1. INTRODUCTION
Plastic polymers are a large family that belongs to the monomer unit of ethylene (–CH2–CH2–)n and are used in packaging, electronics, and textiles [1]. Therefore, these are recalcitrants that accumulate in the environment and pollute the sediments immensely around the earth. Due to plastic pollution, a sharp public concern has been seen in a massive quantity that poses an ecological threat such as leaching, fragmentation, additive migration due to quickly colonization by the diversity of microorganisms, and termed as plastisphere. Furthermore, the biodegradation of highly durable polymers such as low-density polyethylene (LDPE) is less suitable for microorganisms due to its higher redox potential, requiring more energy to break the C–C bonds. The worldwide utilization of polyethylene rapidly increases by over 320 million tons every year and seems to be doubled by 2034 [2]. The pollution caused by polyethylene includes blocking drains, water, soil, air contamination, sewage lines around the urban areas due to non-degradability and is not beneficial to the topsoil [3].
LDPE is usually unreactive at room temperature, and with the addition of oxidizing agents, they become reactive. In most cases, plastics decompose aerobically, which produces carbon dioxide and water as a product. However, anaerobically, it produces methane, water, and carbon dioxide. The outcomes of oxidative degradation of polyethylene were studied in a controlled condition and found to consist of harmful chemicals that are dangerous for humans and the environment [4]. Therefore, the bioremediation method boosts the degradation rate without damaging the environment. The bioremediation process removes recalcitrants from their origin and eliminates the ecosystem’s pollutants [5]. This process is influenced by many factors such as the number of microorganisms that participated, contact time, nutrient availability, temperature, pH, consortium formulation, and pollutant characteristics. Various researchers explored polyethylene degrading bacteria such as Bacillus spp., Pseudomonas spp., Acinetobacter sp., Brevibacillus sp., Flavobacterium spp., Ralstonia spp., Micrococcus spp., Microbacterium sp., and Nocardia sp. [6]. Enzymes like dehydrogenases and esterases, the AlkB family of hydrolases, initiate the LDPE oxidation process [7]. The laccase enzyme (copper-binding bacterial enzyme) can also degrade LDPE. This enzyme reduces the LDPE polymer’s molecular weight (40,000 Daltons) with a simultaneous increase in a keto-carbonyl index [8]. A well-known example, Pseudomonas aeruginosa E7, releases alkane hydroxylases, which showed 30% of LDPE degradation [9]. The depolymerases enzymes, which are extracellular, break the complex synthetic polymers into monomers and dimers [10]. Thus, the microbes utilize these short-structured oligomers as carbon sources [11]. The PETase enzyme secreted by Ideonella sakaiensis hydrolyzes Polyethylene terephthalate (PET) into mono hydroxyethyl terephthalate, terephthalate, and ethylene glycol, which are further metabolized by bacteria through the pathway of β-oxidation [12]. Bacillus and Brevibacillus sp. secrete enzyme protease, which catalyzes the proteolysis of LDPE [13]. The bacteria Streptomyces viridosporus T7A, Streptomyces badius 252, and Streptomyces setonii 75Vi2 secretes enzymes like monooxygenase, dioxygenase dehydrogenase, which break down complex polymer LDPE [14]. Various techniques such as X-ray diffraction (XRD), scanning electron microscopy (SEM), and Fourier-transform infrared spectroscopy (FTIR) analysis are often used to detect LDPE structure changes [15]. In the present study, the ability to degrade LDPE has been investigated by a bacterium SARR1 isolated from waste disposal sites of Haryana, India. The LDPE degradation was studied using SEM, XRD, FTIR, and gas chromatography-mass spectrometry (GC-MS).
2. MATERIALS AND METHODS
2.1. Sample Collection and LDPE Used
In the current study, LDPE was procured from the local market, Rohtak, Haryana. LDPE thickness was 10 μ, and thin strips were cut into 3 × 3 cm. These LDPE strips were disinfected and washed with 75% ethanol and air-dried. It was used as the sole carbon source in the media for the growth of bacteria. The bacterial culture was isolated from the soil sample collected from the waste disposal site, Ismailabad, Pehowa (Haryana), India. The soils samples were then processed within 24 hours of collection and stored at 4°C.
2.2. Isolation and Cultivation of Bacterial Isolates
The enrichment culture technique was implemented to isolate bacteria that can degrade LDPE. In this technique, 10 g of soil sample was suspended in 90 ml of mineral salt broth media, amended with LDPE strips, and incubated for 30 days at 37°C under static conditions, respectively. After incubation, 100 μl inoculums were spread on agar plates and purified using the streak plate method [16].
