1. INTRODUCTION
Soybean (Glycine max) is an important crop globally due to its high percentage of proteins and oil contain [1,2]. Various food products are made from soybean seeds [3] and substantial efforts have been placed on increasing soybean yield in order to fulfill the requirement of larger population worldwide [4,5]. Pests, pathogens, and weeds cause detrimental effect resulting in low production of soybean crop [6]. Microorganisms such as fungi, viruses, bacteria, oomycetes, and nematodes contribute to economic damage every year [7]. The most important diseases of soybean are soybean rust, bacterial blight, and soybean mosaic which affect soybean production.
Plants need to protect themselves against attack from viruses, microbes, invertebrates, and even other plants [8]. It has been speculated that plants have two different types of disease resistance system called as basal defense and R gene involving defense [9-11]. The R genes are considered to be specificity determinants of the plant immune response [12]. The R genes are the fundamental elements of genetic interactions between biotrophic bacteria and plants which are known to control resistance against bacterial attack [13].
The majority of the sequences of cloned plant disease resistance genes, which provide resistance to the bacterial, viral, or fungal pathogens show that these consist of similar structural motifs and sequences [14]. The Arabidopsis genes, RPM1 and RPS2, provide resistance against bacterial blight caused by pathogen Pseudomonas syringae; the rice gene Xa2l confers resistance to Xanthomonas oryzae; the tomato gene Cf-9 confers resistance to the fungal pathogen Cladosporium fulvum; the flax gene L6 confers resistance to a rust fungus and the tobacco gene N confer resistance to tobacco mosaic virus; all these contain leucine-rich repeats (LRR) that encode protein motifs often linked with protein-protein interactions or ligand binding [15]. The resistance genes of Arabidopsis thaliana and G. max, RPM1 and Rpg1b, respectively, mediate recognition of the same Type III effectors protein, AvrB, from P. syringae. RPM1 and Rpg1b both belong to the coiled-coil nucleotide-binding sites (non-TIR or CC-NBS) class of resistance gene and they share only limited sequence similarity outside the conserved domains. Kanazin et al. [16] suggested that the similarities among resistance genes make it feasible to take advantage of sequence homologies for identification of other resistance genes. Resistance (R) genes cloning is the most recent method for obtaining disease resistant in legume crops [17].
Genetic and physical localization of G. max (Williams 82) Rpg1b disease resistance gene showed a complex locus consisting of various tightly linked families of NBS-LRR gene. 1 Mega base (Mb) region of soybean cultivar Williams 82 centered on the Rpg1b disease resistance gene on chromosome 13, which is effective against certain races of bacterial blight. This region also contains numerous NBS-LRR type genes, out of which 52D1_8 gene is also there, which belongs to non-TIR class like Rpg1b gene and hence it has similarity with Rpg1b gene of Williams 82. This is a putative disease resistance gene; whose sequence is devoid of introns [18-22]. The genome of Williams 82 has been completely sequenced [23]. However, very little is known about the genome of Indian soybean varieties at molecular level. Identification of resistance gene analogues in the Indian varieties is expected to aid significantly in the breeding programs of soybean. This study aims to amplify the disease resistance genes from the Indian varieties of G. max and to determine variations in sequences and functions of genes among these varieties.
2. MATERIALS AND METHODS
2.1. DNA Extraction from Selected Genotypes of Different Legumes
2.1.1. Plant material/tissue
Seeds of 11 different varieties of soybean (NRC-37, JS-20-69, JS-20-98, NRC-86, JS-20-29, NRC-7, RVS-2001-18, NRC-121, JS-20-34, JS-97-52, and NRC-12) of soybean (G. max) were collected from the Indian Institute of Soybean Research, Indore (IISR). After 22–25 days of sowing, when seedling developed into healthy plants, young unopened leaves were plucked for DNA isolation.
