1. INTRODUCTION
Piperine, a major bioactive compound from the fruits of pepper (Piper nigrum), has anti-oxidant, anticarcinogenic, anti-inflammatory, and antimicrobial properties [1]. Black pepper is used in traditional medicine to treat health problems, such as intermittent fever, muscular pain, and migraine [2]. Research conducted to investigate the cytotoxicity effect of piperine on cell lines of epithelial origin such as cervical, oral, colon, and prostate cancer concluded that the piperine compound shows toxicity to these cell lines by increasing the level of reactive oxygen species (ROS), induces DNA fragmentation, and triggers apoptosis reaction [3–6].
Evasion of programmed cell death (apoptosis) by cancer cells via inhibition of caspase activity, upregulation of the antiapoptotic proteins, mutation of the proapoptotic proteins, and loss of the apoptotic activity is amongst the hallmarks of cancer [7]. In fact, the anti-inflammatory nature of apoptosis is crucial for the eradication of virally infected potential cancer cells [8]. Therefore, targeting apoptosis is an effective nonsurgical treatment for cancer. Apoptosis is regulated by two main pathways, the intrinsic and extrinsic pathways. Basically, the intrinsic pathway is controlled by the Bcl-2 family member of proteins which comprises antiapoptotic and proapoptotic proteins that cointeract [9,10]. Proapoptotic proteins, such as Bad and Nova, are apoptosis effectors that work in concert with antiapoptotic proteins (Bcl-2, Bcl-xL, and Mcl-1) and other proapoptotic proteins (Bax, Bak, and Bok) to mediate caspase-associated apoptotic mechanisms [9,10]. The extrinsic pathway, on the other hand, is stimulated by extracellular signals from death ligands from the tumor necrosis factor receptor superfamily (TNFRSF) where TNF-α, TNFR1/CD95, and Fas are amongst the common players in the caspase-related events of apoptosis [11,12]. Both intrinsic and extrinsic pathways cooperatively regulate cellular integrity [13] and prevent cancer onset. Hence, identifying the drug/chemical compounds that target these pathways is essential in the precision chemotherapy of the cancer condition.
Conventional experimental approaches for drug discovery are laborious and expensive and thus warrant the necessity of computational screening in drug repurposing to determine the feasibility of the protein–ligand binding [14]. This in silico method complements and hastens the development and rationalization of drug candidate choice. In this respect, computational docking of small-molecule ligands to macromolecular targets which considers bound conformations and free binding energies is a relevant structure-based drug design strategy. A standard docking and virtual screening platform for this purpose is the AutoDock suite of programs that allows the basic docking of a drug molecule with an anticancer target including docking with selective receptor flexibility and explicit dehydration and active site prediction [15]. The usability of AutoDockTools is evidenced by multiple studies. These include the in silico docking studies to identify the binding activity of carnosine against a subunit of Caspase-3 in human cervical carcinoma models [16], the discovery of strong binding affinities of phytocompounds such as harpagoside, bromelain, and afzelin against key transcriptional factors (p53, AP-1, c-Myc, β-catenin, and HIF-1α) involved in liver cancer [17], and the computational drug–protein interaction analysis affirming mitochondrial complex III as a likely target of metformin in head and neck squamous cell cancer [18]. Similarly, the AutoDock suite was effective in simulating molecular docking between the 1,2,3-triazole moiety and pyrimidine derivatives against cognate receptors in test models of human esophageal cancer revealing hydrogen bond interactions of the ligands with the amino acid residues in the receptor binding sites [19]. Finally, this structure-based virtual docking approach was essential in the screening of several thousand compounds to identify candidate ULK1 inhibitors and identified a single compound, XST-14, with strong binding affinity at the Lys46, Tyr94, and Asp165 amino acid of the ULK1 protein to block its physiological activity [20].
