1. INTRODUCTION
In Maijdee, a small town of Noakhali in Bangladesh, potable water is scarce due to the high salinity and iron level, making underground water unlikely to drink without treatment. Consequently, people have to rely on centrally processed water supplied by the local government. However, it is insufficient to fulfill the large population’s requirements. In some areas, people have to use untreated pond water, which is even supplied by the same drinking water pipeline. There is a significant concern regarding the quality of water from these unsafe sources. Moreover, because of the faulty drainage system, these unprotected water ponds are flooded during the rainy season, and there is a distinct possibility that those water could mix up with the surroundings wastewater.
Wastewater from hospital-based or other sources is a reservoir of pathogenic microorganisms, including multiple antibiotic-resistant (MAR) bacteria, and can be a potential risk factor for public health and ecological balance [1]. Drug-resistant diseases cause around 700,000 deaths globally a year, which could increase to 10 million deaths globally per annum by 2050 if no action is taken. Nearly 2.4 million people may die in high-income countries between 2015 and 2050 without a sustained effort to contain antimicrobial resistance [2]. The economic damage of uncontrolled antimicrobial resistance may be like the shocks experienced during the 2008-2009 global financial crisis due to dramatically increased health care expenditures, food and feed production, trade, and livelihoods, and increased poverty and inequality [2]. These MAR organisms can often cause severe and sometimes lethal infections such as enteric diarrheal disease, urinary tract infections (UTIs), sepsis, and meningitis in humans. Drug-resistant bacteria caused infections, resulting in worse clinical outcomes with consuming more health-care resources [3]. Excessive and inappropriate way of using antibiotics is the main reasons which lead to the development of antibiotic-resistant bacteria (ARB). Furthermore, human treatments, excessive use of antibiotics in animal husbandry, and aquaculture can also increase antimicrobial resistance in the environment [4]. Initially, it was believed that antibiotic-resistant bacteria could be found only in care giving settings such as the hospital environment, but now they are everywhere [5].
Hospitals are the source of ARB, but it has been reported that these are potentially passed down to the community by the colonized patient and through hospital waste disposal facilities, especially wastewater [6,7]. Several reports have been focused on that Gram-negative isolates are frequently recovered from hospital wastewater among all pathogens and showed multidrug resistance [6,8,9]. The mentioned Gram-negative ARB are Escherichia coli, Klebsiella, Pseudomonas, Serratia, and Citrobacter sp. They are usually the most common causes of the hospital and community-acquired infections [8,10]. In the Gram-positive group, S. aureus, especially methicillin-resistant Staphylococcus aureus (MRSA), is one of the leading causes of infections in both hospital and community-acquired cases [11]. The distribution of MAR bacteria is alarmingly high in wastewater, and different human activities like discharging of hospital effluents to the sewer system without prior treatment are the main reason for the dissemination of MAR bacteria in the environment. It has been reported that only few countries recommend or implement the primary treatment of hospital effluents before discharge into the main wastewater flow. However, in developing countries such as Vietnam and Bangladesh, there are no fixed guidelines for hospital effluent discharge [12]. This untreated wastewater gets mixed up with other reservoirs and contributes to MAR bacteria spread in non-hospital-based water [8].
We have conducted a cross-sectional study to observe the distribution of multiple antibiotic-resistant bacteria in different wastewater sources of Maijdee at Noakhali and determine their resistance pattern to different clinically used antibiotics. Those findings will eventually help local authorities be aware of disseminating resistant bacteria in the water and providing information to implement appropriate guidelines for the proper management of hospital effluents.
2. MATERIALS AND METHODS
2.1. Study Area and Sample Collection
Six sampling sites of wastewater have been selected, including three hospital-based and three non-hospital-based wastewater sources covering around 6 km2 area located in the Maijdee, a small town of around 12.61 km2 , in Noakhali district, Bangladesh. The hospital-based wastewater sampling sites were Noakhali 250 Bed General Hospital (GH-1), Mamoni General Hospital (Pvt.) Ltd. (MMH-1), and Metro Hospital (Pvt.) Ltd. (MH-1), and other sources were Waste Dump of Krishnarampur (GH-2), Maijdee Housing Estate Dump Pond (MMH-2), and Noakhali Zilla Mosque Dump Pond (MH-2). Wastewater samples collected from each site immediately taken to the lab in a cool bag and processed within 2 h of collection.