2.3. Molecular Characterization and Bacterial Identification
The bacterial strain SARR1 was identified using 16S rRNA gene sequencing. The 16S rRNA gene was amplified using universal primers, i.e., fD1 (5?-AGAGTTTGATCCTGGCTCAG-3?) and rP2 (5?-ACGGCTACCTTGTTACGACTT-3?) [17]. The universal primers were complementary to the conserved regions at the beginning of the gene sequence and either of the whole sequence (1,550 bp). Therefore, the sequence of the variable region was used as comparative taxonomy. The polymerase chain reaction (PCR) composition of 50 μl total volume was 50 ng genomic DNA, primer fD1 and rP2 each (20 pmol), Deoxynucleotide triphosphates (dNTPs) (Sigma) (200 M each), Taq polymerase buffer (10×), and Taq DNA polymerase (Sigma) (2.5 U) and PCR grade water. The PCR reaction was carried out using Thermal Cycler (Applied biosystems model 2720), and the amplification was done for 35 cycles [18]. The PCR amplified product was resolved on the 1.5% agarose gel electrophoresis and then purified using a genetix nucleo-pore PCR clean-up kit as per the manufacturer’s instructions.
The nucleotide sequence of 16S rDNA was done by Biokart India Pvt. Ltd., Bangalore, Karnataka, India. The obtained nucleotide sequences were then analyzed on the NCBI Basic Local Alignment Search Tool (BLAST) tool. A phylogenetic tree of the obtained sequence and the retrieved 16S rRNA sequences was constructed using the neighbor-joining method [19]. MEGA X software was used for the evolutionary analysis [20,21].
2.4. Biodegradation Assay of LDPE and Kinetic Model
The growth kinetics of SARR1 was studied at a regular time interval both with and without LDPE strips. The MS broth with and without LDPE strips was inoculated with SARR1 and incubated at 37°C for 30 days, and Optical Density (OD) was measured at 600 nm [22]. The weight-loss method was used to determine LDPE degradation’s rate constant by the first-order kinetics model [23]. The rate constant for the removal of LDPE was followed through half-life (t½) [24]. The biomass growth kinetics of well-grown isolate SARR1 was studied using the Logistic model equation.
2.5. Enzyme Activity
The cell-free supernatant was used to demonstrate the enzyme activities of Esterase and lipase by UV visible spectrophotometer. The samples were extracted and centrifuged at 3,427 × g at 30°C to derive cell-free supernatant from biodegradation assay at 10, 20, and 30 days [25].
2.5.1. Esterase and lipase enzyme assay
To check the activity of esterase and lipase enzyme, the substrate p-nitrophenyl butyrate and palmitate were used respectively, added in 1 ml acetonitrile, and made up the volume of 100 ml with addition 0.1 M phosphate buffer solution (pH 7.0) [25]. The reaction mixture was prepared by adding 0.8 ml phosphate buffer solution (0.1 M, pH 7.0), 0.066 ml supernatant devoid of cells, and the substrate of 0.134 ml incubated at 30°C for 30 minutes. The sample changed its color and was measured with respect to control as a blank using a UV visible spectrophotometer at 410 nm [26].
2.6. LDPE Degradation Analysis
The evaluation of biodegradation and structural changes of LDPE by the bacterial isolate SARR1 was analyzed using SEM, FTIR, and XRD.
2.6.1 LDPE degradation analysis using SEM
After incubation of 30 days, the treated and untreated LDPE strips were rinsed with 2.0% (v/v) Sodium dodecyl sulphate (SDS), followed by distilled water and 70% (v/v) ethanol. The LDPE strip was coated with gold for the 40 seconds (Ion sputter JFC-1100) and analyzed under a high-resolution SEM (JSM-6100SEM) [27].
2.6.2. LDPE degradation analysis using FTIR
The treated and untreated LDPE strip was rinsed with 70% ethanol and air-dried overnight [28]. The samples were analyzed under the attenuated total reflectance-FTIR spectrophotometer (Model Alpha, Bruker, Germany) for the chemical and structural modifications induced by isolate SARR1 on the LDPE strip scanned from the scan 4,000 to 500 cm−1 at room temperature.
2.6.3. LDPE degradation analysis using XRD
The XRD analysis of LDPE was done using an X-ray diffractometer and operated using Cu (Kα) radiation (λ = 1.5406 Å). The radiation scattered and registered in the angular interval (2θ) was from 0° to 80°. A current of 15 mA and a voltage of 30 kV were used (MiniFlex 2 goniometer). The XRD diffraction peaks were investigated under constant operating conditions on the XRD diffractometer, and the data were recorded. The crystallinity percentage was calculated using the following formula [29].
Ic is the total area of crystalline peaks, and Ic + Ia is the total area of all peaks.
2.6.4. LDPE degradation analysis using GC-MS
The degraded LDPE products were analyzed by GC-MS using a Thermo Trace 1300GC coupled with Thermo TSQ 8000 Triple Quadrupole MS, equipped with BP 5MS (30 m × 0.25 mm, 0.25 μm) column. Helium gas as a carrier and di-ethyl ether as a solvent were used [30]. The LDPE strips were extracted from the inoculated broth culture. The cell culture was centrifuged at 6,093 × g for 5 minutes at room temperature, and the cell debris was removed. The supernatant was filtered by Whatman filter paper, and LDPE products were collected using diethyl ether as a solvent. 10 ml filtrate was then dissolved in an equal volume of diethyl ether and separated using a separating funnel [30]. Finally, 1.0 ml of dissolved LDPE degraded products were injected in GC-MS. The metabolites were identified based on mass spectra and National Institute of Standards and Technology (NIST) library 2.0 [31].
