1. INTRODUCTION
β-lactamases are the type of macromolecular enzymes that hydrolyze the β-lactam ring of most β-lactam antibiotics which can inactivate the antibiotic and confer resistance. The increasing prevalence of multidrug-resistant bacterial infections could be diminished by the development of new antibiotics and β-lactamase inhibitors. Streptomyces genus is recognized as an extraordinary natural source of antibiotics and β-lactamase inhibitors such as clavulanic acid, which is extensively applied in clinical practice [1-3]. The introduction of β-lactamase inhibitors introduced new ways to reverse antibiotic resistance through its unique mechanism [4]. Clavulanic acid was first isolated from Streptomyces clavuligerus, later on few other Streptomyces, namely, S. jumonjinensis and S. katsurahamanus also reported to produce clavulanic acid. Diversity of Streptomyces strains showed the ability to produce clavam metabolites with similar structures to clavulanic acid such as valclavam, clavaminic acid, and clavamycins [5]. Many research programs have focused on isolation of marine natural products and their derivatives for medicinal applications including β-lactamase inhibitors [6,7]. In a recent study, our group has reported a β-lactamase inhibitor molecule kalafungin from marine Streptomyces sp. SBRK1 [8].
The intracellular mechanisms of these actinomycetes control the accumulation of bioactive metabolites, types of nutrients, their concentrations in cellular level, and physiological conditions in the fermentation medium. The selection of appropriate fermentation medium components is particularly important to ensure correct productivity with economically low costs of the production method and to obtain the final product at the highest level of purity and yield [9,10]. It is well known that cell free extracts containing secondary metabolites are significantly influenced by major medium compositions, especially with carbon and nitrogen sources [11,12], salts composition [13], and pH value [14]. Various statistical and non-statistical methods can be used to influence the fermentation process [15]. In the initial stages of classical media optimization process the easy and convenience method, one-factor-at-a- time (OFAT) analysis has been employed, whereas only single component is varied at a time while other variables keeping constant [16,17] However, the regular methods often fail to identify the optimal production conditions because molecular interactions among the factors are neglected. Moreover, they involve an extensive amount of work and time [18]. To overcome these disadvantages, nowadays an effective Response Surface Methodology (RSM) is widely preferred for the optimization of target metabolites production [19]. It can also be used to evaluate the relative significance of several variables simultaneously [20]. Placket–Burman Design (PBD) is a two-level design useful for economically detecting the major effects of all the other interactions are negligible when comparing the important effects, that is, when there are no interactions, the observed effect of a factor can be superior or under estimated by other factors [21]. Optimization of fermentation medium is very complex because any substances that enhance microbial growth and metabolite production can be considered as key variable [22].
The main objective of the present work was to apply the statistical Plackett–Burman Design, Box–Behnken design and non-statistical OFAT methods to optimize the medium composition for the maximum production of β-lactamase inhibitor kalafungin from Streptomyces sp. SBRK1.
2. MATERIALS AND METHODS
2.1. Production and Extraction of β- lactamase Inhibitor
Kalafungin producing strain Streptomyces sp. SBRK1 was isolated from marine sponge Spirostella sp. and cultured in Actinomycetes Isolation Medium (AIM) (GeneBank: KX151717). Agar plate fermentation was employed for secondary metabolite production in AIM medium comprised soluble starch 20 g, KNO3 1 g, NaCl 0.5 g, K2HPO4 0.5 g, MgSO4 0.5 g, FeSO4 20 μM, and distilled water 1000 ml at 28°C for 7 days. 1 × 107 CFU/mL of spore suspension was used as inoculum in the agar plate fermentation and the metabolites were extracted with ethyl acetate by cold percolation method described earlier by Iniyan et al. [23]. β-lactamase inhibition was assessed spectrophotometrically by the hydrolysis of nitrocefin (Merck), a chromogenic substrate using microplate assay method [24].
