1. INTRODUCTION
Bacterial wilt disease is a major cause of yield loss in the Solanaceae family caused by phytopathogen Ralstonia solanacearum having a wide variety of host ranges of 200 plant species and more than 54 families, infecting mainly potato, tomato, chilly, eggplants, and banana [1,2]. Direct crop yield losses accounted by R. solanacearum vary widely according to the host on the basis of cultivar, climate, types of soil, crop rotation, and strain type. For example, Elphinstone et al. [2] reported that yield losses vary from 0% to 91% in the tomato, 33% to 90% in the potato, 10% to 30% in tobacco, 80% to 100% in the banana, and up to 20% in the groundnut.
Ralstonia solanacearum is a soil-borne, motile, non-spore forming, Gram-negative rod. Ralstonia solanacearum strains represent a heterogeneous group divided into four phylotypes (I–IV) on the basis of geographical source, subdivided into five races using host range, and further into five biovars based on biochemical and physiological tests. Ralstonia solanacearum species complex (RSSC) as a destructive phytopathogen has been ranked 2nd in the top 10 plant pathogens [3,4].
The pathogens enter the plant system through mechanical wounds and damaged root hairs to form dense populations (109 CFU/g fresh weight) resulting in biofilm formation in the intercellular spaces of the xylem tissues. Vascular dysfunction is caused by pathogen-derived hydrolytic enzymes degrading the cell wall as well as exopolymeric substances (EPS1) that block water transportation [5–7]. Some direct infections have been seen on potato tubers caused by R. solanacearum.
In India, R. solanacearum has been found to be endemic across the nation and is the cause of 30%–70% loss in potato tuber yields. A study from the cold storage of potato tubers in Ghajipur, Uttar Pradesh, also showed the presence of R. solanacearum wilt [8].
In this study, we report isolation and pathogenicity characterization of bacterial phytopathogens around Kanpur and Fatehpur districts of Uttar Pradesh which initiated with specific microbial isolation strategy for R. solanacearum. Out of 57 isolates, 22 were found to be Ralstonia on the basis of biochemical characterization. Furthermore, isolates were confirmed as R. solanacearum based on sandwich-enzyme-linked immunosorbent assay (ELISA) results. Sugar utilization test was carried out for biovar determination. Biofilm formation, hydrolytic enzymes production, motility, siderophore and hydrogen cyanide (HCN) production, potassium solubilization, as well as infectivity studies of R. solanacearum isolates in tomato seedlings and potato slices are described. It is important to understand local biodiversity of wilt phytopathogens for early detection and better management to prevent food losses.
2. MATERIALS AND METHODS
2.1. Sampling and Isolation of R. s olanacearum Bacteria
Bacteria were isolated from ooze in the collar regions of stems from wilted tomato and potato plants as well as infected tubers, collected from cultivars of solanaceous crops from Kanpur (26.44°N and 80.33°E) and Fatehpur regions (25.92°N and 80.79°E) of Uttar Pradesh, India. Plant surfaces were washed firstly with tap water, followed by sterilized doubled distilled water (ddH2O), to remove soil and external microflora. To release bacterial ooze, the stem was cut and placed into 10 ml sterilized ddH2O, which was streaked on triphenyl tetrazolium chloride (TZC) medium. Brown discolorations of vascular system of tomato plants was directly placed on TZC medium (Kelman TZC agar) and incubated at 28°C–30°C for 48 hours for further bacteria isolation.
2.2. Morphological, Physical, and Biochemical Characterization of Isolates
Bacterial isolates of potato and tomato wilt were characterized by using the following tests: Gram’s staining, potassium hydroxide (KOH) solubility test, oxidase, catalase, oxidation/fermentation, starch and gelatin liquefaction, levan production, arginine dihydrolase, and fluorescent pigment production on King’s B medium [9–11]. Additionally, thermostability (37°C and 40°C) and halotolerance (2.5% NaCl) were tested in Tryptone and Soya peptone agar for each of the isolates.
2.3. Sugar Utilization Test for Biovar Determination
For biovar determination of isolates, the sugar utilization test was carried out by using disaccharides (maltose, lactose, and cellobiose) and hexose alcohols (Mannitol, Dulcitol, and Sorbitol) based on semisolid basal medium some minor modification in microtiter plate. Log-phase isolates were inoculated in triplicate, incubated for 3–5 days at 28°C [12]. Qualitative results were recorded based on color change due to pH indicator bromothymol blue dye that was olivaceous green color at neutral pH and changed to yellow upon lowering of pH due to sugar utilization/alcohol oxidation.