2.1.2. Reagents and chemicals
All the chemicals used in this study were of high-quality analytical grade reagents from HiMedia (MB502-HiPurA™ Plant DNA Isolation Kit, CTAB Method).
2.1.3. DNA extraction
Genomic DNA was extracted from the young unopened/fresh leaves collected directly from the plants and brought over the ice from the pots to the laboratory. Total DNA was extracted using CTAB method as described by Doyle and Doyle [24] with some modifications.
2.1.4. DNA purification
Purification of the extracted DNA was done to remove RNA, proteins, and polysaccharides, the major contaminants of the DNA precipitate. However, inclusion of CTAB in the DNA extraction buffer helps in the elimination of polysaccharides [25]. RNA was removed by RNase treatment and proteins were removed by phenol-chloroform extraction.
2.2. Polymerase Chain Reaction (PCR) Primers were designed from Selected Disease Resistance Genes
2.2.1. Tools used
Different web-based resources/tools were used for primer design such as Primer3, Primer-BLAST, and ClustalW software.
2.2.2. Primer design
The DNA sequences of disease resistant gene, Rpg1b (G. max), were retrieved from NCBI [26] and multiple sequence alignment (MSA) was performed using ClustalW [27]. After the alignment results, the conserved region was taken and both the primers (forward and reverse) were designed using online tool/software, namely, Primer3.
Set of primers (forward and reverse) for both NBS and LRR regions were constructed for disease resistance gene (Rpg1b) of G. max. The primers sequence in 5’–3’ direction of both the regions of genes is shown in Table 1a and another primer set were used from bacterial artificial chromosomes (BAC) libraries of Rpg1b in Table 1b. These primers were selected from the NBS-LRR genes 21-F22_29, 52-D1_8, and 42i-18_2 from soybean BAC 21-F22, 52-D1, and 42i-18, respectively. Standardization of NBS and LRR of Rpg1b for G. max and 5 more primer sets, labeled with A, B, C, D, and F, was done. PCR amplification was carried out according to the reaction mixture and conditions described below using these primers.
2.3. PCR Amplifications and Sequencing
2.3.1. PCR and purification of its amplified products
For PCR amplification, the reaction mixture was consisted of 2.5 μl of 10× polymerase buffer, 100 μM of each of the four dNTPs, 2.5 mM MgCl2, 25 ng of each of the forward and backward primers, 20 ng of genomic DNA, and 1.2 units of Taq DNA polymerase in a total of 25 μl volume. A master mix was prepared using PCR Recipe [28]. The genomic DNA was added separately after transferring master mix in PCR tubes. The reaction mixture after addition of all the components was mixed gently and PCR was carried in the thermal cycler (Amplitron make) using the thermal cycling conditions, as shown in Table 2.
 | Table 1a: Nucleotide sequences of selected primers. [Click here to view] |
 | Table 1b: Second sets of standard primers with their sequence. [Click here to view] |
 | Table 2: Thermal cycling conditions for polymerase chain reaction amplification. [Click here to view] |
Repeat step 2 from Table 2 for 35 times at 4°C. PCR products were checked on 2.0% agarose gel along with appropriate size marker to check desired amplicon; PCR amplicon was purified by column purification to remove contaminants, present if any.
2.3.2. Gene sequencing
DNA Sanger sequencing method was used for sequencing of selected genes:
Rpg1b_NBS – NRC-37 (S1), NRC-86 (S4), JS-20-29 (S5) and JS-20-34 (S9)
Rpg1b_LRR – NRC-37 (S1), NRC-86 (S4), JS-20-29 (S5) and JS-20-34 (S9)
DNA sequencing reaction of PCR amplicon was carried out with same PCR primers (F and R) using BDT v3.1 Cycle sequencing kit on ABI 3730 × 1 Genetic Analyzer. Obtained sequences were analyzed for its quality in Applied Biosystems sequencing software at the genomics facility of GeneXplore, Ahmedabad. Thereafter, MSA, Boxshade of the DNA sequences was performed for confirming the presence of Rpg1b gene and to check the sequence variations among them.