The study reported herein describes the work and findings from the deployment of AutoDockTools (and associated software) in a systematic endeavor of structure-based drug design strategy to carry out novel simulated molecular docking of piperine (ligand) with apoptotic proteins (receptors). Our study will provide vital conceptual insights into the anticancer potentials of piperine.
2. MATERIALS AND METHODS
In this study, PyMOL (version 1.8) [21], AutoDockTools, AutoDock Vina (version 1.5.6) [22], and LigPlot + [23] programs were used to investigate the possible conformation of the binding between piperine and the different apoptotic proteins (Bax, Bcl-2, TNF-α, TNFR-1, Caspase-3, Caspase-8, and Caspase-9).
2.1. Selection and In Silico Preparation of Protein Target
Bax and Bcl-2 proteins were selected due to their major role in activating the intrinsic pathway of apoptosis [24], which is commonly used as a biomarker to determine the fate of cells and would be a potential therapeutic target for cancer treatment [25]. TNF-α and TNFR1 were also chosen as they are valid biomarkers for the extrinsic pathway [26]. Both initiator caspases, Caspase-9 and Caspase-8, were selected due to their role in regulating apoptosis reaction in cancer and their association with sensitivity to chemotherapeutic agents [27]. The executioner caspase, Caspase-3, was selected based on the fact that it is the only protein that is closely associated with DNA fragmentation—a prerequisite for apoptosis [28].
The 3D crystallographic protein structure for the respective proteins was downloaded from the Research Collaboratory for Structural Bioinformatics Protein Drug Bank website in the Protein Databank (PDB) format. The protein chosen was of human origin with a high-resolution structure ranging from 1.0 to 3.0 Å and was identified using the X-ray diffraction method. Ligands found in proteins structure were removed via PyMOL software prior to preparation. Then, the crystallographic water molecule of each protein was removed to reduce interference using AutoDockTools. After this, polar hydrogen molecules and Kollman United Atom charges were assigned to each protein molecule. All proteins were converted into the Protein Data Bank format with partial charges (‘Q’) and AutoDock 4 atom types (‘T’) (PBDQT) file format and saved for later use.
2.2. In Silico Preparation of Ligand Molecule
The piperine compound was used as the ligand for molecular docking in this study. The 3D conformer of the piperine compound was downloaded in an SDF file from the PubChem databases. Then, the ligand was converted into a PDB file format via PyMOL software and minimized by computing Gasteiger charges. In the final processing, piperine was converted to PDBQT file format using AutoDockTools.
2.3. Molecular Docking Simulation
The prepared protein and ligand were called out via AutoDockTools to compute suitable grid maps for each protein–ligand combination. Grid size, grid spacing, and grid center (Table 1) were generated using AutoDockTools based on the predicted binding pockets generated by the Computed Atlas of Surface Topography of proteins (CASTp) website [29]. A configuration file that consists of the grid box properties was created and saved as .txt file format. Then, docking was carried out using AutoDock Vina by inserting command lines in the Command Prompt application to generate the output score and the best fit model (mode 1) was selected from the nine different conformations generated. Interacting amino acid residues that were found in the binding site were visualized using LigPlot + software. Amino acid residues exhibiting hydrogen bonding and hydrophobic interactions with piperine are summarized and tabulated in Tables 2 and 3, respectively. A bar chart showing the docking score for each protein–piperine complex was constructed.
4. RESULTS
Binding affinities between piperine and target proteins show varied score values across the seven different receptors (Table 2). The score values range from −5.2 to −7.2 kcal/mol, and thus the binding complexes can be arranged in decreasing binding affinity order as follows: TNFR-1 > Caspase-3 > TNF-α > Caspase-8 > Bcl-2 > Caspase-9 > Bax. The lower the score value, the higher the binding affinity. Among all the protein/receptor–ligand pairs, the lowest binding score of −7.2 kcal/mol was recorded between TNFR-1 protein and piperine, and the highest involved Bax protein (−5.2 kcal/mol). The docking score from the predicted best fit model was selected for all ligand–receptor binding combinations and a graphical (bar chart format) comparison of the binding affinity is illustrated in Figure 1.
