1. INTRODUCTION
Cyamopsis tetragonoloba is an edible crop of annual legume plants belonging to the family of Fabaceae. It is cultivated in different regions of India like Rajasthan, MP, Haryana, and Punjab. It is an excellent source of guar gum and has high protein content [1,2]. It is used in cosmetics, textiles, paper industry, drilling, exploration mining, petroleum industry, and beverages [3]. It is known for its drought and high-temperature tolerance ability due to its deep roots [4].
Proteases of the tropical plants show high stability toward high temperature compared with proteases from many other plants since they tend to adapt and cope with differences in environmental temperature that is pivotal for survival of plants [5,6]. Higher temperature tends to denature enzymes by breaking hydrogen bonds [7].
Plants represent an excellent source of enzymes due to various characteristics. The leguminous plant seeds contain a high amount of protein. Legumes accumulate a high level of protease and protease inhibitors that regulate the overburden of protein [8]. Proteases are involved in the physiological process of plants including germination, plant growth, development, chloroplast synthesis, ubiqutination of misfolded protein, and programmed cell death [9–12].
Protease contributes 65% of the world enzyme market to the annual sale of enzymes [13]. Plant proteases are also currently used as therapeutic enzymes in wound healing, treatment of cancer, digestion disorder, infection, and food industry [14,15]. Plant proteases are also used in feather processing, bioremediation, and biotransformation.
This study was carried out to purify proteases from seeds of C. tetragonoloba. Because of its thermostable nature, purified proteases from C. tetragonoloba may have potential industrial applications.
2. METHODOLOGY
2.1. PMaterials and Methods
Cyamopsis tetragonoloba seeds were procured from the local market in Indore (India), Gulmohar Company. Ammonium sulfate, sodium dodecyl sulphate (SDS), was purchased from SRL. Ethylenediaminetetraacetic acid (EDTA) and ethylene glycol bis(2-aminoethyl ether)-N,N,N,N-tetraacetic acid (EGTA) were purchased from HiMedia. Phenyl Methyl Sulfonyl Fluoride (PMSF), l-trans-epoxysuccinyl olamide(4-guanidino)butane (E-64), pepstatin A, soybean trypsin inhibitor (SBTI), Iodoacetamide (IAA), aprotinin, N-αBenzoyl-DL-arginine ?-nitroanilide (BAPNA), N-succinyl-Phe-p-nitroanilide, bovine serum albumin (BSA), benzamidine HCl, casein, gelatin, Diethylaminoethyl (DEAE)-cellulose 52, and prestained markers were purchased from Sigma-Aldrich. N,N,N,N-Tetramethylethylenediamine (TEMED), coomassie brilliant blue (CBB), Trichloroacetic acid (TCA) sulfuric acid, β-mercaptoethanol (β-ME), bromophenol blue, acrylamide, N, N-methylenebisacrylamide, phenol, Dimethyl sulfoxide (DMSO), isopropanol, ethanol, methanol, butanol, Triton X-100, and glacial acetic acid used were of analytical grade.
2.2. Preparation of Seed Extract
Cyamopsis tetragonoloba seeds were washed with distilled water to remove coated copper sulfate and inhibitors. Seeds were soaked in a 10 mM phosphate buffer of pH 7.5 for 48 hours. Homogenates were prepared in the same buffer using a homogenizer (Remi auto mix blender) at low speed for 5 minutes. The homogenates were stirred for proper mixing for 1 hour followed by centrifugation at 10,000 rpm for 30 minutes for the removal of insoluble particles. The supernatants were heated at 80°C for 20 minutes and then centrifuged at 10,000 rpm for 30 minutes. The supernatant was collected and stored at −20°C for further use for the purification of protease.