 | Table 3: Varieties of different Indian soybean leaf samples. [Click here to view] |
 | Table 4: Primer optimization status. [Click here to view] |
3. RESULTS AND DISCUSSION
3.1. DNA Isolation
Genomic DNA from 11 different Indian cultivars of G. max was isolated by CTAB method, purified and amplified using Rpg1b_NBS and Rpg1b_LRR primers in PCR. Variety name of different Indian soybean leaf samples is mentioned in Table 3 and genomic DNA was extracted from sample S1 to S10 and S13 of soybean (G. max).
 | Figure 1: Gel description of samples S1 to S9 with primer Rpg1b_NBS where well 1: NRC-37, well 2: JS-20-69, well 3: JS-20-98, well 4: NRC-86, well 5: JS-20-29, well 6: 100 bp ladder, well 7: NRC-7, well 8: RVS-2001-18, well 9: NRC-121, and well 10: JS-20-34 (Band approx. 900 bp). [Click here to view] |
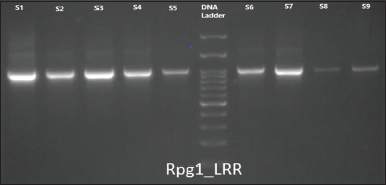 | Figure 2: Gel description of samples S1 to S9 with primer Rpg1b_LRR where well 1: NRC-37, well 2: JS-20-69, well 3: JS-20-98, well 4: NRC-86, well 5: JS-20-29, well 6: 100 bp ladder, well 7: NRC-7, well 8: RVS-2001-18, well 9: NRC-121, and well 10: JS-20-34 (Band approx. 900 bp). [Click here to view] |
 | Figure 3: Gel description of the samples S10 (JS-97-52) and S13 (NRC-12) with primers Rpg1_LRR and Rpg1_NBS. [Click here to view] |
 | Figure 4a: Multiple sequence alignment using ClustalW and Boxshade of NBS region of four Indian soybean varieties and Williams 82 (NM_001353616.1). Here, S1 is NRC-37, S4 is NRC-86, S5 is JS-20-29, and S9 is JS-20-34. [Click here to view] |
 | Figure 4b: Multiple sequence alignment using ClustalW and Boxshade of LRR region of four Indian soybean varieties and Williams 82 (NM_001353616.1). Here, S1 is NRC-37, S4 is NRC-86, S5 is JS-20-29 and S9 is JS-20-34. [Click here to view] |
3.2. PCR Optimization
Optimizing PCR amplifications, the results obtained are described in Table 4, where primers of Rpg1b NBS and LRR are showing amplification at annealing temperature 55°C and the approx. band size obtained is up to 900 bp. In primer sets, Rpg1b_NBS and Rpg1b_LRR amplification were obtained at all three annealing temperature used 52, 55, and 58°C. The primer optimization results as shown in Figures 1-3 for Rpg1b_NBS and Rpg1b_LRR, at 55°C temperature revealed clear and sharp band so it has been considered (55°C) for all remaining samples, that is, S1 to S13 of G. max except S11 and S12 as there is no DNA obtained.
3.3. Gene Sequencing
Out of 11 different varieties, four local varieties which are of economic importance and good yield were selected for further sequencing. DNA of ~1200 base pairs was successfully amplified from the four sample of G. max and sequenced. Sequence was closely matching with Williams 82 cultivar. These sequences were submitted in NCBI with GenBank accession no. MH700732.1 to MH700739.1.
Four Indian soybean varieties sequence were aligned using MSA tool with reference to Rpg1b gene (disease resistance gene consisting NBS-LRR regions) from Williams 82. Variations exist in Williams 82 region and Indian varieties, as shown in Figure 4a and b. In NBS region, variety S5 (JS-20-29) revealed maximum variations, while varieties S1 (NRC-37) and S4 (NRC-86) show the lowest variations. In LRR region, variety S9 (JS-20-34) has shown maximum significant variation and the variety S4 (NRC-86) shows minimum variations.