 | Table 1: The grid spacing, grid size and grid center used for protein-ligand docking based on CASTp prediction. [Click here to view] |
 | Table 2: Binding score and the bond length of hydrogen bond formed between piperine and amino acid residue in each protein. [Click here to view] |
 | Table 3: Results of the amino acid residues in binding pockets that interact with piperine for all the protein-ligand combinations as displayed via LigPlot + assessment. [Click here to view] |
According to the 2D structural model generated via LigPlot + (Fig. 2A–G), the hydrophobic interaction in the analyzed shows the involvement of a relatively varied number of interacting amino acid residues across the different ligand–receptor complexes. Bcl-2, Caspase-9, TNFR-1, and Caspase-8 demonstrated eight interacting amino acid residues each with piperine and were the biggest numbers compared to other piperine–apoptotic protein complexes. The ligand–receptor binding predicted for piperine and TNF-α had the lowest number (six) of interacting amino acid residues. Of all studied protein–ligand combinations, a single hydrogen bond was found in TNFR-1 (Fig. 2E) and Caspase-8 (Fig. 2F) protein–ligand complexes with bond lengths of 2.98 and 3.05 (Fig. 2, Table 2), respectively. On the other hand, a double hydrogen bond was shown between piperine and amino acid residues of TNF-α (Fig. 2F) protein with bond lengths of 2.85 and 3.24 (Fig. 2, Table 2). The piperine molecule binds with the studied apoptotic proteins mainly by hydrophobic interactions with the amino acid residues listed in Table 2. A complete rendition of the results on hydrophobic interaction and hydrogen bond visualized using the LigPlot + application is provided in both Tables 2 and 3.
 | Figure 1: Binding score (kcal/mol) of the predicted best fit model generated via AutoDock Vina for docking of ligand (piperine) and the apoptotic proteins (Bax, Bcl-2, Caspase-9, TNF-α, TNFR-1, Caspase-8, and Caspase-3). [Click here to view] |
5. DISCUSSION
Virtual screening and computational drug–target binding simulation are common in silico approaches and supportive tools for drug discovery and drug repurposing. This is especially pertinent and relevant considering the fact that conventional experimental laboratory strategies are technically demanding, time-consuming, and expensive and involve countless trial and error endeavors. In this study, the docking and interaction of the piperine compound with proteins involved in apoptotic pathways were studied and compared using open-source software. AutoDockTools (specifically, AutoDock Vina) was deployed in our analysis. The working paradigm of this analytical tool/resource allows flexible ligand docking where the target protein (receptor) can be set as a rigid molecule and ligand as a flexible molecule in order to generate all possible binding combinations based on available conformations [30]. This is relevant to our study design and hence the use of AutoDock Vina as our application of choice.
Based on the binding simulation results (Table 2), negative binding free energy was generated from all the docked pairs suggesting the strong likelihood of binding affinity between each protein–ligand combination. While negative binding free energy value, in terms of kcal/mol, provides information on the degree of affinity between receptor and ligand, the molecular interaction between the amino acid residues from targeted protein and ligand (piperine, in our case) would be another essential factor to consider in stimulating logical favorable docking conformation [31].
Our findings reveal that piperine exhibits strong hydrophobic interaction toward all seven apoptosis-related proteins studied. This has a few pertinent indications. Firstly, hydrophobic contacts are the most essential type of protein–ligand binding that is stereotypical of drug–receptor interaction [32] and our simulated data substantiate the calculated plausibility of the targeting of key apoptotic proteins by piperine. To the best of our knowledge, this has never been reported before. Secondly, apart from hydrophobic interactions, TNF-α, TNFR-1, and Caspase-8 proteins also showed hydrogen bonding with the piperine compound. A noncovalent hydrogen bond plays a critical role in the integrity and stability in the formation of receptor–ligand complexes [33].