2.7. Phytotoxicity Analysis of LDPE Degraded Products
Seed germination bioassay is the most common method used to evaluate the phytotoxicity of any treated samples [32]. The phytotoxicity tests were performed to assess the toxicity of the treated LDPE sample. The experiment was performed using sterilized Petri plates lined with circular pieces of Whatman no.1 filter paper. The 1.0 ml of the treated and untreated LDPE sample was poured into the Petri plates, respectively. The seeds of Vigna radiata were disinfected with 70% ethanol, soaked for 30 minutes in respective LDPE samples. Afterward, seeds were placed on the moistened filter papers at room temperature for 48 hours [33]. The phytotoxicity of seeds was estimated by comparing the germination of seeds in both treated and untreated samples, respectively [34].
3. RESULTS AND DISCUSSION
3.1. Isolation of LDPE Degrading Bacteria
Bacterial cultures were isolated and screened out based on the weight loss method. An isolate Bacillus licheniformis SARR1 showed 33.32% ± 1.39% biodegradation efficiency at 37°C and pH 7.0. The isolate SARR1 was more capable of consuming and assimilating the LDPE strips as a sole carbon source and thus showed biodegradation in aerobic conditions. However, after utilization, the conversion of the polymeric compound into simple oligomers occurred, and byproducts such as CO2 and H2O also might be released. Researchers have earlier reported various isolates, but in contrast, to present isolate SARR1, the capability of degradation was less in prior reported cultures. Enterobacter cloacae AKS7 sp. was the effective strain for LDPE degradation from an agricultural site inoculated in mineral salt broth medium amended with LDPE strips as a sole carbon source [35].
3.2. 16S rRNA Gene Sequence and Phylogenetic Analysis
3.2.1. 16S rRNA gene sequence analysis
For the identification of potent bacterial isolate SARR1, the 16S rRNA gene sequencing method was used. The amplification of the PCR product of the 16S rRNA gene of the isolate SARR1 was approximately 1,210 bp (Fig. 1). The obtained 16S rRNA nucleotides were then subjected to BLAST analysis for the identification of an isolate. The partial sequence of the 16S rRNA gene was submitted to NCBI with the accession number MT974150. The 16S rRNA gene sequences of isolate SARR1 and the retrieved sequences from the NCBI database and Remote Desktop Pro (RDP) database showed sequence homology to B. licheniformis strain DSM 13 and strain BCRC 11702 with a sequence similarity of 99.92%. The phylogenetic tree of the isolate SARR1 was constructed using the neighbor-joining method with the closest neighbor strain (Bacillus piscis strain 16MFT21 and Bacillus paralicheniformis strain KJ-16), which showed the isolate belonged to B. licheniformis (Figs. 1 and S1). Similarly, LDPE degrading bacterial isolates had earlier been isolated and identified as Pseudomonas citronellolis EMBS027 [36], Microbulbifer hydrolyticus IRE-31 [37], and P. aeruginosa ISJ14 [6].
3.3. Biodegradation Assay of Isolate and Its Growth Kinetic Model
The biodegradation assay of LDPE was studied for 30 days, and percent weight loss was recorded for growth kinetic analysis at a time interval of 5 days of incubation (Fig. 2). The bacterial growth was increased in the MS media amended with LDPE compared with the presence of limited glucose concentration (0.5 g/l) in MS broth media. The altered growth rate was observed in the limited glucose concentration, which its metabolite pool might control compared to environmental conditions. In this way, the strain SARR1 showed the potential to grow and consume LDPE as a sole carbon source. The maximum degradation of LDPE was then evaluated as 33.32% ± 1.39% under optimized conditions. Similarly, the isolate P. aeruginosa ISJ14 degraded only 6.5% LDPE within 2 months of incubation [38] while Brevibacillus borstelensis 707 degraded 2.5% after 1 month of incubation [39]. The Streptomyces sp. growth was observed in the presence of LDPE as a carbon source showed the potential to degrade LDPE [22]. The isolate SARR1 had an LDPE removal rate (K) of 0.069 g day−1 through a longer half-life (t½) approximately required 335.32 days to degrade LDPE strips into its half in contrast to P. aeruginosa ISJ14 that showed a removal rate of LDPE of 0.0012 g day−1 and degrade into its half in 577.5 days [38]. Similarly, Bacillus sp. and Rhodococcus sp. showed the removal rate of microplastic 0.0019 g day−1 with a half-life of 363 days [24].
 | Figure 1: The phylogenetic relationship of isolate SARR1 with the retrieved 16S rRNA sequences from the NCBI BLAST was constructed using the neighbor-joining method. The evolutionary analysis was conducted in MEGA X. [Click here to view] |
The biomass production of strain SARR1 in the LDPE enriched medium was 0.98 ± 0.006 gl−1 (Xm) with a weight loss percentage of 32.15% ± 2.27% after incubation of 30 days. Montazer et al. [40] reported biomass production of 0.15 ± 0.007 gl−1 for Micrococcus luteus IRN20 and 0.37 ± 0.05 gl−1 of biomass production for Cupriavidus necator H16 within 21 days on LDPE containing media.