2.2. Isolation and Identification of Bacteria
The samples were processed following the American Public Health Association guidelines [13]. The temperature, pH, and odor were measured and documented. Serial dilutions were made up to 10−6 . Then, 1 mL of each dilution was plated on different selective and differential media, such as on MacConkey agar, Mannitol salt agar (MSA), Eosin methylene blue (EMB), Cetrimide agar, and Thiosulfate-citrate-bile salts-sucrose (TCBS) agar and incubated them at 37°C for 24 h. After obtaining pure colonies, the single random colonies were picked up and proceeded to identify based on their microscopic, physiological, and biochemical characteristics following the “Bergey’s Manual of Determinative Bacteriology,” 8th edition [14].
2.3. Screening of Multiple Antibiotic-resistant Bacteria by Antibiotic Susceptibility Test (AST)
Standard Kirby–Bauer disk diffusion method was performed to determine the antimicrobial susceptibility profiles of the isolates following the CLSI-2018 guidelines [15]. The antimicrobial sensitivity testing was performed against 16 antibiotic disks, azithromycin, AZM (15 μg); gentamicin, GEN (10 μg); ampicillin, AMP (25μg); tobramycin, TOB (10 μg); cefepime, FEP (30 μg); doxycycline, DOX (30 μg); nitrofurantoin, NIT (300 μg); netilmicin, NET (30 μg); ciprofloxacin, CIP (5 μg); cefotaxime, CTX (30 μg); chloramphenicol, CHL (30 μg); amikacin AMK (30 μg); oxacillin, OX (1 μg); streptomycin, STR (10 μg); cefixime, CFM (5 μg); and ceftazidime, CAZ (30 μg) for all the isolates, and additional methicillin, MET (5 μg), disk for only Staphylococcus aureus. The zones of inhibition for each antibiotic were measured in millimeters. Based on the inhibition zone, the isolates were categorized as resistant (R), intermediate (I), and sensitive (S).
2.4. Multiple Antibiotic-resistance (MAR) Indexing
Isolates showed resistance to three or more than three antibiotics are called multidrug resistant (MDR) or multiple antibiotic resistant (MAR) [16]. MAR index value of all the isolates was determined to evaluate the health risk of the environment. The value was calculated according to the formula: Number of antibiotics to which all isolates were resistant (a)/[number of antibiotics tested (b) × number of isolates recovered from a sample (c)] [8].
2.5. Statistical Analysis
To determine the variation of risk among or between hospital and non-hospital wastewater sample groups, we have performed the two-way analysis of variance (ANOVA) using Statistical Package for the Social Sciences (SPSS) software version 25 (SPSS Inc, Chicago, IL, USA). All statistical test values of p < 0.05 were considered statistically significant.
3. RESULTS AND DISCUSSION
Sixty-three bacteria were isolated from wastewater samples of the six selected sites of the Maijdee of the Noakhali district in Bangladesh. The range of pH and temperatures of the wastewater samples was recorded as 6.69 to 8.04 and 28.5 to 36.4°C, respectively. All samples from hospital-based sites were found with malodorous, and the average bacterial load of all sites was found between 3.5×108 and 12.8×108 CFU/mL [Table 1]. Among the bacterial isolates, we found that 30 (47.62%) isolates were resistant to at least six antibiotics and designated as multiple antibiotic-resistant bacteria (MAR) and thus considered for further analysis in this study. Moreover, among the MAR bacteria, 14 (46.67%) from hospital-based sources, and 16 (53.33%) were from non-hospital-based sources. The predominant MAR bacteria were Staphylococcus aureus (n = 6.20%), Escherichia coli (n = 6.20%), Enterobacter spp. (n = 6.20%), and Klebsiella spp. (n = 5, 16.67%). We also found Pseudomonas spp. (n = 2, 6.67%), Proteus spp. (n = 2, 6.67%), Vibrio spp. (n = 1, 3.33%), Shigella flexneri (n = 1, 3.33%), and Staphylococcus epidermidis (n = 1, 3.33%) in relatively lower abundance [Figure 1].