It is shown in Figure 1 that Caspase-3 possesses the second lowest binding affinity, indicating a stronger affinity of the proteins toward the piperine compound. However, a lower number (seven) of hydrophobic interactions and no hydrogen bonding was shown in the protein–ligand complex. This may be explained by the presence of other types of protein–ligand interactions such as salt bridges and amide stacking [32], which contribute to a higher binding affinity (−6.9) from the docking of Caspase-3 and piperine, as the 2D structure obtained from LigPlot + consists of hydrophobic interaction and hydrogen bonding only.
The docking of piperine with TNF-α, TNFR-1, and Caspase-8 demonstrated higher binding affinity compared to Bax, Bcl-2, and Caspase-9. This, therefore, indicates a stronger interaction between piperine and proteins from the extrinsic pathway and suggests the potency of piperine in the regulation of cell signaling via the extrinsic pathway of apoptosis. Again, this phenomenon has never been systematically explored prior to our study. In a study on the leukemic cell line, piperine was found to exert cytotoxic properties with the constitutive deregulation of Bax, Caspase-3, Caspase-9, and Bcl-2 [34]. In silico docking analysis of piperine with a limited number of apoptosis-associated proteins (Bax, Caspase-3, and Caspase-9) has been demonstrated before [35] but our data reveals a larger repertoire of apoptosis-associated proteins predicted to bind with piperine. Taken together, these data reinforce the notion of the physiologic involvement of piperine in apoptosis.
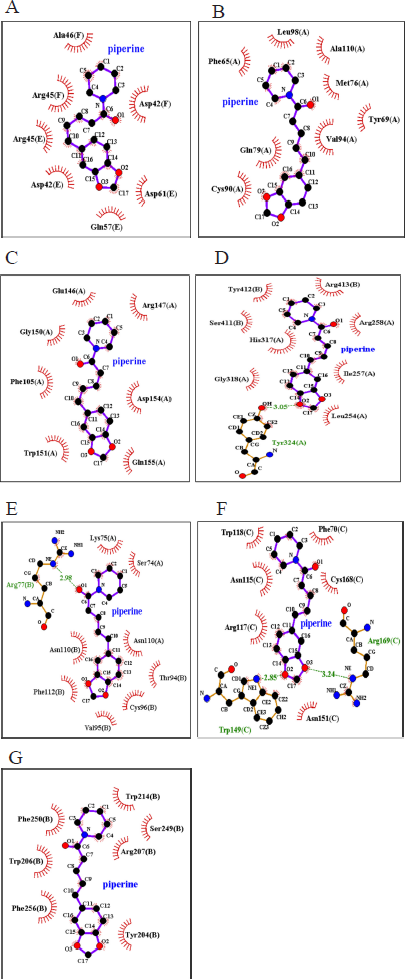 | Figure 2: 2D representation results for LigPlot+ analysis of the piperine compound interacting with (A) Bax, (B) Bcl-2, (C) Caspase-9, (D) TNF-α, (E) TNFR-1, (F) Caspase-8, and (G) Caspase-3. Hydrophobic interactions are represented by red spiky arcs, while hydrogen bonds are indicated by green dashed lines along with calculated bond lengths. [Click here to view] |
The anticancer role of TNF-α can be explained by the activation of the death domain in TNFR-1 leading to the recruitment of certain adaptor proteins, such as Caspase-8, and TNFR1-associated death domain [Tumor necrosis factor receptor type 1-associated DEATH domain protein (TRADD)]; stimulating Caspase-3 thus triggers apoptosis reaction within the cells [36]. Alternative TNF-α activity may happen at times which leads to chronic inflammation resulting from the activation of NF-κB that would serve as an effective promoter of carcinogenesis by inducing inhibitors of apoptotic proteins (IAPs) [37]. These IAPs would inhibit apoptotic reactions in cancer cells by interfering with the activation of the caspase cascade reaction. In a clinical study conducted by Jin et al. [38], TNF-α was shown to be overexpressed among Nasopharyngeal carcinoma (NPC) patients compared to healthy individuals and had decreased significantly after cancer treatment, suggesting the important role of inflammatory cytokines, TNF-α in NPC progression. Furthermore, several studies suggested that the expression level of TNF-α is useful in the prediction of distant metastasis and overall survival rate in NPC patients, demonstrating the possibility of TNF-α as a protumor factor and potential therapeutic target [39,40]. Another study also suggested the strong relationship of TNF-α protein expression with the clinical staging of NPC and that cancer cells are more dependent on the transcription factor stimulated by TNF-α as compared to normal cells in cancer progression [41].