2.2. Determination of Kalafungin Production by HPLC
The aerial mycelia was collected from the fermentation plates and dried at 60°C. The dried mycelia were weighed to determine the growth of biomass. The collected biomass was extracted using ethyl acetate. The crude extract was filtered and concentrated in a vacuum concentrator (Eppendorf 5301) at 30°C. Kalafungin production in optimization process was quantified with a standard graph by analytical HPLC and the fraction was eluted up to 1 mg. The peak of the compounds was detected at 275 nm. Regression equations and R2 values were calculated from the standard plot to quantify the presence of kalafungin. Twenty microliters of the sample extracted from each optimization step were injected into the chromatographic column. The growth in terms of mycelial biomass formation and kalafungin production was expressed as mg/L of the culture medium.
2.3. Media Optimization by OFAT
Kalafungin yield and biomass production were assessed by substituting the soluble starch in the production medium with different carbon sources such as glucose, sucrose, fructose, galactose, lactose, dextrose, maltose and glycerol. Yeast extract, beef extract, malt extract, sodium nitrate, ammonium nitrate, peptone, ammonium sulfate, and sodium nitrate are the different nitrogen sources used in this study [25]. Tolerance of the strain to NaCl was studied using different concentrations such as 0%, 0.1%, 0.5%, 1%, 2%, 3%, 5%, and 10%, respectively, in the AIM media for 7 days incubation at 28°C. Different incubation period such as 3, 5, 7, 9, 14, and 21 days were used to study the mycelial growth and kalafungin production. To find the optimum pH, the AIM media was adjusted with different pH such as 4, 5, 6, 7, 8, and 9 separately and the culture was inoculated. The strain Streptomyces sp. SBRK1 was incubated in different temperatures (24°C, 28°C, 37°C, and 45°C) and observed for the growth and production of kalafungin using AIM media for 7 days incubation.
2.4. Screening of Medium Ingredients by Plackett–Burman Design (PBD)
Plackett–Burman Design (PBD) was used to screen the ingredients in media for their influence on kalafungin production by Streptomyces sp. SBRK1 [26]. Using the PBD, 9 medium components including chemical and physical parameters (independent variables) were screened by representing them at two levels [Table 1], low (−) and high (+) in 16 trials [Table 2]. All the experiments were carried out in triplicate and the average activity were taken as the response (R1). The high confidence level showing variables were considered to have a significant kalafungin production.
2.5. Optimization of Selected Ingredients by RSM
2.5.1. Box–Behnken design
Box–Behnken design was used to improve the kalafungin production by Streptomyces sp. SBRK1. The design expert trial package (version 11) was used to design the experiment and regression analysis of the obtained data. The three medium conditions that is starch, incubation, and temperature (independent variables) were selected as major input variables at three different levels for low (−), intermediate (0), and high (+) concentrations [Table 3]. The concentrations of the other ingredients and conditions were fixed unchanged at zero level. The experiment were carried out with 20 trials as shown in Table 4 with five replicates at the center point and the results taken with the mean values. Further to predict the optimal value, a second order model was built-in to compare the relationship between the selected variables and response. For the three factors, the equation is
Y = β0+β1 X1+β2 X2+β3 X3+β12 X1 X2+β13 X1 X3+β23 X2 X3+β11 X12+β22 X22+β33 X32
Where Y is the predicted response, β0 model constant; X1, X2, and X3 are independent variables; β1, β2, and β3 are linear coefficients; β12, β13, and β23 are cross product coefficients; and β11, β22, and β33 are the quadratic coefficients [27]. The Box–Behnken designs were further subjected to first order and second order multiple regression analysis by least-square regression analysis to obtain the mathematical model parameters. Significance of experimental values was derived from the software results from model coefficients, R2 values, and F values.
Finally, the combination of different optimized variables identified from the response was experimentally validated by culturing Streptomyces sp. SBRK1 in optimized and unoptimized fermentation medium and the production was quantified.