The data of the antibiotic susceptibility test (AST) revealed that 100% (n = 30) of the MAR bacteria were resistant to ampicillin, oxacillin, and cefixime, and 97% (n = 29) of isolates were resistant to cefotaxime. Altogether, above 50% of the isolates were resistant to cefepime, ciprofloxacin, ceftazidime, azithromycin, chloramphenicol, and streptomycin. If we consider the clinically significant pathogenic gram-negative bacteria separately, then E. coli, Enterobacter spp., and Klebsiella spp. showed 100% resistance to ampicillin, oxacillin, cefotaxime, and cefixime, whereas over 50% of them found resistant to azithromycin and ciprofloxacin [Table 2].
S. aureus isolates were highly antibiotic-resistant; out of the 17 treated antibiotics 16.67% and 33.37% of the isolates showed resistance to 14 and 13 antibiotics, respectively. In contrast, 50% Pseudomonas spp., 50% Proteus spp., and 16.67% of S. aureus isolates were resistant to 12 antibiotics. Furthermore, 16.67% E. coli, 50% Enterobacter, 20% Klebsiella spp., and 16.67% of S. aureus were resistant to 11 antibiotics with different combinations. In contrast, 66.66% of E. coli, 20% of Klebsiella spp., and 16.67% of S. aureus were resistant to 10 antibiotics. Simultaneously, 16.67% of E. coli and Enterobacter spp., 20% of Klebsiella spp., 100% of Vibrio spp. and 16.67% of S. aureus showed resistance to 9 antibiotics [Figure 2].
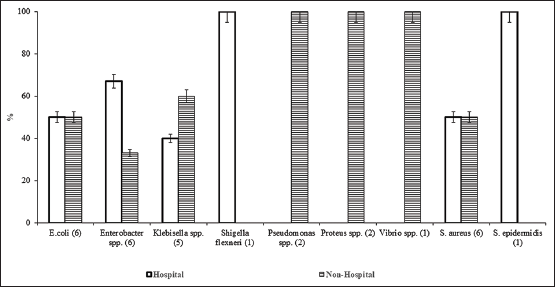 | Figure 1: Distribution of multiple antibiotic-resistant bacteria in hospital and non-hospital-based wastewater. [Click here to view] |
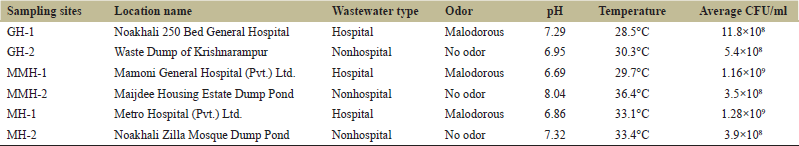 | Table 1: The physical and biological properties of wastewater samples were collected from different sites. [Click here to view] |
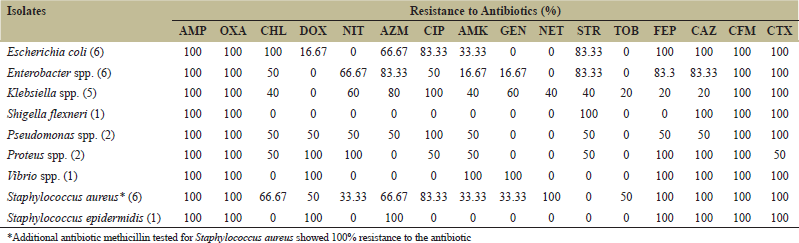 | Table 2: High levels of multiple-antibiotic resistance (MAR) by isolated bacteria against 16 commonly used antibiotics. [Click here to view] |
The multiple antibiotic resistance (MAR) index showed that, the MAR values of all the sites were above 0.20 except for the one site (GH-1), which implied that the isolates were from the high-risk environment. The range of MAR index of hospital source GH-1, MH-1, and MMH-1 were 0.16–0.63, 0.28–0.68, and 0.25–0.62, respectively. On the contrary, the range of MAR index values of non-hospital sources of GH-2, MH-2, and MMH-2 was 0.28–0.75, 0.210.68, and 0.31–0.56, respectively [Table 3]. Two-way analysis of variance (ANOVA) of the MAR data did not find any significant difference in environmental risk between or within the hospital and non-hospital-based wastewater.
