreased gradually. The higher the molds addition does not mean the higher the enzyme activities at the end of the koji process [19]. Additionally, mycelium formation and enzyme activities were closely related. Amylase and protease activities increased strongly when the medium began to form mycelium (from 24 to 30 hours) and decreased when spores appeared. This result is similarly demonstrated in the study of [20]. This was due to either the asexual cycle or the spore formation of A. oryzae that was involved in the production of secondary metabolites, such as enzymes or organic acids. Spore formation is undesirable because of reducing enzyme activities. The process of maximizing enzyme production usually ends when molds begin to form spores [17]. In addition, the lack-of-fit values of the models that were 0.06 and 0.43 (>0.05), respectively, showed the model’s compatibility with the experiment. The optimal amylase activity was 61.77 U/g dry matter when 0.03% molds and 30.75 hours of incubation time were applied. Protease activity was optimal at 12.37 U/g dry matter when the percentage of molds was 0.03 and it was fermented for 30.41 hours.
 | Figure 3: Changes in the moisture content of koji according to incubation time. [Click here to view] |
 | Figure 4: The response surface and contour plots showing the effect of mold addition and koji-making time on (a) Amylase activity and (b) Protease activity. [Click here to view] |
Regression equations showing the relationship of the molds addition and koji-making time to amylase and protease activities were described in Table 3. All equations met the conditions with a high coefficient of determination (R2 > 0.84). The compatibility between the predictive and experimental data of amylase and protease activities was shown in Figure 5 and there was a high correlation between predictive and experimental data (R2 > 0.84).
The results of simultaneous optimization of multiple response surfaces showed optimal amylase and protease activities when 0.03% of mold and 30 hours for incubation time were applied (Fig. 6). Amylase and protease activities were 61.62 and 12.52 U/g dry matter, respectively.
Thus, the parameters selected for the next studies were the addition of molds at the rate of 0.03% and the time of koji making was 30 hours.
3.2. The Effects of Temperature and pH of Koji-Making on Enzyme Activities
The response surface models showing the correlation between the incubation temperature and pH of the medium to amylase and protease activities were built up (Fig. 7). The temperature and pH of koji-making affected the quadratic on enzyme activities. When the pH of the medium increased from 5.5 to 6.0, the amylase and protease activities increased; however, the pH continued to increase to 6.5 and the enzyme activities decreased. Likewise, amylase and protease activities increased with increasing temperature (up to 30°C) and a further increasing in the koji-making temperature decreased. The results showed that amylase production with optimal activity was 61.43 U/g dry matter when it was fermented at 30.14°C and pH 6.02. Meanwhile, the optimal protease activity was 12.37 U/g dry matter when the temperature and pH of koji making were 27.76°C and 5.89°C, respectively.
 | Table 3: Predictive regression equations of the relationship between the molds and koji-making time. [Click here to view] |
 | Figure 5: Correlation between experimental and theoretical data (from Y1 and Y2) for (a) Amylase activity and (b) Protease activity. [Click here to view] |
 | Figure 6: Overplay plots of enzyme activities according to molds addition and koji-making time. [Click here to view] |
The temperature of koji-making is a strong influence on the reaction rate. The speed of reaction will increase rapidly under the effect of temperature. If the optimal threshold is exceeded, it will inhibit the incubation [21]. Temperature not only affects the growth of microorganisms but also affects their biological activity. The effects of incubation temperature on mold growth and enzyme activities need to be studied to understand the role of temperature control in koji-making. In addition, the initial pH of the koji medium is one of the factors that have an important influence on the ability of enzyme biosynthesis. The initial pH of the medium affects the metabolism of microorganisms, leading to a change in the absorption of food. The influence level of pH also depends on the substrate, buffer properties, temperature, and so forth [22].
 | Figure 7: The response surface and contour plots showing the effect of temperature and pH of koji on (a) Amylase activity and (b) Protease activity. [Click here to view] |
 | Table 4: Predictive regression equations of the relationship between temperature and pH on enzyme activities. [Click here to view] |
 | Figure 8: Correlation between experimental and theoretical data (from Y3 and Y4) for (a) amylase activity and (b) protease activity. [Click here to view] |
 | Figure 9: Overplay plots of enzyme activities according to temperature and pH of koji. [Click here to view] |
 | Table 5: Results of the experiment and optimization model. [Click here to view] |
In addition, the lack-of-fit values were not statistically significant (p > 0.05) and correlation coefficients of regression equations showing the relationship of temperature and pH to amylase (Y3) and protease (Y4) were high (R2 > 0.85) (Table 4), so it confirmed the more significance and reliability of the regression models established. The compatibility between the predictive and experimental data of amylase and protease activities was determined with the high correlation coefficient (R2 > 0.85) (Fig. 8). Furthermore, the results of optimization of multiple response surfaces showed amylase and protease activities reached the highest values at 29.94°C and pH 5.97 (Fig. 9). The experimental results of incubating at 30°C and pH 6.0 were equivalent to the results predicted from the model (Table 5). The previous studies had shown that incubating at 30°C and pH 6.0 was the optimal temperature and pH for A. oryzae to grow and produce enzymes with high activities on solid-state fermentation (SSF) [23–26].
4. CONCLUSION
In traditional fermentation, SSF is suitable for mold growth and enzyme activities are an important indicator of the enzyme-producing ability of A. oryzae. The optimal conditions were found at 0.03% of molds addition, pH 6.0, and incubation at 30°C in 30 hours to obtain the highest enzyme activities for oyster mushrooms koji.