3.4. Enzymatic Assay During Degradation of LDPE
The production of esterase and lipase enzymes was determined and revealed during the degradation of LDPE by an isolate SARR1. The isolate SARR1 showed the production and activity of esterase and lipase enzymes on different days (Fig. 3a and b). The earlier studies demonstrated that lipase and esterase enzymes were the lipolytic enzyme that belonged to the hydrolases class of enzymes that hydrolyzed fatty acid and glycerol. They also participate in the catabolic activities of hydrocarbon degradation [25]. The action of enzymes connected with respiration and energy production pathways demonstrates the metabolic state of bacteria [26]. Kumar et al. [41] identified that Bacillus sp. was the best producer of lipase and esterase enzyme for LDPE degradation.
 | Figure 2: (a) Biodegradation assay demonstrating weight loss percentage of LDPE at regular intervals of time (b) Growth kinetics of the isolate SARR1 with LDPE and with limited glucose concentration (0.5 g/l). [Click here to view] |
 | Figure 3: Enzyme activity at different days was optimized by UV visible spectroscopy (a) Esterase activity (b) Lipase activity. [Click here to view] |
3.5. Evaluation of Structural Changes of LDPE
3.5.1. LDPE analysis using SEM
The SEM analysis of the surfaces of treated and untreated LDPE strips was done at different resolutions (500× and 2,500×) after 30 days of incubation. The surface erosion cracks, folds, and big holes were seen on the surface of LDPE strips (Fig. 4). Hence, the disintegration of the smooth LDPE surface was observed and recorded. Several researchers have well studied the microbial degradation mechanism of LDPE. The bacteria utilized the oxidized region and solubilized it, leading to pits formation on the surface of LDPE [42,43]. It was depicted that due to enzymatic activity, oxidation of LDPE occurred, which may break the amorphous regions of LDPE. Mukherjee et al. [44] also observed a disintegrated surface of LDPE after treatment with Pseudomonas fluorescens. Microbial attachment on the LDPE surface indicates its strong adherence and LDPE utilization capacities. The cracks and pits on the LDPE surface after treatment with Microbulbifer hydrolyticus IRE-31 and Streptomyces albogriseolus LBX-2 confirmed LDPE degradation [37].
3.5.2. LDPE analysis using FTIR
As the LDPE consists of ethylene groups, the major infrared (IR) band has –CH2– stretching and bending vibrations. The change in the peak values and functional groups supported the conformational change of the polymer surface. The IR spectra of the treated LDPE surfaces showed peaks at 2,845.08, 1,744.36–1,463.13, and 718.84–626.59 cm–1 for C–H deformation vibration CH2 scissoring vibration, –CHO stretching, and strong alkyl structure, respectively. The strong bond pattern was observed at 2,913.98, which showed C–H stretching vibration asymmetrically (Fig. 5). The new peaks with a marked shift at 1,500–1,540 cm−1 (C=C stretching vibration, O–H stretching: carboxylic acid) and 1,000–1,100 cm–1 (–CHO stretching, C=O stretching) was observed in the IR spectra of LDPE degradation. The disappearance of the peaks in the range of 2,000 to 2,500 cm−1 was also observed and depicted that the N–H and C–H vibrations represented the amide group addition. This proved that bacteria are capable of utilizing long, hydrophobic polyethylene chains. The formation of new peaks at 2,236.00–2,269.00 cm–1 (O–H stretching: carboxylic acid), 1,555.44 cm−1 (C=C stretching vibration), 788.01–788.86 cm−1 (C–H rocking vibration: CH3) on IR spectra of degraded LDPE were also observed by Kunlere et al. [43]. Similarly, the peak at 2,660 cm−1 (-CHO stretch) was disappeared in Bacillus amyloliquefaciens treated LDPE, while the new peak was demonstrated at 939 cm−1 (O–H bend) which showed LDPE de-polymerization [45].