Globally, antibiotic resistance in bacteria is a serious health threat, particularly in developing countries [17]. Moreover, antibiotic residues and hospital effluents containing antibiotic-resistant bacteria (ARB) are emerging pollutants of the aquatic environment [18]. These pollutants are excreted into the water reservoirs through the hospitals’ wastewater discharge process and spread antimicrobial resistance on the ecosystem and human health. In particular, wastewater from hospitals, especially if untreated, could play a role in the ARB emerging in the environment. A study by Asfaw et al. [10] reported that out of 84 identified isolates from untreated wastewater samples, 76.2% (64/84) of them were multi-drug-resistant. Similarly, Moges et al. [19] reported that out of 113 identified isolates, the rate of multidrug-resistant bacteria from the hospital and non-hospital sources were 46.9% and 23%, respectively. Our study reported that 47.62% of the recovered isolates as MAR from the hospital and non-hospital sources, and there was no significant difference in MAR isolates from both sources. However, Moges et al. [19] reported that the distribution of MAR isolates in the hospital environment was higher than in non-hospital environments. One possible reason behind this, maybe the area was flooded with rain, and both types of wastewater get mixed up.
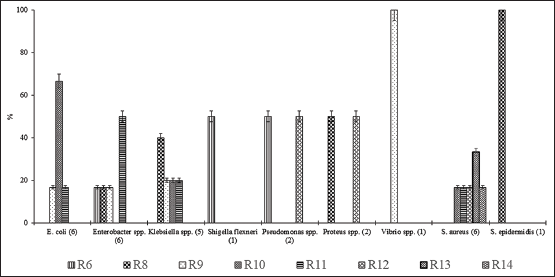 | Figure 2: All multiple antibiotic-resistant bacteria from wastewater showed resistance to 6-13 antibiotics (R5, R7, R8, R9, R10, R11, and R12 denoted that the bacteria were resistant to five, seven, eight, nine, ten, eleven, and twelve antibiotics, respectively). [Click here to view] |
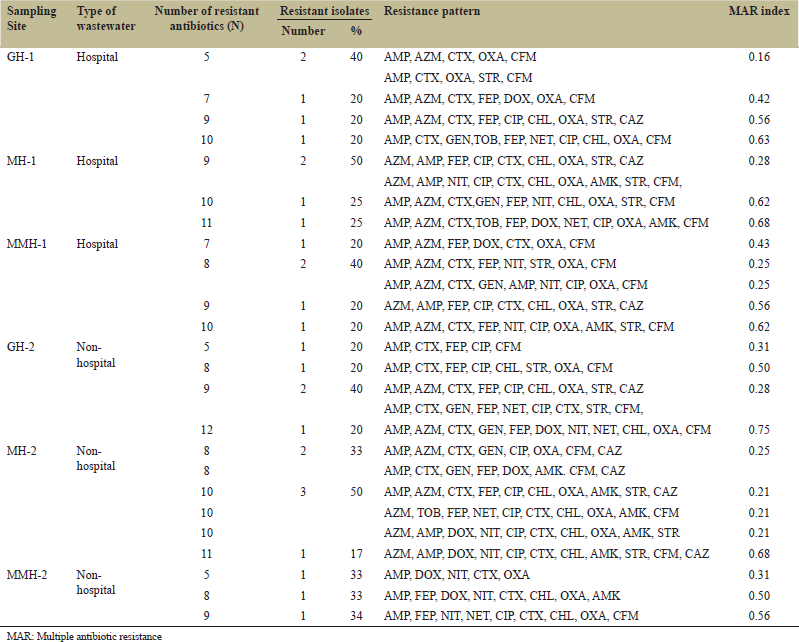 | Table 3: MAR index of isolated bacteria. [Click here to view] |
Furthermore, the higher distribution of such MAR strains in hospital wastewater is due to gram-negative enteric bacteria, which acquires several mechanisms against β-lactams antibiotics [20]. Likewise, two studies in Ethiopia, one in the northwest part of the country, reported Klebsiella spp., followed by Pseudomonas spp., E. coli, Citrobacter spp., and S. aureus as predominant isolates [19], and another from the southern part reported Staphylococcus, E. coli, Klebsiella, and Shigella were most frequently detected isolates in hospital wastewater [21]. A study from another developing country, Nigeria, also reported that bacterial genera, E. coli, Klebsiella, Pseudomonas, and Shigella were the most frequently distributed isolates in wastewater [7]. On the other hand, the most frequent MAR bacteria found in this study were S. aureus (20%), E. coli (20%), Enterobacter spp. (20%), and Klebsiella spp. (16.67%).