2.3. Purification of the Protease
A protease was purified from the seed of C. tetragonoloba to homogeneity, using ammonium sulfate precipitate followed by DEAE-cellulose column chromatography. The heated supernatant was first subjected to 20% ammonium sulfate precipitation; then gradually 40%, 60%, and 80% ammonium sulfate was added and allowed to mix thoroughly for 1 hour, followed by centrifugation at 10,000 rpm for 30 minutes for the removal of insoluble material. The 60% ammonium sulfate precipitate was resuspended in a minimum amount of 10 mM sodium phosphate of pH 7.5 and dialyzed in the same buffer. The dialyzed suspension was loaded on DEAE-cellulose columns preincubated with the same buffer. The unbound protein was washed with a sodium phosphate buffer until no unbound protein was eluted. The bound proteins were eluted with 60 ml each of 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 M NaCl in the sodium phosphate buffer. About 3 ml fractions of each molar (M) concentration were collected at the flow rate of 2 ml per minute. The total protein content and proteolytic activity of all the fractions were assayed.
2.4. Protein Concentration
Protein concentration was monitored at every stage of purification according to the method of Lowry using BSA as a standard, as well as by absorbance at 280 nm [16].
2.5. Protease Activity Assay
The proteolytic activity of protease was assayed using natural substrate gelatin, hemoglobin (Hb), and casein. The protease activity was monitored using 1% casein as a substrate with slight modifications of the method described by Kembhavi et al. [17]. The proteolytic reaction mixture contained protease 10 μg in total volume, 500 μl of 10 mM phosphate buffer of pH 7.5 and 500 μl of 1% casein solution were added and incubated at 37°C for 30 minutes. To stop the reaction, 1 ml of 10% (w/v) TCA was added to the reaction mixture and kept for 10 minutes at room temperature. The reaction mixture was centrifuged at 10,000 rpm for 10 minutes. To 1 ml of supernatant, 3 ml sodium carbonate solution and 1 ml of Folin–Ciocalteu were added and mixed thoroughly. The color development after 30 minutes of incubation at 37°C was measured at 660 nm in the Shimadzu UV-visible spectrometer. Protease activity with the Hb, gelatin, and BSA was measured in the same way as 1% casein. The unit of protease activity was defined as the amount of enzyme that yielded a 1 μmol tyrosine per minute at 37°C [18]. The amount of tyrosine released was calculated from the tyrosine standard curve. The specific activity of protease was expressed in Unit/mg.
2.5.1. Calculation
Unit/ml = (μmole tyrosine equivalent released × total vol.) / (protease vol. × Time × colorimetric Assay vol.)
2.6. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Protein profiles of the eluted sample were analyzed by SDS-PAGE according the method of Laemmli [19], and the sample isolated from ion exchange was run at 14% polyacrylamide gel under reducing and non-reducing conditions. SDS-PAGE of purified protease was carried out using 5% stacking gel and 14% polyacrylamide resolving gel. The gel was run using Mini prot (Bio-Rad, CA). Prestained molecular weight markers were used to estimate the molecular weight of purified protease. Sample protease was dissolved in solution of 60 mM Tris-HCl pH.6.8, 14.4 mM β-ME, 2% SDS, 2% glycerol (v/v), and 0.01% (w/v) bromophenol blue and quickly heated at 100°C for 10 minutes. SDS-PAGE was carried out at constant 100 V for 2 hours.
Then the gel was stained with CBB R-250 [20]. The gel was destained with methanol: acetic acid: distilled water in the ratio of 3:1:6.
2.7. Zymography
Zymography was carried out to detect protease activity on a gelatin polyacrylamide gel. 1% gelatin was added in 14% polyacrylamide gel [21]. Zymogram gel was used in the electrophoretic unit (Bio-Rad). The electrophoretic tank was filled with a running buffer. Sample buffer contained 62.5 mM Tris-HCl of pH 6.8, 2% SDS, 25% glycerol (v/v), and 0.01% (w/v) bromophenol blue. The sample was loaded and the gel ran at 100 V for 2 hours.