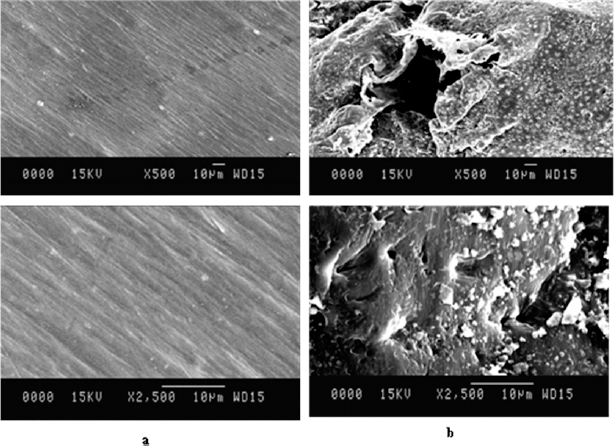 | Figure 4: SEM analysis of LDPE biodegradation at different resolutions (500×, 2,500×) (a) control sample (b) Different resolutions of biodegraded LDPE treated with isolate SARR1 revealing surface changes. [Click here to view] |
3.5.3. LDPE analysis using XRD
The XRD analysis showed that the distinguished highest peak in the control and treated sample chromatogram was observed at 21.7 of the angular position 2θ (Fig. 6a and b). The new peaks were observed in the range of 20–30 of angular position 2θ. After degradation, the minor shift in the peak’s position at 20.7, 20.92, 21.8, and 22.62 was seen. The crystallinity percentage was decreased from 71.69% (control) to 50.78% in the treated sample within 30 days of incubation (Fig. 6c). The amorphous region of LDPE was found disintegrated in the treated sample. Bacillus siamensis and Bacillus wiedmannii decreased the crystallinity percentage of LDPE after 90 days of incubation [46]. The crystallinity of LDPE was decreased by treating with the mixed cultures of Aspergillus sp. and Lysinibacillus sp. [29], Vibrio sp., and Aspergillus sp. [16].
3.5.4. LDPE analysis using GC-MS
The GC-MS analysis showed that compounds like acetone, diazene, dimethyl, and carbamimidothioic acid, 1-methyl ethyl ester were obtained with peak area percentages in the treated sample 23.38%, 65.58%, and 11.04%, respectively (Table S1, Fig. S2). A new peak of Carbamimidothioic acid, 1-methylethyl ester appeared in the treated LDPE while the compounds like acetone, 1,2,4,5-tetroxane, 3,3,6,6-tetramethyl, 1,4-benzenedicarboxylic acid, bis(2ethylhexyl)ester, 1,3-benzenedicarboxylic acid, bis(2ethylhexyl) ester, isophthalic acid, 2-ethylhexyl octyl ester, terephthalic acid, 4 octyl ester, terephthalic acid, and 2-Ethylhexyl octyl ester were obtained with peak area percentages of 3.11%, 6.11%, 1.05%, 1.13%, 1.25%, 1.73%, 2.58%, 2.12%, 25.58%, and 55.35%, respectively in the control sample (Table S1, Fig. S3). The compound Carbamimidothioic acid, 1-methylethyl ester, can act as a potent anti-cancer agent [47]. After further degradation of carboxylic acid by an isolate, SARR1 might produce alkane compounds such as butane, 1,2,4,5-tetroxane, and methane, shown in the chromatogram of the treated sample. The production of such compounds was confirmed by observing the IR spectra obtained after the degradation of LDPE. Similarly, new peaks with retention times 7.46 and 17.28 of 2-butene, 2-methyl, and ethane were earlier observed in Acinetobacter baumannii treated LDPE [48]. However, P. aeruginosa treated LDPE film showed a long chain of esters, hydrocarbons, aldehydes, oxygenated chemical compounds, ketones, fatty acids, ether groups, and unknown compounds by gas chromatography [30]. On the other hand, the Lysinibacillus fusiformis strain VASB14/WL treated LDPE film produced 1- trimethylsilyl methanol, hexadecanoic acid, and 1,2,3,4- tetra methylbenzene, whereas Bacillus cereus strain VASB1/TS resulted in the formation of 1-trimethylsilyl methanol, 1,2,3-trimethyl benzene, and hexadecanoic acid [49].
 | Figure 5: FTIR spectra of LDPE biodegradation after 30 days of the incubation period (a) Control sample (b) LDPE sample treated with isolate SARR1. [Click here to view] |
3.6. Phytotoxicity Analysis of LDPE Degraded Products
The primary aim was to assess the LDPE degradation’s phytotoxicity and metabolites after degradation by bacterial isolate SARR1. The phytotoxicity analysis showed that the seed germination in V. radiata was high (100% ± 0%) when irrigated with SARR1 treated LDPE samples compared to untreated (44.49% ± 2.47%). Similarly, the germination speed was also quite high in the treated LDPE samples (Fig. 7). So, it indicated that the phytotoxicity of the treated samples was reduced and showed no inhibition on seed germination, and hence became eco-friendly. Therefore, the isolate B. licheniformis SARR1 showed a positive effect on the germination of V. radiata. Pathak and Kumar [50] also reported a positive effect on the seed germination, shoot height, and root length of Vigna mungo when irrigated with Bacillus sp. V8 and Pseudomonas sp. C25 treated LDPE samples. The phytotoxicity standard tests have also been performed to determine the toxicity of polyethylene on Lepidium sativum, which resulted in a potential threat to the growth of plants [51]. The pollutant causes oxidative stress, and as a result, plants produce a high concentration of reactive oxygen species in their tissues and ultimately leads to cell damage [52].