Gram-negative bacterial isolates from the hospital and non-hospital water sources were found to be highly resistant to ampicillin (100%), cefixime (100%), oxacillin (100%), cefotaxime (94%), and ceftazidime (84%). Previously, Moges et al. [19] also reported that maximum isolates (97%) of the study showing high resistance to ampicillin from hospital wastewater sources and from non-hospital water sources. Similar results were seen as highly ampicillin, amoxicillin/clavulanic acid-resistant Enterobacteriaceae were reported by Galler et al. [22]. Simachew Dires (2018) also found a higher proportion of ampicillin-resistant Salmonella, E. coli, Shigella, and Klebsiella from hospital wastewater samples [21].
The MAR bacteria in this study showed a high level of resistance to third-generation cephalosporins. Maximum isolates except Proteus spp. were resistant to cefixime, ceftazidime and cefotaxime, whereas Proteus spp. showed total resistance to all but cefotaxime (50%). Surprisingly, 100% of E. coli, Proteus spp., Vibrio spp., S. aureus, and S. epidermidis were even resistant to a fourth-generation cephalosporin (cefepime). Similar to our findings, a study by Rabbani et al. [23] reported a high level of cefotaxime, ceftazidime resistance in E. coli and Klebsiella isolated from two hospital wastewater sources located in Bangladesh. Similar results were seen from non-hospital water sources as cefuroxime, cefotaxime, and ceftazidime resistant Enterobacteriaceae reported by Galler et al. [22]. Undoubtedly, the most successful antibiotic group against which most bacteria established their resistance is β-lactam derivatives such as penicillin, cephalosporin, and ampicillin [24]. Over the time bacteria acquires resistance by several mechanisms such as producing β lactamase enzyme, efflux pump, and producing an altered PBP (penicillin-binding protein) with a lower affinity β-lactam antibiotics [24,25].
Among the aminoglycosides group, the highest resistance was observed by Klebsiella against amikacin (40%) and gentamicin (60%). Interestingly, all E. coli isolates were sensitive against gentamicin. In contrast, Fekadu et al. [26] from Ethiopia reported that E. coli isolated from wastewater showed maximum resistance against gentamicin. However, low levels of gentamicin resistance were also reported in hospital wastewater of India as they noticed that all the Gram-negative isolates from different hospital wastewater sources were less resistant toward gentamicin; the range varied from 2% to 20% [8]. Dires et al. [21] and Ferreira et al. [27] also reported relatively lower resistance among bacterial isolates to gentamicin. Furthermore, against streptomycin, E. coli, and Enterobacter (83.33%) showed high resistance in this study. Similarly, another study in Ethiopia also reported Enterobacter (30%), Klebsiella (29%), and E. coli (8%) isolates showing resistance against streptomycin [19]. Thompson et al. [28] also reported gentamicin and amikacin-resistant S. aureus isolated from two different hospital wastewater sources.