The gel was removed from the apparatus and incubated at 37°C for 30 minutes in 100 ml of renaturation buffer (2.5% Triton X-100, 50 mM Tris-HCl of pH 7.5, 1 μM ZnCl2, and 5 mM CaCl2) on an orbital shaker. The renaturation buffer was removed and soaked in a development buffer (1% Triton X-100, 50 mM Tris-HCl2, and 5 mM CaCl2) overnight. The development buffer was washed off with water. The gel was stained with 0.5% CBB R-250. The gel was destained with methanol: acetic acid: distilled water (3:1:6) until the clear band appeared against the dark blue background.
2.8. Effects of Substrate Concentration on the Rate of Reaction
The effect of the increasing concentration of substrate on the rate of reaction was monitored by employing casein as substrate in 0.05 mM sodium phosphate buffer of pH 7.5 at 37°C. The effect of substrate concentration was studied with casein in the range of 1–500 μM and 10 μg of protease.
2.9. Effects of pH on the Protease Activity
The effect of the pH of the buffer on the purified protease activity was examined to determine the optimum pH of the protease. The proteolytic activity of protease was examined in the pH range from 2.5 to 12.5 to determine the optimum pH. The buffers used were 0.05 M glycine-HCl (pH 2.5), 0.05 M sodium citrate (pH 4.5), 0.05 M sodium phosphate (pH 6.5), 0.05 M sodium phosphate (pH 7.5), 0.05 M Tris-HCl (pH 9.5), 0.05 M Tris-Base (pH 10.5), and 0.05 M glycine-NaOH (pH 12.5). Protease activity was monitored with BAPNA at 410 nm and each assay was carried out in triplicate.
2.10. Effects of pH on the Protease Stability
The effect of the pH of the buffer on the purified protease stability was examined. The proteolytic activity of protease was examined in the pH range from 2.5 to 12.5 to determine stability. The buffers used were 0.05 M glycine-HCl (pH 2.5), 0.05 M sodium-citrate (pH 4.5), 0.05 M sodium phosphate (pH 6.5), 0.05 M sodium phosphate (pH 7.5), 0.05 M Tris–HCl (pH 9.5), 0.05 M Tris-Base (pH 10.5), and 0.05M glycine-NaOH (pH 12.5). Protease activity was monitored with BAPNA at 410 nm, and each assay was carried out in triplicate. For analyzing the pH stability, the protease sample was incubated before the assay with the respective buffer for up to 1 hour. The reaction was initiated by adding a substrate at 37°C in the same buffer.
2.11. Effects of Temperature on Proteases Activity
The effect of temperature on the purified protease activity was examined to determine the optimum temperature of the protease. The protease activity was examined at different temperatures ranging from 20°C to 100°C with the 10 mM buffer. Protease activity was monitored with BAPNA at 410 nm, and each assay was carried out in triplicate. The highest activity of the protease was considered 100%.
2.12. Effects of Temperature on Proteases Stability
The effect of temperature on the purified protease stability was examined. For studying the heat stability, the protease sample was incubated before the assay at different temperatures ranging from 20°C to 100°C for up to 1 hour. The reaction was initiated by adding a substrate at 37°C. Protease activity was monitored with BAPNA at 410 nm, and each assay was carried out in triplicate.
2.13. Effects of Various Protease Inhibitors on Protease Activity
The effects of protease inhibitors on protease activity were examined. The protease inhibitors used were PMSF, SBTI, EGTA, EDTA, pepstatin A, IAA, aprotinin, and E-64 at different concentrations from 10 to 100 μm. The proteases were incubated for 20 minutes before the assay with the different protease inhibitors. The reaction was initiated by adding a substrate at 37°C. The control activity is considered as 100% that is devoid of protease inhibitors. The protease activity was measured using BAPNA at 410 nm. The assay was carried out in a similar way to the above mentioned method.
2.14. Effects of Various Solvents and Surfactants on Protease Stability
The stability of purified protease was monitored in the presence of a solvent and surfactants. The purified protease was mixed with different concentrations of the solvent. The mixture was incubated for 24 hours with constant mixing on an orbital shaker. The reaction was initiated by adding a substrate at 37°C. The control activity is considered as 100% that is devoid of solvents or surfactants. The results were expressed as percent residual activity.