2.4. Pathogenicity Studies and Plant Bioassay
The pathogenicity test was carried out in tomato seedlings and potato tuber slice bioassays as described below.
2.4.1. Tomato seedling assay
Tomato seeds (S-22 variety from local Kanpur farmer’s market) were sterilized in 70% ethanol, and germinated on a sterilized wet cotton bed in plastic trays. The growth of the seedlings appeared on day 6. About 4–5 cm in height tomato plantings with two cotyledon leaves were taken for the plant bioassay study. The experiment was carried out in 2 ml microcentrifuge tubes containing 1.5 ml phosphate buffer with 108 cells per ml of Ralstonia isolates. Pathogenicity was determined by wilt symptoms observation of incubated tomato seedlings compared with positive controls (reference strains) and negative (buffer) control seedlings. Furthermore, fresh weight was calculated for tomato seedlings. Reference strains R. solanacearum (NAIMCC-F01629) procured from National Bureau of Agriculturally Important Micro-organisms, Kushmaur, Uttar Pradesh, India, and R. solanacearum F1C1 provided by Dr Suvendra Roy, Department of Molecular and Biotechnology, Tezpur University, Assam, were used as positive controls.
2.4.2. Potato tuber bioassay
The experiment was designed for direct inoculation method on potato tuber slices to check pathogenicity ability of bacterial isolates [10]. Fresh potato tubers were washed with double distilled water and the surface was sterilized with 70% ethanol. Potato tuber slices of 5 × 4 × 0.5 cm3 were inoculated with a single colony and incubated at 28 ºC under daily observation in triplicates. Uninoculated tuber slices were used as control. Diseased symptoms included vascular browning, bacterial ooze, and tuber necrosis were recorded daily up to day 10 post-incubation. Grading scale used for qualitative measure of tuber disease symptoms was as follows: Grade 0: no symptoms; Grade 1: yellow discoloration, no ooze, and no necrosis; Grade 2: brown discoloration, no ooze, softening of tissue at center of lesion, but hard to scoop or pick with inoculating loop; Grade 3: brown discoloration with ring formation, whitish ooze from tuber, necrotic tissue softening which is easy to scoop; and Grade 4: dark brown-blackish discoloration within the ringed lesion, copious whitish ooze from lesion, and total tissue necrosis which is easy to scoop.
2.5. Pathogenic Traits Studies of Ralstonia Isolates
2.5.1. Production of hydrolytic enzymes
Hydrolytic enzyme production by Ralstonia isolates was carried out on minimal salt agar supplemented with 1% carboxymethyl cellulose salt agar for β-1,4-endoglucanase activity and with 1% polygalaturinic acid for polygalacturinase activity with some modification [13]. After 72 hours incubation, β-1,4-endoglucanase activity was determined by 1% Congo red solution staining for 30 minutes, followed by 1 M NaCl for 15 minutes, and polygalacturinase activity was determined by staining with iodine solution. The appearance of clear zones on the inoculated region of R. solanacearum was detected.
2.5.2. Secretary molecules production
The production of siderophores on chroma azurol sulfonate (CAS) medium was carried out following the method of Lynne et al. [14]. Siderophore production by the isolates was tested by inoculating log cultures on CAS media. Phosphate solubilization tests of isolated strains were carried out on Pikovskaya medium [15]. Plates were incubated at 28°C for 48–72 hours. Potassium solubilizing activity by all isolates was studied qualitatively on modified Aleksandrov medium plates by the spot test method [16].
HCN production of bacterial isolates was tested qualitatively as per the protocol of Bakker and Schipper (1987). Bacteria were streaked on Kings B medium supplemented with glycine at 4.4 g/l and covered with sterile filter paper saturated with picric acid on Petri plate lid. Plate was incubated for 3–4 days to observe for brown coloration that was recorded as positive test.
2.5.3. Motility tests
Motility test was carried out in 0.3% agar for swimming motility and 1.0% agar for twitching motility [17,18]. For twitching motility, fresh log culture was taken by the inoculation needle and prickled into agar completely and for swimming motility test log cultures were just touched on agar surface in a single point and incubated at 28°C for 24–48 hours.