In the case of macrolides (azithromycin), E. coli (66.67%), S. aureus (66.67%), Enterobacter spp. (83%), Klebsiella spp. (80%), Pseudomonas spp. (50%), and S. epidermidis (100%) were found to be resistant. A study conducted in Ethiopia by Simachew Dires [21] reported more than 40% of E. coli, Shigella, Klebsiella, and Salmonella to be erythromycin-resistant where Salmonella showed maximum resistance (75%). In contrast, Gram-positive Staphylococcus spp. showed the lowest resistance (20%) against the drug in our study.
On the other hand, our study’s maximum number of isolates showed resistance toward fluoroquinolones (ciprofloxacin) such as E. coli (83.33%), Enterobacter spp. (50%), and Klebsiella spp. (100%). Several studies in other parts of Bangladesh also reported ciprofloxacin-resistant E. coli and K. pneumoniae from hospital wastewater [23,29,30]. Similar findings were also reported by a study from Portugal that the prevalence of ciprofloxacin resistance was significantly higher in the hospital effluents [31].
Against chloramphenicol, all E. coli isolates showed resistance, whereas more than 50% of Enterobacter spp., Pseudomonas spp., Proteus spp., S. aureus, and 40% of Klebsiella spp. showed resistance to the antibiotic. Dires et al. [21] also reported the isolation of chloramphenicol resistant Enterobacteriaceae from wastewater sources, whereas Alam et al. [8] reported low levels of resistance against chloramphenicol by hospital wastewater isolate. In non-hospital wastewater isolates, Everage et al. [32] reported high level of chloramphenicol resistance among E. coli, S, aureus, and E. cloacae isolated from six different sewage samples collected from the City of Thibodaux (USA) Sewage Treatment Plant.
Among Gram-positive bacteria, S. aureus was predominating isolate in this study. Thompson et al. [28] reported the presence of methicillin-resistant S. aureus (MRSA) (68%, 131/192) from hospital wastewater and water from sewage treatment plants. The study also mentioned MRSA dissemination possibilities from the sewage treatment plant to normal water sources if sewage water was left untreated. Moges et al. [19] also reported S. aureus from wastewater resistant to 11 antimicrobials, including ampicillin (100%) and methicillin (100%). Our data also showed that all MAR S. aureus were resistant to methicillin and oxacillin, which illustrated that the wastewater could be spreading MRSA into the normal water sources. Although, we did not perform further tests to distinguished MRSA or MSSA (methicillin-resistant Staphylococcus aureus).
Multiple antibiotic resistance (MAR) index can indicate a contamination source, especially from high-risk environments such as hospital environments, human, commercial poultry farms, swine, and dairy cattle [8,33]. The MAR value above 0.20 is said to have originated from high-risk areas and originate from an environment where several antibiotics are used [33]. The MAR index of this study showed that all of the isolates were from high-risk environments. These highly resistant isolates in the environment might work as a potential reservoir for transferring resistant genes into other highly infectious pathogens present in the wastewater [23]. Irrational use of antibiotics facilitated the increasing rate of drug resistance among bacteria. The situation is getting worse, especially in developing countries like Bangladesh [17], where all liquid waste originated from hospitals, veterinaries, and other sources directly or through municipality drainage system is discharged into ecological water bodies resulting serious pollutions of the environment with resistant bacteria [34]. The reduction of selective pressure by regulating antibiotic use is a key step to control the spread of resistance in hospital-associated wastewater, not to favor resistant strains [35].