2.15. Assays for Amidolytic Activity toward the Synthetic Substrate
The amidolytic activity of the synthetic substrate by the purified protease was determined by the method of a microtiter plate [22]. BAPNA and N-succinyl-Phe-p-nitroanilide were used as substrates. Substrate concentrations in the range of 0.01–1.2 mM were prepared in DMSO. The final reaction mixture consists of 20 μg of protease in 170 μl of 100 mM phosphate buffer of pH 7.5 and 30 μl substrate of 1 mM. The reaction mixture was incubated at 25°C for 20 minutes. The amount of p-nitroanilide released was measured by absorbance change at 410 nm (extinction coefficient = 8.8 ml μmol-1 cm−1). The unit of protease activity was defined as the amount of protease that yielded a 1 μmol p-Nitroanilide per minute at 37°C.
Unit/ml = [?Abs × Reaction Volume (ml)]/ [EC × Time ×Protease volume (ml)]
where ?Abs = increase in absorbance and EC = extinction coefficient.
2.16. Sugar Determination
The sugar content of protease was determined according to the method of DuBois [23]. First, 1 ml of 5% phenol solution was added to 1 ml of protease solution, followed by the addition of 4 ml concentrated sulfuric acid. The solution was allowed to cool at room temperature then the color intensity was measured at 490 nm against a blank of BSA. A standard curve was obtained with dextrose. The sugar content of protease was calculated from the standard curve.
3. RESULTS AND DISCUSSION
3.1. Purification of Protease
Ammonium sulfate precipitation was carried out as the initial step for purification according to the method of Englard [24]. The heated supernatant was subjected to 20%, 40%, 60%, and 80% ammonium sulfate precipitation. The 60% precipitate showed protease purification of 6.31-fold. The bound protease was eluted at 0.3M NaCl with 27.61-fold purification (Table 1).
3.2. SDS-PAGE and Zymography
Protease was purified from the seeds of C. tetragonoloba to homogeneity. The single band on SDS-PAGE showed that protease was monomer (Fig. 1). The molecular weight of the protease was estimated at 69.9 kDa by plotting the standard curve of standard molecular weight versus relative mobility (Fig. 2). The molecular weight of the serine protease isolated from the plant’s source falls between 19 and 120 kDa. Most of the plant serine protease falls between 60 and 80 kDa [25]. The molecular weight of the purified protease was similar to that of best characterize plant serine protease crinumin [26]. The molecular weight is similar to plant serine protease from Cucumis trigonus and Taraxcum officinale is 67 kDa [27,28]. The molecular weight is also similar to coconut endosperm serine protease 68 kDa [29].
The proteolytic activity of protease was also confirmed by gelatin zymography in which gelatin was degraded by protease and a clear band appeared against the dark blue background.
3.3. Rate of Reaction
The effect of increasing substrate concentration on the velocity of the enzyme-catalyzed reaction was studied using casein as the substrate at pH 7.5 at 37°C. The effect of increasing substrate concentration on the velocity of the enzyme obeys the Michaelis–Menten equation. The Lineweaver–Burk plot was plotted with 10 μg of protease and the substrate was in the range of 1–500 μM (Fig. 3). The value of Km was 56.56 μM/minute (Fig. 4). Wrighten protease isolated from Wrighten tinctoria showed that Km was 50 μM/minute.
3.4. Effects of pH on the Protease Activity
The purified protease from C. tetragonoloba was active under a wide range of pH. There was an increase in protease activity from pH 6.5 up to 7.5, and then it gradually decreased when the pH was increased above the optimum pH. Therefore, the results showed that the optimum pH for the protease was 7.5 (Fig. 5). Wrighten protease purified from W. tinctoria showed maximum activity at pH 7.5 [29].