2.5.4. Biofilm formation
Biofilm formation property of microbial isolates was carried out by two different assays: Congo red and crystal violet. Briefly, in Congo red binding assay, 0.8% Congo red amended in tryptone and soya agar (TSA) with 1% sucrose was streaked with R. solanacearum strains and incubated at 28°C for 48 hours. Plates were observed for the appearance of blackish red color around the streak area indicating sugar binding ability.
Crystal violet biofilm assay was carried out on 96-well polystyrene microtiter plate for static biofilm formation as described previously by O’Toole et al. [19] with some minor modification. Briefly, R. solanacearum strains were grown on the microtiter plate containing 200 μl TSB for 24 hours at 37°C. After incubation, planktonic cells were washed thoroughly with phosphate buffer saline (PBS) and stained with 0.2% crystal violet for 20 minutes. Subsequently, the adherent biofilm was washed with saline, dissolved in 200 μl of dimethyl sulfoxide and solubilized biofilm was measured by using ELISA microreader (Thermo Fisher Multiscan, USA) at A600.
2.5.5. Antibiotic resistance
Antibiotic susceptibility test of all isolates was carried out by Kirby–Bauer disk diffusion method and their resistance was determined as per CLSI standard [20]. Briefly, 24 hours log-phase cultures were spread on the Mueller Hinton agar plates; antibiotic disks were placed using sterilized forceps on the inoculated plates and incubated at 37°C for 24 hours. Inhibition zone sizes were compared with the standard chart. The antibiotics tested were ampicillin (A: 25 μg), erythromycin (E: 15 μg), streptomycin (STR: 30 μg), tetracyclin (TE: 30 μg), and vancomycin (V: 10 μg).
2.5.6. Serological detection using double antibody sandwich (DAS)-ELISA
Serological confirmation was carried out by using monoclonal antibodies of DAS-ELISA kit (catalog no. SRP 33900), purchased form Life TechnologiesTM (India) Pvt. Ltd. ELISA test was carried out as per protocol of Umrao et al. [21]
2.6. Molecular Identification of Bacterial Strains
Molecular identification of bacterial isolates were carried out by 16S (recombinant deoxyribonucleic acid) rDNA sequence analysis. DNA isolation from microbial samples were carried out using the EXpure Microbial DNA isolation kit developed by Bogar Bio Bee stores Pvt Ltd. Genomic DNA isolates RS1, RS2, RS3, RS4, PS1, PS5, TS3, and TS5 were amplified by polymerase chain reaction (PCR) using consensus primers 27F/1492R (27F 5'AGAGTTTGATCTGGCTCAG3'/ 1492R 5'TACGGTACCTTGTTACGACTT3') with Taq Master Mix (Taq DNA polymerase is supplied in 2 × Taq buffer, 0.4 mM dNTPs, 3.2 mM MgCl2, and 0.02% bromophenol blue) in 25 μl of PCR reaction solution (1.5 μl of forward primer and reverse primer, 5 μl of deionized water, and 12 μl of Taq Master Mix) and PCR was carried out using the following thermal cycling conditions: initial denaturation of 95°C for 2 minutes, followed by 25 cycles of 95°C for 30 seconds, 55°C for 1 minute and 72°C for 2 minutes and a final extension of 72°C for 10 minutes and sequenced (Agri Genome Labs Pvt Ltd., Kochi, India).
Single-pass sequencing was carried out on each template using 16S ribosomal ribonucleic acid (rRNA) universal primers by using automated DNA sequencer (ABI 3730×1 Genetic Analyser from Applied Biosystems Inc.). Partial DNA sequence obtained was analyzed in the National Center for Biotechnology Information server using BLAST tool and corresponding sequences downloaded (www.ncbi.nih.nlm.in). The phylogeny analysis of sequences with the closely related sequence of blast results was performed by multiple sequence alignment (MUSCLE 3.7) [22]. The resulting aligned sequences were cured using the program Gblocks 0.91b [23]. Phylogenetic tree was constructed using PhyML 3.0 aLRT. Program tree Dyn 198.3 was used for tree rendering [24].
3. RESULTS AND DISCUSSION
3.1. Isolation of R. solanacearum
Whitish ooze and vascular tissues from collar regions of the infected tomato and potato plants including infected potato tuber from fields around Kanpur and Fatehpur area were collected (Fig. 1A). Bacterial colonies that appeared with fluidal red center, white margins, and irregular forms were further isolated using standard pure culture isolation techniques on TZC media at 30°C after 48 hours of incubation (Fig. 1B). A total of 57 independent bacterial colonies were isolated and used for further microbiological, biochemical, and pathogenicity characterization.