Whether piperine acts as an agonist or antagonist in the activation or inhibition of cellular apoptotic factors remains to be further characterized. Thus, the molecular dynamic technique may be carried out on the ligand–protein complexes studied following molecular docking simulation in order to improve the drug discovery process by providing a more specific representation of protein motion [42].
6. CONCLUSION
Our in silico analysis of simulated receptor–ligand binding between the piperine compound and key proteins from apoptotic pathways reveals the occurrence of strong binding affinity. Piperine showed a greater affinity for proteins of the extrinsic apoptosis pathway (TNF-α, TNFR-1, and Caspase-8) compared to those of the intrinsic pathway suggesting their physiologic involvement in cellular health. Irrevocably, our computational data provide novel insights into the drug–target mechanisms and are considered a preliminary investigation that supports further validation on the anticancer potentials of a natural and purified compound (piperine) extracted from the black pepper plant (P. nigrum) via in vivo or in vitro experimentation.
7. ACKNOWLEDGMENTS
The authors acknowledge the Sarawak Government for funding this study via the Sarawak Research Development Council under the Catalyst Fund Scheme (Grant no. RDCRG/CAT/2019/20).
8. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
9. CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
10. FUNDING
This project was funded by the Sarawak Government via the Sarawak Research Development Council under the Catalyst Fund Scheme (Grant. no. RDCRG/CAT/2019/20).
11. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Tiwari A, Mahadik KR, Gabhe SY. Piperine: a comprehensive review of methods of isolation, purification and biological properties. Med Drug Discov 2020;7:100027. doi:10.1016/j.medidd.2020.100027 CrossRef
2. Shityakov S, Bigdelian E, Hussien AA, Hussain MB, Tripathi YC, Khan MU, et al. Phytochemical and pharmacologiacal attributes of piperine: a bioactive ingredient of black pepper. Euro J Med Chem 2019;176:149–61. CrossRef
3. Siddiqui S, Ahamad MD, Jafri A, Afzal M, Arshad M. Piperine triggers apoptosis of human oral squamous carcinoma through cell cycle arrest and mitochondrial oxidative stress. Nutr Cancer 2017;69(5):791–9. CrossRef
4. George K, Thomas NS, Malathi R. Piperine blocks voltage gated K+ current and inhibits proliferation in androgen sensitive and insensitive human prostate cancer cell lines. Arch Biochem Biophys 2019;667:36–48. CrossRef
5. Jafri A, Siddiqui S, Rais J, Ahmad MS, Kumar S, Jafar T, et al. Induction of apoptosis by piperine in human cervical adenocarcinoma via Ros mediated mitochondrial pathway and caspase-3 activation. EXCLI J, Int Online J Adv Sci 2019;18:154–64.
6. Song L, Wang Y, Zhen Y, Li D, He X, Yang H, et al. Piperine inhibits colorectal cancer migration and invasion by regulating STAT3/snail-mediated epithelial mesenchymal transition. Biotechnol Lett 2020;42(10):2049–58. CrossRef
7. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74. CrossRef
8. Yanumula A, Cusick JK. Biochemistry, extrinsic pathway of apoptosis [Internet]. StatPearls Publishing LLC, Treasure Island, Florida, 2020. Available via https://www.ncbi.nlm.nih.gov/books/NBK560811/ (Accessed 27 March 2021).
9. Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol 2018;8:1–1. CrossRef
10. Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 2019;20(3):175–93. CrossRef
11. Zhu J, Jin M, Wang J, Zhang H, Wu Y, Li D, et al. TNFα induces Ca2+ influx to accelerate extrinsic apoptosis in hepatocellular carcinoma cells. J Exp Clin Cancer Res 2018;37(1):43. doi:10.1186/s13046-018-0714-6 CrossRef
12. Jan R, Chaudhry, G. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv Pharm Bull 2019;9(2):205–18. CrossRef
13. Wang Y, Hu S, Tuerdi M, Yu X, Zhang H, Zhou Y, et al. Initiator and executioner caspases in salivary gland apoptosis of Rhipicephalus haemaphysaloides. Parasit Vectors 2020;13(1):288. doi: 10.1186/s13071-020-04164-5 CrossRef
14. Neves BJ, Braga RC, Melo-Filho CC, Moreira-Filho JT, Muratov EN, Andrade CH. QSAR-based virtual screening: advances and applications in drug discovery. Front Pharmacol 2018;9:1275. doi: 10.3389/fphar.2018.01275 CrossRef
15. Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoco 2016;11(5):905–19. CrossRef
16. Pandurangan M, Enkhtaivan G, Kim DH. Therapeutic efficacy of natural dipeptide carnosine against human cervical carcinoma cells. J Mol Recognit 2016;29(9):426–35. CrossRef
17. Murthy SS, Narsaiah TB. Molecular docking studies of phytocompounds with transcriptional factors in hepatocellular carcinoma. Rasayan J Chem 2019;14(4):2030–8. CrossRef
18. Kuo SZ, Honda CO, Li WT, Honda TK, Kim E, Altuna X, et al. Metformin results in diametrically opposed effects by targeting non-stem cancer cells but protecting cancer stem cells in head and neck squamous cell carcinoma. Int J Mol Sci 2019;20(1):193. doi:10.3390/ijms20010193 CrossRef
19. Adegoke RO, Oyebamiji AK, Semire B. Dataset on the DFT-QSAR, and docking approaches for anticancer activities of 1, 2, 3-triazole-pyrimidine derivatives against human esophageal carcinoma (EC-109). Data Brief 2020;31:105963. doi:10.1016/j.dib.2020.105963 CrossRef
20. Xue ST, Li K, Gao Y, Zhao LY, Gao Y, Yi H, et al. The role of the key autophagy kinase ULK1 in hepatocellular carcinoma and its validation as a treatment target. Autophagy 2020;16(10):1823–37. CrossRef