 | Figure 1: (A) Zymography of purified protease. Clear band of CBB of 1% gelatin zymography of 10 μg of protease showing proteolytic activity under non-denaturing condition. (B) SDS-PAGE of purified protease. Lane 1 crude extract and Lane 2 prestained marker of different molecular weight from 11 to 250 kDa. (C) SDS-PAGE. Lane 1 crude extract, Lane 2 purified protease, and Lane 3 molecular marker. [Click here to view] |
 | Figure 2: Estimation of molecular weight of purified protease by SDS-PAGE. A standard curve of the log of molecular weight versus relative mobility was plotted. The relative mobility value calculated from the graph is 0.18644. [Click here to view] |
 | Figure 3: The effect of the increasing concentration of the substrate on the rate of reaction was monitored by employing casein as the substrate in the range of 1–500 μM and 10 μg of protease. [Click here to view] |
 | Figure 4: Effect of the substrate concentration on enzyme kinetics. Lineweaver–Burk plots were plotted to determine protease affinity toward the substrate. [Click here to view] |
 | Figure 5: Effects of pH on the protease activity of C. tetragonoloba. 15 ug of protease was used to determine protease activity. The effect of pH was determined at 37°C in different buffers of different pH values (2.5–12.5). The activity of the protease at pH 7.5 was determined to be 100%. [Click here to view] |
3.5. Effects of pH on the Protease Stability
The purified protease from C. tetragonoloba was stable under a wide range of pH. There was an increase in protease activity from pH 6.5 up to 7.5, and then it gradually decreased when the pH was increased to above the optimum pH. Therefore, the results show that there was an increase in protease stability from pH 4.5 up to 10.5. The protease showed 40% and 65% activity at pH 4.5 and 12.5, respectively. The protease showed 80%–100% activity between pH 6.5 and 10 (Fig. 6), and this profile is similar to plant serine protease from W. tinctoria [29].
3.6. Effects of Temperature on Protease Activity
The purified protease from C. tetragonoloba was active under a wide range of temperatures. There was an increase in protease activity from 20°C to 60°C and then a gradual decrease when the temperature was increased above the optimum temperature. Therefore, the results show that the optimum temperature for the purified protease is 60°C (Fig. 7). Most of the plant serine proteases show optimum temperature from 20°C to 80°C.
3.7. Effects of Temperature on Proteases Stability
The purified protease from C. tetragonoloba was stable under a wide range of temperatures. There was an increase in protease activity on storage from 20°C to 70°C and then a gradual decrease when the temperature was increased above the optimum temperature. The protease showed 60% stability at 80°C. Therefore, the results showed that the protease shows 80%–100% stability between 20°C and 70°C (Fig. 8). This profile is similar to plant serine protease from subtilisin. Canavalia ensiformis has reported the optimum temperature between 30°C and 60°C and was stable from 30°C to 70°C, as reported by Tomar et al. [30].
 | Figure 6: Effects of pH on the protease stability of C. tetragonoloba. The stability of the protease was determined by incubating protease in different buffers of different pH for 1 hour at 37°C and the protease activity was measured at pH 7.5. [Click here to view] |
 | Figure 7: Effects of temperature on the protease activity of C. tetragonoloba. 15 ug of protease was used to determine the activity. The activity of the protease at 60°C was determined to be 100%. [Click here to view] |
 | Figure 8: Effects of temperature on the protease stability of C. tetragonoloba. The stability of the proteins was determined by incubating protease at different temperatures for 1 hour at pH 7.5 [Click here to view] |
3.8. Effects of Various Protease Inhibitors on Protease Activity
The effects of various protease inhibitors on protease activity are shown in Table 2. The purified protease retains 106% and 104% residual activity in the presence of EGTA and EDTA, respectively. It is reported that metalloprotease from Bacillus licheniformis RP1 in the presence of EGTA and EDTA shows 90% inhibition of activity [31]. The aspartic protease inhibitor (pepstatin A) shows no effect on protease activity. It is reported that aspartic protease from C. ensiformis was also not inhibited by pepstatin A [32].