3.2. Morphological, Physiological, and Biochemical Characterization of Bacterial Isolates
All 57 strains were found to be Gram-negative small rods, further confirmed by KOH solubility test (Table 1). In order to distinguish the strains from Pseudomonas, fluorescent pigmentation test on King’s B agar was carried out in which 35 fluorescent positive strains were considered as Pseudomonas, and only 22 bacterial isolates were found to be non-fluorescent Gram-negative rods. All 22 strains were positive for oxidase and catalase. 82% strains were positive in Levan production and negative for arginine dihydrolase. None of the strains hydrolyzed starch nor liquefied gelatin. All 22 strains showed 2.5% halotolerant and growth at 37°C, but not at 40°C. One of screened bacterial strains RS1 was morphologically characterized as rod-shaped of length 1.4 μm possessing polar tuft flagella under transmission electron microscope (Fig. 1C).
 | Figure 1. Isolation and morphological study of R. solanacearum. A. Wilt symptoms in potato plants. B. Mucoid white margin red centered colony of bacteria Tub1 isolated from infected potato plant bacterial ooze streaked on TTC medium. C. Rod-shaped, size 1–1.5 μ, possess lophotricous flagella of R. solanacearum under transmission electron microscope. [Click here to view] |
 | Table 1. Isolation of R. solanacearum from infected plants collected from different cultivars of tomato and potato. [Click here to view] |
3.3. Biovar Determination of Isolated Strains
Three disaccharides (lactose, cellobiose, and maltose) and three monosaccharides hexose alcohols (Mannitol, Sorbitol, and Dulcitol) were used in sugar utilization test to determine biovar of isolated bacterial strains using sugar disks. Strains PS1–PS10, TR3, PR4, TS11, and TS13 showed utilization of all three disaccharides and monosaccharides except Dulcitol. TS5 and TS9 fermented only one sugar Mannitol, while TS3 strain showed fermentation of Mannitol and Sorbitol. RS1, RS2, RS3, and RS4 including potato tuber strains Tub1 and Tub2 with reference strains (R1629 and F1C1) were found to utilize all sugars tested, considered as biovar III (Table 2).
3.4. Pathogenicity Tests Using Plant Bioassay
All 22 bacterial isolates showed pathogenicity on potato slices and tomato seedlings causing wilt disease. Pathogenicity assay was carried out using tomato seedlings about 4–4.5 cm in length grown on a wet cotton bed (Fig. 2A) carried out in 2 ml microcentrifuge tubes containing 108 cells per ml in PBS in triplicates along with reference R. solanacearum F1C1 and R1629 pathogen controls (positive control) as well as buffer control (negative control) (Fig. 2B). Pathogen-treated tomato seedlings showed wilting and weight loss 72 hours post-treatment in comparison to uninfected control seedlings. In this study, all strains also observed to be pathogenic in pathogenicity test on potato tuber slice by secreting slimy ooze-like substance and were graded as per material and methods shown in Figure 2C. Strains were re-isolated from the ooze in pure culture and confirmed as Ralstonia as per Koch’s postulates. Pathogenicity demonstrated in tomato seedlings exhibited 18% isolates are found to be highly pathogenic, 64% having moderate pathogenic ability, and 18% show poor pathogenicity (Fig. 3).
3.5. Pathogenicity Traits of R. solacearum Isolates
The virulence factors of isolated strains of R. solanacearum are characterized as presented in Table 3.
3.5.1. Biofilm formation
Using crystal violet assay, strains PS1, PS2, PS3, PS5, PS6, PS8, PS9, TS5, TS9, PR4, Tub1, and Tub2 including reference strain F1C1 were characterized as strong biofilm formers (54.5%), while RS1, RS2, RS4, RS4, PS4, PS10, TS3, TS11, TS13, TR3, and reference strain R1629 were found to be moderate biofilm formers (45.4%) (Fig. 4). Black colored colonies on Congo red agar media were classified as biofilm former and red colored colonies as non-biofilm formers (Fig. 5A). 14 isolates out of 22 were concluded as strong biofilm former (63.6%) in addition to reference strain R1629.
 | Table 2. Sugar utilization test on basal medium for biovar determination. [Click here to view] |
 | Figure 2. In vitro pathogenicity test on tomato seedlings and potato tuber slices. A. Tomato seedlings germinated on a sterilized wet cotton bed on plastic tray. B. Pathogenicity cauterization of all Ralstonia isolates in tomato seedlings carried out in 2 ml microcentrifuge tubes confirmed on the basis of biochemical test. C. Pathogenic ability on potato tuber slice bioassay and potato slices inoculated with isolated strains secreted slimy ooze-like substances. [Click here to view] |
Ralstonia biofilm formation comprising ralfuranone sugars and extracellular nucleases contributes to virulence by different mechanisms, including tissue specific colonization, EPS blockage of xylems, and increased resistance to plant antimicrobial compounds [25–27].