 | Figure 6: XRD spectra of the LDPE films after incubation of 30 days (a) control (b) treated with isolate SARR1 (c) % Crystallinity. [Click here to view] |
 | Figure 7: Phytotoxicity analysis of LDPE on seed germination of V. radiata (a) control (b) treated sample (c) germinated speed and % germination of seeds. [Click here to view] |
4. CONCLUSION
Plastic waste and polyethylene are generally major ecological threats to the environment due to their non-biodegradable behavior. Therefore, significant attention has been placed on the biodegradation of polymers and the identifications of microorganisms with great potential. The current investigations demonstrated that the bacterium SARR1 develops substantially enhanced biodegradation over the surface of LDPE by exploiting the hydrophobicity of the cell surface. The biodegradation efficiency (%) of the strain SARR1 was 33.32% ± 1.39% at temp. 37°C and pH 7.0. The biomass production by isolate SARR1 in the LDPE enriched medium was 0.98 ± 0.006 gl−1 (Xm) after incubation of 30 days, and the LDPE removal rate (K) was 0.069 g day−1 with approximately 335.32 days of a half-life to degrade LDPE strips.
Moreover, the GC-MS analysis showed a new compound, carbamimidothioic acid, 1-methylethyl ester, formed during the degradation process, which has been reported as anticancer property. The SEM analysis of treated and untreated LDPE samples showed clear degradability of LDPE. Enzymatic study reveals that the esterase and lipase enzymes are supposed to be a part of LDPE degradation. The degraded products released from SARR1 treated LDPE showed no phytotoxic effect and are hence eco-friendly. This study would enable to develop more efficient microbial consortium having LDPE degradation capability. Moreover, to understand the mechanism of LDPE biodegradation, investigations towards the metabolic pathways and their enzymatic reactions are needed.
5. ACKNOWLEDGMENT
The grant received from DST-SERB (EEQ/2018/000892) to ARS is highly acknowledged. In addition, RR acknowledges Maharshi Dayanand University, Rohtak, India, for University Research Scholarship.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
REFERENCES
1. Sridharan R, Vetriselvan M, Krishnaswamy VG, Rishin H. Integrated approach in LDPE degradation–an application using Winogradsky column, computational modeling, and pathway prediction. J Hazard Mater 2021;412:125336. CrossRef
2. El-Sayed MT, Rabie GH, Hamed EA. Biodegradation of low-density polyethylene (LDPE) using the mixed culture of Aspergillus carbonarius and A. fumigates. Environ Dev Sustainability 2021;23:1–29. CrossRef
3. Dey A, Dhumal CV, Sengupta P, Kumar A, Pramanik NK, Alam T. Challenges and possible solutions to mitigate the problems of single-use plastics used for packaging food items: a review. J Food Sci Technol 2020;58:1–19. CrossRef
4. Sojak L, Kubinec R, Jurdakova H, Hájeková E, Bajus M. GC-MS of polyethylene and polypropylene thermal cracking products. Petrol Coal 2006;48(1):1–14.
5. Lee DH, Kim Y, Fearing RS, Maboudian R. Effect of fiber geometry on macroscale friction of ordered low-density polyethylene nanofiber arrays. Langmuir 2011;27(17):11008–16. CrossRef
6. Gupta KK, Devi D. Characteristics investigation on biofilm formation and biodegradation activities of Pseudomonas aeruginosa strain ISJ14 colonizing low density polyethylene (LDPE) surface. Heliyon 2020;6(7):e04398. CrossRef
7. Yoon MG, Jeon HJ, Kim MN. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J Biorem Biodegrad 2012;3(4):1–8.
8. Montazer Z, Najafi MBH, Levin DB. Challenges with verifying microbial degradation of polyethylene. Polymers 2020;12:123. CrossRef
9. Jeon HJ, Kim MN. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int Biodeterior Biodegrad 2015;103:141–6. CrossRef
10. Ghosh S, Qureshi A, Purohit HJ. Microbial degradation of plastics: biofilms and degradation pathways. Contaminants in agriculture and environment: health risks and remediation, Agriculture and Environmental Science Academy, Haridwar, India. vol. 1, pp 184–99, 2019. CrossRef
11. Koutny M, Lemaire J, Delort AM. Biodegradation of polyethylene films with prooxidant additives. Chemosphere 2006;64(8):1243–52. CrossRef
12. Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, et al. Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation. Nat Commun 2018;9(1):1–12. CrossRef
13. Deep A, Bhatt D, Shrivastav V, Bhardwaj SK, Malik P. Synthesis, characterization and applications of polyolefin based eco-friendly polymer composites. In: Sustainable polymer composites and nanocomposites, pp 65–103, 2019. CrossRef
14. Pathak VM, Navneet. Review on the current status of polymer degradation: a microbial approach. Bioresour Bioprocess 2017; 4:1–31. CrossRef
15. Sangale MK, Shahnawaz M, Ade AB. Gas chromatography-mass spectra analysis and deleterious potential of fungal based polythene-degradation products. Sci Rep 2019;9:1–6. CrossRef
16. Sarkhel R, Sengupta S, Das P, Bhowal A. Comparative biodegradation study of polymer from plastic bottle waste using novel isolated bacteria and fungi from marine source. J Polym Res 2019;27(1):1–8. CrossRef
17. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991;173:697–703. CrossRef
18. Santal AR, Singh NP, Saharan BS. Biodegradation and detoxification of melanoidin from distillery effluent using an aerobic bacterial strain SAG 5 of Alcaligenes faecalis. J Hazard Mater 2011;193:319–24. CrossRef
19. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406–25.