This protease was also not inhibited by cysteine protease inhibitors (IAA, E-64). Recently, it is reported that cystein protease and porcein B from Calotropis procera showed maximum inhibition of activity in the presence of IAA and E-64, respectively [32,26]. However, in the presence of serine protease inhibitors, purified protease showed 2%, 12%, 2.8%, and 53% activity with benzamidine HCl, aprotonine, PMSF, and SBTI, respectively (Table 2). This inhibition by specific serine protease inhibitors as PMSF, SBTI, benzamidine HCl, and aprotinin revealed that protease belongs to the serine protease class. It is reported that protease streblin from Streblus asper was inhibited by PMSF by 95.5% [33].
3.9. Effects of Various Solvent and Surfactant on Protease Activity
The purified protease retained 90%–95% residual activity in the presence of 50% v/v of methanol, isopropanol, and DMSO, while in the presence of ethanol and butanol, up to 34% and 60% residual activity was retained, respectively. 81% activity was retained in presence of SDS, also indicating extreme stability of this protease (Table 3).
3.10. Substrate Specificity of the Protease
The substrate specificity of protease was assayed using high-grade (1% w/v) gelatin, Hb, BSA, and casein. The purified protease was showing cleavage of gelatin, Hb, BSA, and casein (Table 4) with equal efficiency which is similar to other serine proteases like cuminisn and Q-SP [34,35]. The specific activity of purified protease for chromogenic substrate BAPNA was calculated as 1.5101 U/mg. Purified protease does not show any significant result for the N-succinyl-Phe-p-nitroanilide.
3.11. Sugar Determination
Glycosylation is ubiquitous and sugar residues are attached to the side chain. Glycosylation plays an important role in the influencing of protease activity [36]. Cyamopsis tetragonoloba protease contains about 35 μg/ml sugar determined by the method of DuBois (Fig. 9). Benghalensin showed 10%–12% sugar content from what was isolated from Ficus benghalensis [37].
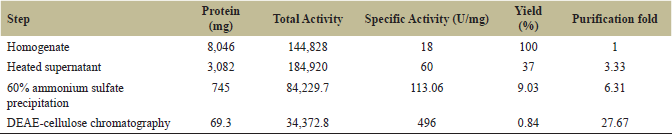 | Table 1: Summary of protease purification from the seeds of C. tetragonoloba. [Click here to view] |
 | Table 2: Effects of various protease inhibitors on protease activity. [Click here to view] |
 | Table 3: Effects of various solvents and surfactants on protease activity. [Click here to view] |
 | Table 4: Relative activities of protease with different substrates at different pH. The activity of protease with casein was considered 100% activity and the relative activities with the other substrate were represented in percentage relative activity to that of casein. [Click here to view] |
 | Figure 9: Determination of sugar. Estimation of the sugar content of purified protease was carried out by DuBois method. A standard curve of the absorbance versus dextrose concentration as standard was plotted. [Click here to view] |
4. CONCLUSION
In the present study, purification and biochemical characterization of protease from the seeds of C. tetragonoloba were carried out. The Vmax value of the protease was 102.04 μM/minute with casein as the substrate and Km value was 56.56 μM/minute. The purified protease was completely inhibited by serine proteases inhibitors PMSF, SBTI, benzamidine HCl, and aprotinin. This protease was not inhibited by IAA, E-64, pepstatin A, EGTA, and EDTA. Thus, the protease inhibitor assay results concluded that the purified protease was serine protease, inhibited by serine proteases inhibitors. The results shown in Table 4 determine that gelatin shows the highest proteolytic activity when compared to the casein, Hb, and BSA. BAPNA shows 1.5101 U/mg specific activities. The present study found that purified protease was a highly thermal stable serine protease that can function in harsh conditions in the presence of detergents and solvents and has a wide range of substrate specificity, so it could be used more in a biotechnological and industrial application. However, research is under progress with regard to the characterization of protease and application.