3.5.2. Antibiotic resistance
All 22 strains were found to be resistant to ampicillin, Vancomycin, and erythromycin. Potato isolates RS1, RS2, RS3, RS4, PS1, PS2, PS3, PS4, PS5, PS6, and PS8 as well as tomato isolates TS3, TS5 including R1629 exhibited sensitivity to antibiotics streptomycin and tetracycline. Four strains TR3 (tomato strain), PR4 (potato strain), and tuber isolated strains Tub1 and Tub2 were sensitive to only streptomycin. Furthermore, PS9, PS10, and TS13 were sensitive to only tetracycline. Only the TS11 strain which is highly resistant did not show sensitivity in any tested antibiotics ampicillin, erythromycin, streptomycin, and tetracycline (Fig. 5B). Multidrug resistance (MDR) bacteria are defined as bacteria which are resistant to three or more different classes of antibiotics. Emerging MDR is a serious problem globally, hence it needs an alternative for biocontrol of plant pathogens, such as phage therapy and PGPR antagonists [28,29].
 | Figure 3. Data of pathogenicity test on tomato seedling. The graph shows fresh weight of tomato seedlings before and after inoculation with isolated strains of Ralstonia isolates. [Click here to view] |
3.5.3. Production of hydrolytic cell wall degrading enzymes and other secretory molecules
All the isolates have the ability to produce enzymes polygalacturonase (pectinase) and cellobiohydrolases (cellulase) that can help in pathogens entry and colonization into host plants. Positive test is indicated as clear zone around bacterial growth on pectin and cellulose agar media, respectively (Fig.5C and D). Potato-isolated strains PS1, PS2, PS3, PS5, PS6, PS8, RS2, and tuber isolates Tub1 and Tub2, and positive control F1C1 exhibit potassium utilization (Fig. 5E). None of the isolates was found to solubilize phosphate (Fig. 5F). In addition, a volatile compound HCN was produced by 16 strains out of 21 strains RS1, RS2, RS3, TS3, TS5, TS9, PS3, and tuber isolates Tub1 and Tub2 including reference strain F1C1 which may cause lethality for plant metabolism. Some R. solanacearum isolates utilize potassium from their surrounding medium possibly creating nutritional deficiency by depletion of potassium in plant tissue system.
All isolated strains show the ability to produce siderophores on CAS media (Fig. 5G). Many bacteria encode iron acquisition system and produce siderophores to scavenge iron from their surroundings as virulence factors [30].
3.5.4. Motility
All 22 strains including reference strains F1C1 and R1629 showed swimming motility on 0.3% agarose media as well as twitching motility on 1.0% agarose, respectively (Fig 5H). Ralstonia isolates possess polar flagellar tuft for motility as seen in SEM imaging of bacterial morphology (Fig. 1D). Ralstonia solanacearum possess flagellar movement required for pathogen movement, colonization and invasion and type IV pilli-driven twitching motility which is directly related to virulence; however, R. solanacearum lose their motility in xylem in plants [31].
3.5.6. Extracellular polysaccharide I (EPSI) quantification using DAS ELISA
Serological confirmation was carried out by using monoclonal antibodies specific to EPS1 of R. solanacearum by DAS-ELISA. EPS1 is a heterogeneous N-acetylated polymer that plays an important role in R. solanacearum virulence, causing wilt by blocking water movement in plant xylem [32]. All strains were confirmed as R. solanacearum producing copious amount of EPS. Spectrophotometric readings of most isolates were found higher than positive control (F1C1) (Fig. 6). PS1 strain showing highest EPS production was also characterized as strong biofilm former. Isolate PS10, which shows no EPS1 production in DAS ELISA but was characterized as low to moderate biofilm formed as per Congo red and crystal violet assay. Results of biofilm formation and EPS as per DAS ELISA are highly corroborated for all other isolates. EPS1 mutants do not colonize plant xylem vessels as efficient as wild-type [5]. Similar response has been observed of wild-type and EPS1 mutant in susceptible tomato plants, but in resistant tomato plants, the wild-type induced significantly greater defense responses than the EPS1 mutants [33].