21. Schrödinger LLC. The PyMOL molecular graphics system, Schrödinger Inc., New York, NY, USA, version 1.8. 2015.
22. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimisation and multithreading, J Comput Chem 2010;31:455–61. CrossRef
23. Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51(10):2778–86. CrossRef
24. Edlich F. BCL-2 proteins and apoptosis: recent insights and unknowns. Biochem Biophys Res Commun 2018;500(1):26–34. CrossRef
25. Warren CFA, Wong-Brown MW, Bowden NA. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis 2019;10(3):177. doi:10.1038/s41419-019-1407-6 CrossRef
26. Kominami K, Nakabayashi J, Nagai T, Tsujimura Y, Chiba K, Kimura H, et al. The molecular mechanism of apoptosis upon caspase-8 activation: quantitative experimental validation of a mathematical model. Biochimica Biophysica Acta 2012;1823(10):1825–40. CrossRef
27. Raudenska M, Balvan J, Masarik M. Cell death in head and neck cancer pathogenesis and treatment. Cell Death Dis 2021;12(2):192. doi:10.1038/s41419-021-03474-5 CrossRef
28. McComb S, Chan PK, Guinot A, Hartmannsdottir H, Jenni S, Dobay MP, et al. Efficient apoptosis requires feedback amplification of upstream apoptotic signals by effector caspase-3 or -7. Sci Adv 2019;5(7):eaau9433. doi:10.1126/sciadv.aau9433 CrossRef
29. Tian W, Chen C, Lei X, Zhao JL, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 2018;46(W1):W363–7. doi:10.1093/nar/gky437 CrossRef
30. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review, Biophys Rev 2017;9(2):91–102. CrossRef
31. Pantsar T, Poso A. Binding affinity via docking: fact and fiction. Molecules 2018;23(8):1899. doi:10.3390/molecules23081899 CrossRef
32. Freitas RF, Schapira M. Asystemic analysis of atomic protein-ligand interactions in the PDB. Med Chem Commun 2017;8(10):1970–81. CrossRef
33. Afriza D, Suriyah WH, Ichwan SJA. In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. J Phys Conf Ser 2018;1073(3):032001. doi:10.1088/1742-6596/1073/032001 CrossRef
34. Banerjee S, Katiyar P, Kumar V, Saini SS, Varshney R, Krishnan V, et al. Black pepper and piperine induce anticancer effects on leukemia cell line. Toxicol Res 2021;10(2):169–82. doi:10.1093/toxres/tfab001 CrossRef
35. Kirubhanand C, Selvaraj J, Rekha UV, Vishnupriya V, Nalini D, Mohan SK, et al. Molecular docking data of piperine with Bax, Caspase 3, Cox 2 and Caspase 9. Bioinformation 2020;16(6):458–61. doi:10.6026/97320630016458 CrossRef
36. Sethi G, Sung B, Aggarwal BB. TNF: a master switch for inflammation to cancer. Front Biosci 2008;13:5094–107. doi:10.2741/3066 CrossRef
37. Zeligs KP, Neuman MK, Annunziata CM. Molecular pathways: the balance between cancer and the immune system challenges the therapeutic specificity of targeting nuclear factor-kB signalling for cancer treatment. Clin Cancer Res 2016;22(17). doi:10.1158/1078-0432.CCR-15-1374 CrossRef
38. Jin YB, Zhang GY, Lin KR, Chen XP, Cui JH, Wang YJ, et al. Changes of plasma cytokines and chemokines expression level in nasopharyngeal carcinoma patients after treatment with definitive intensity-modulated radiotherapy (IMRT). PLoS One 2017;12(2):1–9. doi:10.1371/journal.pone.0172264 CrossRef
39. Lu X, Qian CN, Mu YG, Li NW, Li S, Zhang HB, et al. Serum CCL2 and serum TNF-α – two new biomarkers predict bone invasion, post-treatment distant metastasis and poor overall survival in nasopharyngeal carcinoma. Euro J Cancer 2011;47(3):339–46. doi:10.1016/j.ejca.2010.09.025 CrossRef
40. Yu Y, Ke L, Xia WX, Xiang Y, Lv X, Bu J. Elevated levels of TNF-α and decreased levels of CD68-positive macrophages in primary tumor tissues are unfavorable for the survival of patients with nasopharyngeal carcinoma. Technol Cancer Res Treat 2019;18:1–1. doi:10.1177/1533033819874807 CrossRef
41. Farhat RA, Asnir A, Yudhistira, Susilo RR, Daulay ER, Chrestella J. Stem cell oncology: correlation of TNF-α expression to clinical stadium in nasopharyngeal carcinoma (NPC). CRC Press Taylor & Francis Group, Boca Raton, FL, pp 129–32, 2018, vol 4. CrossRef
42. Santos LHS, Ferreira RS, Caffarena, ER. Integrating molecular docking and molecular dynamics simulations. Docking screens for drug discovery. Methods Mol Biol 2019;2053:13-34. doi:10.1007/978-1-4939-9752-7_2 CrossRef