5. ABBREVIATIONS
BAPNA, Nα-Benzoyl-DL-arginine ?-nitroanilide; BSA, Bovine serum albumin; β-ME, β-mercaptoethanol; CBB, Coomassie Brilliant Blue; DEAE, Diethylaminoethyl; DMSO, Dimethyl sulfoxide; E-64, l-trans-epoxysuccinyl olamide(4-guanidino)butane; EDTA, Ethylenediaminetetraacetic acid; EGTA, ethylene glycol bis(2-aminoethyl ether)-N,N,N,N-tetraacetic acid; Hb, Hemoglobin; PMSF, Phenyl Methyl Sulfonyl Fluoride; PAGE, Polyacrylamide gel electrophoresis; Hb, haemoglobin; IAA, Iodoacetamide; M, Molar.
6. ACKNOWLEDGMENT
The authors gratefully acknowledge financial assistance from the University Grant Commission, India, as Rajiv Gandhi National Fellowship to R.K.R and the National OBC fellowship to P.P.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
REFERENCES
1. Rai PS, Dharmatti PR, Shashidhar TR, Patil RV, Patil BR. Genetic variability studies in cluster bean [Cyamopsis tetragonoloba (L.) Taub] Karnataka. J Agri Sci 2012;25:108–11.
2. El Kheir S, Abdalla AH, Nouri SH. Quality assessment of guar gum endosperm of gaur (Cyamopsis tetragonoloba). ISCA J Biol Sci 2012;1:67–70.
3. Bhatt RK, Jukanti AK, Roy MM. Cluster bean [Cyamopsis tetragonoloba (L.) Taub], an important industrial arid legume: a review. Agri Res Commun Center 2015;3599:1–8.
4. Kumar D, Rodge AB. Status, scope and strategies of arid legumes research in India a review. J Food Legum 2012;25:255–72.
5. Khan H, Ali A, Khan U. Purification and biochemical characterization of alkaline serine from Caesalpinia bonducella. Nat Prod Commun 2010;5(6):931–4.
6. Patel S, Ricucci D, Durak C, Tay F. Internal root resorption: a review. J Endod 2010;36(7):1107–21.
7. Vogts G, Woell S, Argos P. Protein thermal stability, hydrogen bonds and ion pairs. J Mol Biol 1997;269:4631–43
8. Hirata A, Hori Y, Koga Y, Okada J, Sakudo A, Ikuta A, et al. Enzymatic activity of a substantially in-home bolo TK SP from thermococcus in detergent and its stability to degrade the normal free on protein. BMC Biotechnol 2013;13:19.
9. Kurepa J, Wang S, Li Y, Smalle J. Proteasomes regulation, plant growth and stress tolerance. Plant Signal Behav 2009;4(10):924–7.
10. Adam Z. Emerging roles for diverse intramembrane proteäses in plant biology. Biochem Biophys Act 2013;1828(12):2933–6.
11. Jashni MK, Mehrabi R, Collemare J, Mesarich CH, de Wit PJGM. The battle in the last further insights into the role of proteus and their innovation in plant pathogens and fractions. Front Plant Sci 2015;6:584.
12. Mosolov VV, Valueva TA. Participation of proteolytic enzymes in the interaction of the plants with phytopathogenic microorganisms. Biochemistry 2006;71(8):838–45.
13. Shine K, Kanimozhi K, Panneerselvam A, Muthukumar C, Thajuddin N. Production and optimisation of alkaline protease by Bacillus cereus RS3 isolated from desert soil. Int J Adv Res Biol Sci 2016;3(7):193–202.
14. Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Dis 2006;5(9):785–99.
15. Lopez Otin C, Bond JS. Proteases: multifunctional enzymes in life and diseases. J Biol Chem 2008;283(45):30433–7.
16. Folin O, Ciocalteu V. On tyrosine and tryptophane determinations in proteins. J Biol Chem 1929;73:627.
17. Kembhavi AA, Kulkarni A, Pant a salt-tolerant and thermostable alkaline protease from Bacillus subtilis NCIM No. 64. Appl Biochem Biotechnol 1993;38:83–92.