3.6. Molecular Characterization using 16S rDNA sequencing
Totally, eight strains were selected for 16S rRNA partial sequencing and phylogenetic analysis carried out as described in materials and methods. Two strains PS1 and PS5 showed 99.69% for Serratia marcescens strain SER1 (JQ439899.1), stain TS5 was identified to be Providencia vermicola strain BAB1 (MF276891.1) with 100% match, strain TS3 showed 99.61% identity to E. cloacae strain I19 (MT186255.1), and RS1, RS2, RS3, and RS4 with 99.92% match with E. cloacae strain R5 (MK064182.1) using nBLAST similarity analysis (Fig. 7; Table 4). Interestingly, there were variations in comparative genomic analysis of 16S rDNA with the biochemical and pathological nature of the results that were obtained. It is hence not conclusive that the isolates in our study were R. solanacearum. It is also likely that these isolates identified play a role in synergistic pathogenicity and enhance R. solanacearum virulence which will require further investigations. It is likely that further genomic level sequencing is required in order to confirm the true classification of the isolates which show all phenotypic and serological characteristics of R. solanacearum.
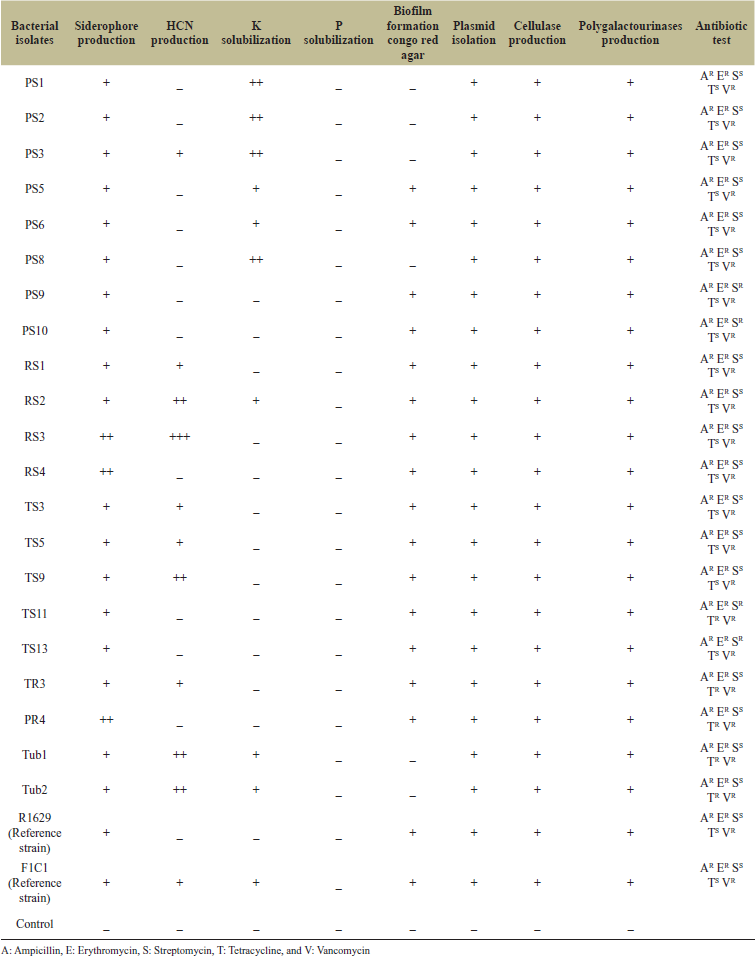 | Table 3. Pathogenicity traits studies of bacterial isolates. [Click here to view] |
 | Figure 4. Biofilm formation using crystal violet microtiter assay. Strains PS1, PS2, PS3, PS5, PS8, PS9, TS5, TS9, PR4, Tub1, and Tub2, including reference strain F1C1 showing strong biofilm formers, while RS1, RS2, RS4, RS4, PS4, PS10, TS3, TS11, TS13, TR3, and reference strain R1629 were found moderate biofilm formers. [Click here to view] |
 | Figure 5. Pathogenicity traits studied in bacterial isolates. A. Biofilm formation using Congo red binding assay. B. Antibiotic sensitivity test. C. Amylase production test. D. Cellulase production test. E. Potassium solubilization test on Aleksandrov medium. F. Phosphate solubilization test on Pikovskaya medium. G. Siderophore production on Chrome azurol S (CAS) medium. H. Swimming motility on 0.3% TSA medium. [Click here to view] |
 | Table 4. Description of 16S rRNA sequence analysis of bacterial isolates. [Click here to view] |
 | Figure 6. EPS1 quantification of isolated strains using DAS-ELISA. Graph represents a decreasing order of EPS1 production, PS1 producing highest EPS1, and PS10 having lowest EPS1 producing capacity. [Click here to view] |
 | Figure 7. 16 S rDNA phylogenetic trees of bacterial isolates. Out of eight, two strains (PS1, PS5) were identified as S. marcescens, one strain (TS5) as P. vermicola, and five strains (TS3, RS1, RS2, RS3, and RS4) were identified as E. cloacae using nBLAST in which strains PS1, TS3, and TS5 phylogenetic trees are represented here. [Click here to view] |
Similar results as our study were recently reported wherein members of Enterobacteriaceae such as Klebsiella oxytoca and Enterobacter cowanii were identified in molecular studies following presumptive R. solanacearum studies in West Bengal, India, as new wilt-causing bacteria [34]. Another study on bacterial wilt of mulberry (Morus alba) caused by E. cloacae in which wilting observed from bottom to top leaves unlike R. solanacearum was reported in China [35]. S. marcescens has been reported to cause yellow wilt disease in sunflowers in Russia [36].
4. CONCLUSION