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547–9. CrossRef
21. Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 2017;67:1613–7. CrossRef
22. Abraham J, Ghosh E, Mukherjee P, Gajendiran A. Microbial degradation of low density polyethylene. Environ Progress Sustainable Energy 2017;36:147–54. CrossRef
23. Alaribe FO, Agamuthu P. Assessment of phytoremediation potentials of Lantana camara in Pb impacted soil with organic waste additives. Ecol Eng 2015;83:513–20. CrossRef
24. Auta HS, Emenike CU, Jayanthi B, Fauziah SH. Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull 2018;127:15–21. CrossRef
25. Kay MJ, McCabe RW, Morton LHG. Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int Biodeterior Biodegrad 1993;31(3):209–25. CrossRef
26. Khandare SD, Chaudhary DR, Jha B. Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic. Biodegradation 2021;32:1–17. CrossRef
27. Al-Salem SM, Behbehani MH, Al-Hazza’a A, Arnold JC, Alston SM, Al-Rowaih AA, et al. Study of the degradation profile for virgin linear low-density polyethylene (LLDPE) and polyolefin (PO) plastic waste blends. J Mater Cycles Waste Manage 2019;21:1106–22. CrossRef
28. Novotný ?, Malachová K, Adamus G, Kwiecie? M, Lotti N, Soccio M, et al. Deterioration of irradiation/high-temperature pretreated, linear low-density polyethylene (LLDPE) by Bacillus amyloliquefaciens. Int Biodeterior Biodegrad 2018;132:259–67. CrossRef
29. Esmaeili A, Pourbabaee AA, Alikhani HA, Shabani F, Esmaeili E. Biodegradation of low-density polyethylene (LDPE) by mixed culture of Lysinibacillus xylanilyticus and Aspergillus niger in soil. PLoS One 2013;8:e71720. CrossRef
30. Kyaw BM, Champakalakshmi R, Sakharkar MK, Lim CS, Sakharkar KR. Biodegradation of low density polythene (LDPE) by Pseudomonas species. Indian J Microbiol 2012;52:411–9. CrossRef
31. Shahnawaz M, Sangale MK, Ade AB. Bacteria-based polythene degradation products: GC-MS analysis and toxicity testing. Environ Sci Pollut Res 2016;23:10733–41. CrossRef
32. Singh S, Tripathi DK, Dubey NK, Chauhan DK. Effects of nano-materials on seed germination and seedling growth: striking the slight balance between the concepts and controversies. Mater Focus 2016;5:195–201. CrossRef
33. Santal AR, Singh NP, Saharan BS. A novel application of Paracoccus pantotrophus for the decolorization of melanoidins from distillery effluent under static conditions. J Environ Manage 2016;169:78–83. CrossRef
34. Kebrom TH, Woldesenbet S, Bayabil HK, Garcia M, Gao M, Ampim P, et al. Evaluation of phytotoxicity of three organic amendments to collard greens using the seed germination bioassay. Environ Sci Pollut Res 2019;26(6):5454–62. CrossRef
35. Sarker RK, Chakraborty P, Paul P, Chatterjee A, Tribedi P. Degradation of low-density poly ethylene (LDPE) by Enterobacter cloacae AKS7: a potential step towards sustainable environmental remediation. Arch Microbiol 2020;202:2117–25. CrossRef
36. Bhatia M, Girdhar A, Tiwari A, Nayarisseri A. Implications of a novel Pseudomonas species on low density polyethylene biodegradation: an in vitro to in silico approach. SpringerPlus 2014;3(1):1–10. CrossRef
37. Li Z, Wei R, Gao M, Ren Y, Yu B, Nie K, et al. Biodegradation of low-density polyethylene by Microbulbifer hydrolyticus {IRE}-31. J Environ Manage 2020;263:110402. CrossRef
38. Amodu OS, Ojumu TV, Ntwampe SKO. Kinetic modelling of cell growth, substrate utilization, and biosurfactant production from solid agrowaste (Beta vulgaris) by Bacillus licheniformis STK 01. Can J Chem Eng 2016;94:2268–75. CrossRef
39. Hadad D, Geresh S, Sivan A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J Appl Microbiol 2005;98:1093–100. CrossRef
40. Montazer Z, Habibi Najafi MB, Levin DB. Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can J Microbiol 2019;65:224–34. CrossRef
41. Kumar D, Kumar L, Nagar S, Raina C, Parshad R, Gupta VK. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch Appl Sci Res 2012;4(4):1763–70.