18. Arnon R. Papain. Meth Enzymol 1970;19:226–44.
19. Laemmli UK. Cleavage of structural protein during the assembly of head of bacteriophage T4. Nature (London) 1970;227:680–5.
20. Bernardino-Niccanor A, Scilingo AA, Anon CM, Davila-Otiz. Guava seed storage: fractionation and characterization. LWT 2006;39:902–10.
21. Heussen C, Dowdle EB. Electrophoresis plasminogen activators acrylamide gel containing sodium dodecyl sulfate and copolymerized substrate. Anal Biochem 1980;102:196–202.
22. Merino-Contreras ML, Sanchez-Morales F, Jimenez-Badillo ML, Alverrez-Gonalez CA. Partial purification of digestive protesa in sheepshead Archosargus probatocephalus (Spariformis: Spaidae). Neotrop Ichthyol 2018;16(4):e180020.
23. DuBois M, Gillies KA, Hamilton JK, Rebes PA, Smith F. Colorimetric method for the determination of sugars and related substances. Anal Chem 1956;2:350–7.
24. Englard S, Seifter S. Precipitation technique. Methods Enzymol 1990;182:285–300.
25. Antao CM, Malcata FX. Plant serine proteases; biochemical, physiological and molecular features. Plant Physiol Biochem 2005;43(7):637–50.
26. Singh KA, Kumar R, Rao JRK, Jagannadham MV. Crinumin, chymotrypsin-like but glycosylated serine protease from Crinum asiaticum purification and physicochemical characterization. Food Chem 2010;119:1352–8.
27. Bogacheva AM, Rudenskaya GN, Preusser A, Tchikileva IO, Dunaevsky YE, Stepano VM. A subtilisin-like a protease from roots of the dandelion Taraxcum officinale Webb S.L. Biochem (Mosc) 1999;64:1030–7.
28. Asif -Ullah M, Kim K, Yu YG. Purification and characterization of serine protease from Cucumis trigonus Roxburghii. Phytochemistry 2006;67:870–5.
29. Panicker LM, Usha R, Roy S, Mandal C. Purification and characterization of a serine protease (CESP)from mature coconut endosperm. BMC Res 2009;2:81.
30. Tomar R, Kumar R, Jagannadham MV. A stable serine protease, wrightin from the latex of the plant Wrightia tinctoria (Roxb.) purification and biochemical. J Agri Food Chem 2008;56:1479–87.
31. Sellami-Kamoun A, Haddar A, El –Hadi Ali N, Ghorbel-Frikha B, Kanoun S,Nasri M Stability of thermostable alkaline protease from Bacillius licheniformis RP1 in commercisl solid laundry detergent formulations. Microbiol Res 2006;163(2008):299–306.
32. Goncalves RN, Gozzini-Barbosa SD, daSilva-Lopez RE. Proteases from canavalia ensiformis: active and thermostable enzymes with potential of application in biotechnology. Biotechnol Res Int 2016;11.
33. Tripathi P, Tomar R, Jagannadham MV Purification and biochemical characterization of novel protease streblin. Food Chem 2011;125:1005–12.
34. Arima K, Uchikoba T, Shimada M, YoneZawa H, Kened M. Cucumisin-like protease from the latex of Euphorbia supina. Phytochemistry 2000;53:639–44.
35. Demartini DR. Wlodawer A, Carlini CR. A comparative study of the expression of serine proteinase in quiescent seeds and in developing Canavalia ensiformis plants. J Exp Bot 2007;58:521–32.
36. Hart W, Kelly WG, Blomberg MA, Roqemore EP, Dong LY, Kreppel L, et al. Glycosylation of nuclear and cytoplasmic protein is as abundant and as dynamic as phosphorylation. In: Wieland F, Reutter W (eds.). Glyco and Cellbiology, Springer, Heidelberg, Germany, pp 90–103, 1994.
37. Sharma A, Kumari M, Jagannadham MV. Benghalnesis, a highly stable serine protease from latex of medicinal plant Ficus benghalensis. J Agric Food Chem 2009;57:11120–6.