In this study, we report 22 R. solanacearum on the basis of morphological, biochemical, and serological characterization out of 57 bacteria isolated from Kanpur and Fatehpur district, Uttar Pradesh, India. Although bacteria were confirmed R. solanacearum on the basis of DAS-ELISA for EPS-1, 16S rDNA sequencing results of eight select isolates did not corroborate with other identification tests. All 22 strains, however, showed bacterial wilt pathogenicity test carried out on tomato seedlings as well as potato tuber slices in laboratory conditions as well as virulence traits required for bacterial wilt diseases. In the future, further genomic-based characterization would be required to confirm the presence of RSSC complex as causative agents of bacterial wilt. Our study also brought about the identification of E. cloacae, S. marcescens, and P. vermicola that may play a role on bacterial wilt disease under certain circumstances.
5. ACKNOWLEDGMENTS
The authors are thankful to SAIF-AIIMS, New Delhi, India, for providing facilities of transmission electron microscopy of bacteria. They are also thankful to NBAIM, Kushmaur, UP, India, for strain R. solanacearum R1629 (NAIMCC-F01629) and Dr Suvendra Roy, Department of Molecular and Biotechnology, Tezpur University, Assam, India, for providing F1C1 strain.
6. CONFLICT OF INTERESTS
The authors declare that they have no conflict of interest.
7. AUTHORS’ CONTRIBUTION
All authors conceived, designed, and analyzed the experiments and contributed to manuscript preparation. Experiments were carried out by PDU.
8. FUNDING
There is no funding to report.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
REFERENCES
1. Hayward AC. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 1991;29:65–87. CrossRef
2. Elphinstone JG. The current bacterial wilt situation: a global overview. In: Allen C, Piror P Hayward AC, (eds.). Bacterial wilt disease and the Ralstonia solanacearum complex. American Phytopathological Society (APS Press), St Paul, MN, pp 9–28, 2005.
3. Álvarez B, Biosca EG, López MM. On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. In: Mendez-Vilas A (ed.). Technology and education topics in applied microbiology and microbial biotechnology. Formatex, Badajoz, Spain, pp 267–79, 2010.
4. Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 2012;13(6):614–29. CrossRef
5. Saile E, Mcgarvey JA, Schell MA, Denny TP. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 1997;87(12):1,264–71. CrossRef
6. McGarvey JA, Denny TP, Schell MA. Spatial-temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology 1999;89(12):1233–9. CrossRef
7. McGarvey JA, Bell CJ, Denny TP, Schell MA. Analysis of extracellular polysaccharide I in culture and in planta using immunological methods_ new insights and implications _ SpringerLink. In: Prior P, Allen C, Elphinstone J, (eds.). Bacterial wilt disease. Springer, Berlin, Heidelberg, pp 157–63, 1998. CrossRef
8. Sagar V, Jeevalatha A, Mian S, Chakrabarti S, Gurjar M, Arora R, et al. Potato bacterial wilt in India caused by strains of phylotype I, II and IV of Ralstonia solanacearum. Eur J Plant Pathol 2014;138(1):51–65. CrossRef
9. Hayward AC. Characteristics of Pseudomonas solanacearum. J Appl Bacteriol 1964;27(2):265–77. CrossRef
10. Lelliott RA, Stead DE. Methods for the diagnosis of bacterial diseases of plants. Blackwell Scientific Publications, Oxford, UK, 1987.