The previous studies have reported the amplification of disease resistance genes using PCR among numerous plant varieties [16,17,29]. In this study, using the primers for the NBS and LRR region of Rpg1b gene of Williams 82 soybean variety, we have amplified the NBS and LRR sequences from different Indian varieties of G. max. Yu et al. [30] reported the amplification and cloning of NBS sequences from soybean using degenerate primers of N and RPS2 genes of tobacco and Arabidopsis, respectively. Chang et al. [31] have also reported the characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide investigations.
Earlier analyses have reported the successful use of primers corresponding to conserved NBS domains in R genes to amplify sequences from soybean [32]. The NBS regions were taken as they are conserved in all resistance genes of legume and LRR were considered, as they are responsible for pathogen recognition. Various forward and reverse primers were designed from the coding region of disease resistance gene (Rpg1b) sequences of G. max using primer3 tool. Using the set theories, permutation, and combinations, different sets of primers were selected. Furthermore, some universal primers of soybean were taken from different BAC libraries and amplified for confirming the amplification of PCR protocol.
Standardization of primers from Williams 82 on different Indian soybean varieties was done, which showed that all these genes (NBS-LRR) were present in Indian soybean cultivars. Both primers show amplification and found to be conserved among those cultivars. After sequencing of Indian varieties of G. max, we have compared the sequence with its analog in Williams 82 and concluded about sequence similarities and variations as shown in MSA, Boxshade results [Figure 4a and b]. The similarities in the sequence of these varieties with Williams 82 showed that disease resistance gene (Rpg1b) is present in these varieties. These variations among the different soybean cultivars revealed the genetic diversity in the disease resistance genes. Development of molecular markers linked to these genes in various legumes has been done recently [33]. Molecular detection and characterization of resistance genes for bacterial blight will serve as an important resource for studying evolution of these genes from the legumes grown in Indian subcontinent. The practical use of this study is development of disease resistance in legumes, as well as in generating basic and fundamental knowledge on the resistance genes. Molecular characterization of these genes would be helpful in genetic modification to produce durable and disease resistance soybean varieties.
4. CONCLUSION
Presence of Rpg1b gene in four Indian soybean cultivars, that is, NRC-37 (S1), NRC-86 (S4), JS-20-29 (S5), and JS-20-34 (S9) was confirmed. Primers were used based on conserved NBS domain and LRR domain. Since Rpg1b is a disease resistance gene, which protects against bacterial blight, therefore, this gene sequences from selected Indian soybean cultivars will be used for developing R gene-based markers and for screening of bacterial blight in soybean. Apart from this, it will greatly facilitate the molecular cloning of disease resistance genes in soybean.
5. ACKNOWLEDGMENTS
Author acknowledge the facilities of the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi (DBT), under the Bioinformatics Subcenter as well as the Golden Jubilee Research Fellowship (GJRF) provided by School of Biotechnology, Devi Ahilya University, Indore., M.P., India.
6. CONFLICTS OF INTEREST
Author declared that he does not have any conflicts of interest.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. ETHICAL APPROVALS
Not applicable.
9. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Singh G, Ratnaparkha M, Kumar A. Comparative analysis of transposable elements from Glycine max, Cajanus cajan and Phaseolus vulgaris. J Exp Biol Agric Sci 2019;7:167-77.
2. Singh G, Kumar A. A review on various aspects of soybean and soybean mosaic virus. Vindhya Bharti 2019a;1:34-44.
3. Dukariya G, Shah S, Singh G, Kumar A. Soybean and its products: Nutritional and health benefits. J Nutr Sci Healthy Diet 2020;1:22-9.