42. Montazer Z, Habibi-Najafi MB, Mohebbi M, Oromiehei A. Microbial degradation of UV-pretreated low-density polyethylene films by novel polyethylene-degrading bacteria isolated from plastic-dump soil. J Polym Environ 2018;26:3613–25. CrossRef
43. Kunlere IO, Fagade OE, Nwadike BI. Biodegradation of low density polyethylene (LDPE) by certain indigenous bacteria and fungi. Int J Environ Stud 2019;76:428–40. CrossRef
44. Mukherjee S, RoyChaudhuri U, Kundu PP. Biodegradation of polyethylene via complete solubilization by the action of Pseudomonas fluorescens, biosurfactant produced by Bacillus licheniformis and anionic surfactant. J Chem Technol Biotechnol 2017;93:1300–11. CrossRef
45. Das MP, Kumar S. An approach to low-density polyethylene biodegradation by Bacillus amyloliquefaciens. 3 Biotech 2015;5:81–6. CrossRef
46. Maroof L, Khan I, Yoo HS, Kim S, Park HT, Ahmad B, et al. Identification and characterization of low density polyethylene-degrading bacteria isolated from soils of waste disposal sites. Environ Eng Res 2021;26(3):109–19. CrossRef
47. Abdel-Kader MS, Ghorab MM, Alsaid MS, Alqasoumi SI. Design, synthesis, and anticancer evaluation of some novel thiourea, carbamimidothioic acid, oxazole, oxazolidine, and 2-amino-1-phenylpropyl-2-chloroacetate derived from L-norephedrine. Russ J Bioorg Chem 2016;42:434–40. CrossRef
48. Pramila R. Brevibacillus parabrevis, Acinetobacter baumannii and Pseudomonas citronellolis – potential candidates for biodegradation of low density polyethylene (LDPE). J Bacteriol Res 2012;4(1):9–14. CrossRef
49. Shahnawaz M, Sangale MK, Ade AB. Analysis of the plastic degradation products. Bioremediation technology for plastic waste, Springer Nature, Switzerland, AG. pp 93–101, 2019. CrossRef
50. Pathak VM, Kumar N. Implications of SiO 2 nanoparticles for in vitro biodegradation of low-density polyethylene with potential isolates of Bacillus Pseudomonas, and their synergistic effect on Vigna mungo growth. Energy Ecol Environ 2017;2:418–27. CrossRef
51. Balestri E, Menicagli V, Ligorini V, Fulignati S, Raspolli Galletti AM, et al. Phytotoxicity assessment of conventional and biodegradable plastic bags using seed germination test. Ecol Indic 2019;102:569–80. CrossRef
52. Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci 2004;9:490–8. CrossRef
SUPPLEMENTARY MATERIAL
 | Table S1: Peak report of GC-MS in control sample and in treated LDPE with SARR1. Peak report of GC-MS in control sample [Click here to view] |
 | Figure S1: The phylogenetic relationship of isolate SARR1 with the retrieved 16S rRNA sequences from the RDP database was constructed using the neighbor-joining method. Evolutionary analyses were conducted in MEGA X. [Click here to view] |
 | Figure S2: GC-MS analysis of LDPE film (a) LDPE film before treatment with the isolate SARR1 as control (b) LDPE film treated with isolate SARR1. [Click here to view] |
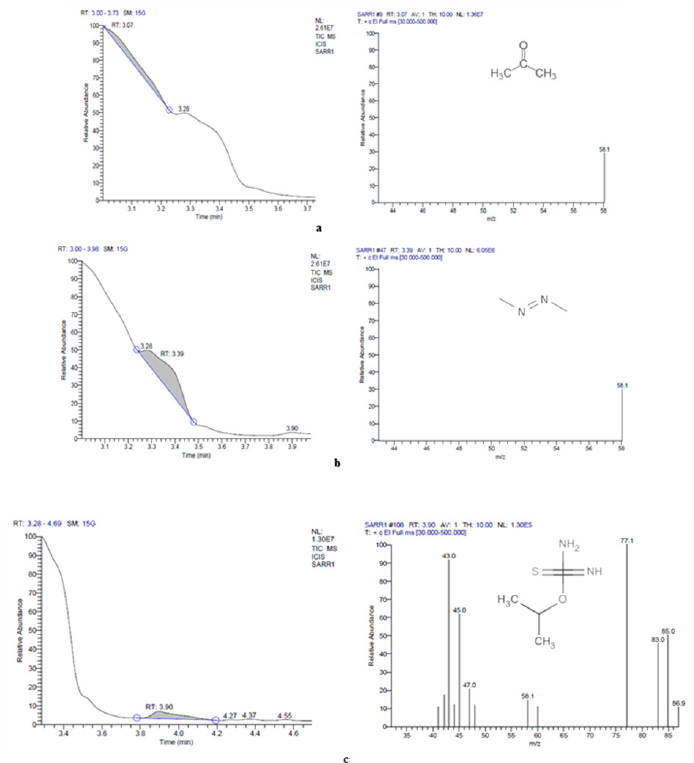 | Figure S3: GC-MS chromatogram of LDPE-degraded compounds synthesized during treatment with isolate SARR1 (a) Acetone (b) Diazene, dimethyl (c) Carbamimidothioic acid, 1-methylethyl ester. [Click here to view] |