11. Schaad NW, Jones JB, Chun W. Laboratory guide for the identification of plant pathogenic bacteria. American Phytopathological Society (APS Press), St Paul, MN, 2001.
12. Ahmed NN, Islam MR, Hossain MA, Meah MB, Hossain MM. Determination of races and biovars of Ralstonia solanacearum causing bacterial wilt disease of potato. J Agric Sci 2013;5(6):86–93. CrossRef
13. Raza W, Ling N, Yang L, Huang Q, Shen Q. Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci Rep 2016;6(1):1–13. CrossRef
14. Lynne AM, Haarmann D, Louden BC. Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ 2011;12(1):51–3. CrossRef
15. Pikovskaya R. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 1948;17:362–70.
16. Setiawati TC, Mutmainnah L. Solubilization of potassium containing mineral by microorganisms from sugarcane rhizosphere. Agric Agric Sci Procedia 2016;9:108–17. CrossRef
17. Meng F, Yao J, Allen C. A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J Bacteriol 2011;193(10):2477–86. CrossRef
18. Liu H, Kang Y, Genin S, Schell MA, Denny TP. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 2001;(2001):3215–29. CrossRef
19. O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 1998;28(3):449–61. CrossRef
20. Hudzicki J. Kirby–Bauer disk diffusion susceptibility test protocol. Am Soc Microbiol 2009;1–13.
21. Umrao PD, Kumar V, Kaistha SD. Enzyme-linked immunosorbent assay detection of bacterial wilt--causing Ralstonia solanacearum. In: Gupta N, Gupta V, (eds.). Experimental protocols in biotechnology. Springer US, New York, NY, pp 1–18, 2020. CrossRef
22. Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(5):1792–7. CrossRef
23. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007;56(4):564–77. CrossRef
24. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36(suppl_2):W465–9. CrossRef
25. Kumar JS, Umesha S, Prasad KS, Niranjana P. Detection of quorum sensing molecules and biofilm formation in Ralstonia solanacearum. Curr Microbiol 2016;72(3):297–305. CrossRef
26. Minh Tran T, MacIntyre A, Khokhani D, Hawes M, Allen C. Extracellular DNases of Ralstonia solanacearum modulate biofilms and facilitate bacterial wilt virulence. Environ Microbiol 2016;18(11):4103–17. CrossRef
27. Mori Y, Hosoi Y, Ishikawa S, Hayashi K, Asai YU, Ohnishi H, et al. Ralfuranones contribute to mushroom-type biofilm formation by Ralstonia solanacearum strain OE1-1. Mol Plant Pathol 2018;9(4):975–85. CrossRef
28. Frampton RA, Pitman AR, Fineran PC. Advances in bacteriophage-mediated control of plant pathogens. Int J Microbiol 2015;2012(2012):11. CrossRef
29. Maji S, Chakrabartty PK. Biocontrol of bacterial wilt of tomato caused by Ralstonia solanacearum by isolates of plant growth promoting rhizobacteria. Aust J Crop Sci 2014;8(2):208–14.
30. Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev 2003;27(2–3):215–37. CrossRef
31. Corral J, Sebastià P, Coll NS, Barbé J, Aranda J, Valls M. Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. mSphere. 2020;5(2):e00740–19. CrossRef
32. Denny T. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, (ed.). Plant-associated bacteria. Springer, Dordrecht, Netherlands, pp 573–644, 2006. CrossRef
33. Milling A, Babujee L, Allen C. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS One 2011;6(1):e15853. CrossRef
34. Sarkar S, Chaudhuri S. New report of additional enterobacterial species causing wilt in West Bengal, India. Can J Microbiol 2015;61(7):477–86. CrossRef
35. Wang GF, Praphat K, Xie GL, Zhu B, Li B, Liu B, et al. Bacterial wilt of mulberry (Morus alba) caused by Enterobacter cloacae in China. Plant Dis 2008;92(3):483. CrossRef
36. Ignatov A, Khodykina M, Polityko V, Sukhacheva M. First report of Serratia marcescens causing yellow wilt disease on sunflower in Russia. New Dis Rep 2016;33:8. CrossRef