4. CONCLUSION
The study’s findings point out the troubling increase of multidrug resistance in the area’s environmental bacteria. Reducing the risk of mortality, morbidity, and healthcare-associated expense of drug-resistant diseases, we strongly suggest that the hospital wastewater required to be treated appropriately before drain out to the environment, and the other wastewater should be drained in a secluded area and must not go to irrigation field, ponds, canals or waterways directly.
5. ACKNOWLEDGMENT
We thankfully acknowledge the Department of Microbiology’s laboratory support at Noakhali Science and Technology University, Bangladesh.
6. DISCLOSURE STATEMENT
None.
7. FUNDING
None.
8. CONTRIBUTION OF AUTHORS
Rahman MM: Designing the study, laboratory works, and data analysis, writing and critical review of the manuscript. Devnath P: Laboratory works, data analysis, and writing manuscript. Jahan R: Laboratory works, data analysis, and writing manuscript. Talukder A: Writing and critical review of the manuscript.
9. CONFLICTS OF INTEREST
The authors of the study declare that they have no conflicts of interest.
REFERENCES
1. Sharpe M. High on pollution: Drugs as environmental contaminants. J Environ Monit 2003;5:42N-6. CrossRef
2. World Health Organization. No Time to Wait: Securing the Future from Drug-resistant Infections. Report to the Secretary-general of the United Nations. Geneva: World Health Organization; 2019.
3. World Health Organization. Global Priority List of Antibiotic Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization; 2017.
4. Du W, Chen H, Xiao S, Tang W, Shi G. New insight on antimicrobial therapy adjustment strategies for gram-negative bacterial infection: A cohort study. Medicine (Baltimore) 2017;96:e6439. CrossRef
5. Vivas R, Barbosa AA, Dolabela SS, Jain S. Multidrug-resistant bacteria and alternative methods to control them: An overview. Microb Drug Resist 2019;25:890-908. CrossRef
6. Belachew T, Mihret A, Legesse T, Million Y, Desta K. High level of drug resistance by gram-negative bacteria from selected sewage polluted urban rivers in Addis Ababa, Ethiopia. BMC Res Notes 2018;11:524. CrossRef
7. Mustapha A, Imir T. Detection of multidrug-resistance gram-negative bacteria from hospital sewage in North East, Nigeria. Front Environ Microbiol 2019;5:1-7. CrossRef
8. Alam M, Imran M. Screening and potential of multi-drug resistance in gram-negative bacteria from hospital wastewater. Iran J Sci Technol Trans A Sci 2018;42:251-9. CrossRef
9. Marinescu F, Marutescu L, Savin I, Lazar V. Antibiotic resistance markers among gram-negative isolates from wastewater and receiving rivers in South Romania. Rom Biotechnol Lett 2015;20:10055-69.
10. Asfaw T, Negash L, Kahsay A, Weldu Y. Antibiotic resistant bacteria from treated and untreated hospital wastewater at Ayder Referral Hospital, Mekelle, North Ethiopia. J Adv Microbiol 2017;7:871-86. CrossRef
11. Klein EY, Mojica N, Jiang W, Cosgrove SE, Septimus E, Morgan DJ, et al. Trends in methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010-2014. Clin Infect Dis 2017;65:1921-3. CrossRef
12. Lien LT, Hoa NQ, Chuc NT, Thoa NT, Phuc HD, Diwan V, et al. Antibiotics in wastewater of a rural and an urban hospital before and after wastewater treatment, and the relationship with antibiotic use-a one year study from Vietnam. Int J Environ Res Public Health 2016;13:588. CrossRef
13. Rice EW, Baird RB, Eaton AD, Clesceri LS. Standard Methods for the Examination of Water and Wastewater. 22nd ed. Washington, DC: American Public Health Association; 2012.
14. Bergey DH, Buchanan RE, Gibbons NE, American Society for Microbiology. Bergey’s Manual of Determinative Bacteriology. 8th ed., Vol. 26. Baltimore: Williams and Wilkins Co.; 1974. p. 1246.
15. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Discs Susceptibility Tests. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
16. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18:268-81. CrossRef
17. Hart CA, Kariuki S. Antimicrobial resistance in developing countries. BMJ 1998;317:647-50. CrossRef
18. Karkman A, Do TT, Walsh F, Virta MP. Antibiotic-resistance genes in waste water. Trends Microbiol 2018;26:220-8. CrossRef
19. Moges F, Endris M, Belyhun Y, Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res Notes 2014;7:215. CrossRef
20. Egbule OS. Detection and transfer of extended spectrum beta lactamase enzymes from untreated hospital waste water. J Adv Microbiol 2016;6:512-20. CrossRef
21. Dires S, Birhanu T, Ambelu A, Sahilu G. Antibiotic resistant bacteria removal of subsurface flow constructed wetlands from hospital wastewater. J Environ Chem Eng 2018;6:4265-72. CrossRef
22. Galler H, Feierl G, Petternel C, Reinthaler FF, Haas D, Habib J, et al. Multiresistant bacteria isolated from activated sludge in Austria. Int J Environ Res Public Health 2018;15:479. CrossRef
23. Rabbani MAG, Howlader MZH, Kabir Y. Detection of multidrug resistant (MDR) bacteria in untreated waste water disposals of hospitals in Dhaka City, Bangladesh. J Glob Antimicrob Resist 2017;10:120-5. CrossRef
24. Worthington RJ, Melander C. Overcoming resistance to β-lactam antibiotics. J Org Chem 2013;78:4207-13. CrossRef
25. Giani T, Antonelli A, Caltagirone M, Mauri C, Nicchi J, Arena F, et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill 2017;22:30583. CrossRef
26. Fekadu S, Merid Y, Beyene H, Teshome W, Gebre-Selassie S. Assessment of antibiotic- and disinfectant-resistant bacteria in hospital wastewater, South Ethiopia: A cross-sectional study. J Infect Dev Ctries 2015;9:149-56. CrossRef
27. Ferreira da Silva M, Vaz-Moreira I, Gonzalez-Pajuelo M, Nunes OC, Manaia CM. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol Ecol 2007;60:166-76. CrossRef
28. Thompson JM, Gündo?du A, Stratton HM, Katouli M. Antibiotic resistant Staphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin-resistant Staphylococcus aureus (MRSA). J Appl Microbiol 2013;114:44-54. CrossRef
29. Adnan N, Sultana M, Islam OK, Nandi SP, Hossain MA. Characterization of ciprofloxacin resistant extended spectrum β-lactamase (ESBL) producing Escherichia spp. from clinical waste water in Bangladesh. Adv Biosci Biotechnol 2013;4:15-23. CrossRef
30. Akter F, Amin MR, Osman KT, Anwar MN, Karim MM, Hossain MA. Ciprofloxacin-resistant Escherichia coli in hospital wastewater of Bangladesh and prediction of its mechanism of resistance. World J Microbiol Biotechnol 2012;28:827-34. CrossRef
31. Varela AR, André S, Nunes OC, Manaia CM. Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system. Water Res 2014;54:327-36. CrossRef
32. Everage TJ, Boopathy R, Nathaniel R, LaFleur G, Doucet J. A survey of antibiotic-resistant bacteria in a sewage treatment plant in Thibodaux, Louisiana, USA. Int Biodeterior Biodegradation 2014;95:2-10. CrossRef
33. M Kurdi Al-Dulaimi M, Abd Mutalib S, Abd Ghani M, Mohd Zaini NA, Ariffin AA. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among Vibrio vulnificus isolates. Antibiotics (Basel) 2019;8:68. CrossRef
34. Hossain MA, Rahman M, Ahmed QS, Malek MA, Sack RB, Albert MJ. Increasing frequency of mecillinam-resistant Shigella isolates in urban Dhaka and rural Matlab, Bangladesh: A 6 year observation. J Antimicrob Chemother 1998;42:99-102. CrossRef
35. Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, et al. Tackling antibiotic resistance. Nat Rev Microbiol 2011;9:894-6. CrossRef