4. Stupar RM. Into the wild: The soybean genome meets its undomesticated relative. Proc Natl Acad Sci 2010;107:21947-8. CrossRef
5. Van K, Hwang EY, Kim MY, Kim YH, Cho YI, Cregan PB, et al. Discovery of single nucleotide polymorphisms in soybean using primers designed from ESTs. Euphytica 2004;139:147-57. CrossRef
6. Powles SB. Gene amplification delivers glyphosate-resistant weed evolution. Proc Natl Acad Sci 2010;107:955-6. CrossRef
7. Gao L, Luo J, Ding X, Wang T, Hu T, Song P, et al. Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol Plant Pathol 2020;21:303-17. CrossRef
8. Hammond-Kosack KE, Jones JD. Plant disease resistance genes. Ann Rev Plant Biol 1997;48:575-607. CrossRef
9. Jones DA, Takemoto D. Plant innate immunity-direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol 2004;16:48-62. CrossRef
10. Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int J Mol Sci 2013;14:7302-26. CrossRef
11. Singh G, Kumar A. Synteny analysis of glycine max and Phaseolus vulgaris revealing conserved regions of NBS-LRR coding genes. Biosci Biotechnol Res Commun 2019b;12:124-33.
12. Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 2004;7:391-9. CrossRef
13. Flor HH. Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology 1955;45:680-5.
14. Tirnaz S, Zhang Y, Batley J. Genome-wide mining of disease resistance gene analogs using conserved domains. In: Legume Genomics. New York: Humana; 2020. p. 365-75. CrossRef
15. Kobe B, Deisenhofer J. The leucine-rich repeat: A versatile binding motif. Trends Biochem Sci 1994;19:415-21. CrossRef
16. Kanazin V, Marek LF, Shoemaker RC. Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci 1996;93:11746-50. CrossRef
17. Reddy AC, Venkat S, Singh TH, Aswath C, Reddy KM, Reddy DL. Isolation, characterization and evolution of NBS-LRR encoding disease-resistance gene analogs in eggplant against bacterial wilt. Eur J Plant Pathol 2015;143:417-26. CrossRef
18. Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, et al. Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol Plant Microb Interact 2003;16:817-26. CrossRef
19. Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 2004;16:309-18. CrossRef
20. Ashfield T, Egan AN, Pfeil BE, Chen NW, Podicheti R, Ratnaparkhe MB, et al. Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 2012;159:336-54. CrossRef
21. Bent AF. Plant disease resistance genes: Function meets structure. Plant Cell 1996;8:1757. CrossRef
22. Tarr DE, Alexander HM. TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res Notes 2009;2:197. CrossRef
23. Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature 2010;463:178. CrossRef
24. Doyle JJ, Doyle JL. Isolation of Plant DNA from fresh tissue. Focus 1990;12:39-40.
25. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 1980;8:4321-6. CrossRef
26. National Centre for Biotechnology Information, U.S. National Library of Medicines: RPG1-B NBS-LRR Type Disease Resistance Protein RPG1-B [Glycine max (Soybean)]. Available from: https://www.ncbi.nlm.nih.gov/gene/100817896.
27. Available from: Multiple Sequence Alignment by ClustalO. Available from: http://www.ebi.ac.uk/tools/clustalw.
28. Singh VK, Kumar A. PCR recipe: A program for setting up PCR reactions. Biotech Softw Intern Rep 2000;1:276-7. CrossRef
29. Ashfield T, Keen NT, Buzzell RI, Innes RW. Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 1995;141:1597-604. CrossRef
30. Yu YG, Buss GR, Maroof MA. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci 1996;93:11751-6. CrossRef
31. Chang HX, Lipka AE, Domier LL, Hartman GL. Characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Phytopathology 2016;106:1139-51. CrossRef
32. Ashfield T, Danzer JR, Held D, Clayton K, Keim P, Maroof MS, et al. Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet 1998;96:1013-21. CrossRef
33. Meziadi C, Richard MM, Derquennes A, Thareau V, Blanchet S, Gratias A, et al. Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Sci 2016;242:351-7. CrossRef