5. ACKNOWLEDGMENT
The authors gratefully acknowledge Can Tho University and An Giang University, Vietnam University, Ho Chi Minh City, Vietnam, for supporting this research.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Zhang YF, Tao WY. Flavor and taste compounds analysis in Chinese solid fermented soy sauce. Afr J Biotechnol 2009;8:673–81.
2. Kataoka S. Functional effects of Japanese style fermented soy sauce (shoyu) and its components. J Biosci Bioeng. 2005;100:227–34. CrossRef
3. Kim KM, Lim J, Lee JJ, Hurh BS, Lee I. Characterization of Aspergillus sojae isolated from Meju, Korean traditional fermented soybean brick. J Microbiol Biotechnol 2017;27:251–61. CrossRef
4. Ward OP, Qin WM, Dhanjoon J, Ye J, Singh A. Physiology and biotechnology of Aspergillus. Adv Appl Microbiol 2006;58:1–55. CrossRef
5. Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005;438:1157–61. CrossRef
6. Liu Z, Gosser Y, Baker PJ. Structural and function studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation. J Am Chem Soc 2009;131(43):15711–6. CrossRef
7. Al-Shehri MA. Production and some properties of protease produced by Bacillus licheniformis isolated from Tihamet Aseer. Saudi Arabica Pak J Biol Sci 2004;7:1631–5. CrossRef
8. Hishiya N, Watanable M, Sakurai M, Fujita K, Noda T. Method for producing rapid fermented type miso-like food material with favorable taste and flavor. Japanese Patent Application, Tokyo, Japan, 2007.
9. Millikan M. Nutritional metals in foods by AAS. In: Farrukh, M.A. (Ed), Atomic Absorption Spectroscopy. InTechopen, London, UK, pp 143–66, 2012. CrossRef
10. Akindahunsi AA, Oyetayo FL. Nutrient and antinutrient distribution of edible mushroom, Pleurotus tuber-regium (fries) singer. LWT Food Sci Technol 2006;39:548–53. CrossRef
11. Okolo BN, Ezeogu LI, Mba CI. Production of raw starch digesting amylase by Aspergillus niger grown on native starch sources. J Sci Food Agri 1995;69:109–15. CrossRef
12. Anson ML. The estimation of pepsin, trypsin, papain and cathepsin with hemoglobin. J Gen Physiol 1938;22:79–89. CrossRef
13. AOAC. Official methods of analysis of association of official analytical chemists. AOAC, Washington, DC, USA, 2010.
14. Biesebeke R, Ruijter G, Rahardjo YSP, Hoogschagen MJ, Heerikhuisen M, Levin A et al. Aspergillus oryzae in solid-state and submerged fermentations progress report on a multi-disciplinary project. FEM Yeast Res 2002;2:245–8. CrossRef
15. Grimm LH, Kelly S, Volkerding II, Krull R, Hempel DC. Influence of mechanical stress and surface interaction on the aggregation of Aspergillus niger conidia. Biotechnol Bioeng 2005;92(7):879–88. CrossRef
16. Paul GC, Thomas CR. Characterisation of mycelial morphology using image analysis. Adv Biochem Eng Biotechnol 1998;60:1–59. CrossRef
17. Chancharoonpong C, Hsieh P, Sheu S. Production of enzyme and growth of Aspergillus oryzae S. on aoybean koji. Int J Biosci Biochem Bioinformatics 2012;2(4):2–5. CrossRef
18. Lubis D, Wina E, Harvanto B, Suhargiantatmo T. Effectiveness of Aspergillus oryzae fermentation culture to improve digestion of fibrous feeds: in vitro. Indones J Anim Vet Sci 2002;7(2):90–8.
19. Jiang X, Xu Y, Ye J, Yang Z, Huang S, Liu Y, et al. Isolation, identification and application on soy sauce fermentation flavor bacteria of CS1.03. J Food Sci Technol 2019;56(4):2016–26. CrossRef
20. Narahara H, Koyama Y, Yoshida T, Pichangkura S, Ueda R, Taguchi H. Growth and enzyme production insolid-state culture of Aspergillus oryzae. J Ferm Technol 1982;60:311–9.
21. Nagel FJ, Tramper J, Bakker MS, Rinzema A. Temperature control in a continuously mixed bioreactor for solid-state fermentation. Biotechnol Bioeng 2001;72:219–30. CrossRef
22. Farid MAF, Shata HMAH. Amylase production from Aspergillus oryzae LS1 by solid-state fermentation and its use for the hydrolysis of wheat flour. Iran J Biotechnol 2011;9(4):267–74.
23. Sangeetha PT, Ramesh MN, Prapulla SG. Production of fructosyltransferase by Aspergillus oryzae CFG 202 in solid state fermentation using agricultural by products. Appl Microbiol Biotechnol 2004;65(5):530–7. CrossRef
24. Zambare V. Solid state fermentation of Aspergillus oryzae for glucoamylase production on agro residues. Int J Life Sci 2010;4:16–25. CrossRef
25. Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandley A. Alpha amylase production by Aspergillus oryzae employing solid state fermentation. Appl Microbiol Biotechnol 2007;66:621–6.
26. Puri S, Aora M, Sarao L. Production and optimization of amylase and glucoamylase using Aspergillus oryzae under solid state fermentation. Int J Res Pure Appl Microbiol 2013;3(3):83–8.