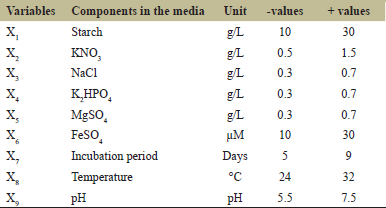 | Table 1: Low and high levels of each variable used in Plackett–Burman design. [Click here to view] |
 | Table 2: Plackett–Burman design and experimental response obtained for the production of kalafungin. [Click here to view] |
 | Table 3: Low, intermediate, and high levels of three independent variables used in RSM. [Click here to view] |
3. RESULTS AND DISCUSSION
3.1. Media Optimization by OFAT
Based on the biomass production and inhibition property, the solid state fermentation with AIM media was utilized for further optimization and production of β-lactamase inhibitor kalafungin from marine sponge symbiotic Streptomyces sp. SBRK1. Initially, the mycelial biomass and kalafungin production was optimized with multiple carbon and nitrogen sources by non-statistical method [28]. In general, the addition of various carbon sources to media may influence cell growth and favors secondary metabolite production [29]. Highest yield of kalafungin was achieved with starch (2%) as a carbon source which gives 28% higher production when compared to other carbon sources tested in the media whereas the growth and active metabolite production was suppressed the media contain dextrose and glucose. In a previous study reported that [30] starch favored for maximum antibiotic production. Glucose has a different role in influencing the antibiotic production by altering the enzymes that are required for metabolite production [31]. Likewise, in this study, it was confirmed that glucose plays the negative effects in kalafungin production. Nitrogen sources (organic and inorganic) always play a vital role in biosynthesis pathway for biologically active compounds production. By this line numerous nitrogen sources such as yeast extract, malt extract, beef extract, sodium nitrate, peptone, ammonium nitrate, ammonium sulfate, and sodium nitrate were tested for improved kalafungin production. Peptone and malt extract were proved as the best nitrogen sources for biomass production, at the same time peptone influences greater production of kalafungin compared to malt extract. S. albovinaceus strain no. 10/2 produces highest ratio of antibacterial metabolite while the basal media was supplemented with malt extract as the nitrogen source [32]. Media supplemented with other organic and inorganic nitrogen source does not favored best growth as well as inhibitor production. Although, it is a marine derived Streptomyces 0.1% NaCl concentration is most suitable for higher yield. While considering pH of the medium, the hydrogen or hydroxyl ion concentration can have a direct effect on cell growth or metabolism in the fermentation medium [33]. The influence of initial medium pH results indicates that the kalafungin production was decreased with decrease in pH of the medium from 4.0 to 6.0; and pH 7.0 was found to be suitable for higher (38.37 ± 0.67 mg/L) kalafungin production. Most of the commercially used Streptomyces species showed optimum pH range from 7 to 8 [34]. The increase in kalafungin production was observed with increase in the incubation time from day 9 to day 14. Due to nutritional depletion and the accumulation of toxic products, the viable cell count and compound production was decreased at 21 days of incubation, the same was reported earlier in the antibacterial metabolite production [35]. The growth and inhibition property was high at 28°C and the growth was arrested at 45°C. The fact of extreme pH and temperature are unfavorable for growth and production is confirmed from the present observations. It is evident that different physiological characteristics are influencing the growth and active biomolecules production of actinomycetes [36]. The effect of a range of selected optimized parameters for biomass production and kalafungin was illustrated in Figure 1. It was observed that production of kalafungin was equivalent with the biomass produced in the fermentation medium.
3.2. Optimization for Essential Medium Ingredients using PBD
Fractional factorial method was used to overcome single-dimensional optimization which is laborious and time consuming. Plackett–Burman design was employed to find the influence of nine different factors on kalafungin production in solid state fermentation. Variation in kalafungin production ranges from 16.6 to 43.4 mg/L in the 16 run and the data obtained from the design were subjected to statistical analysis including regression and analysis of variance (ANOVA). The commercially available β-lactamase inhibitor clavulanic acid production from Streptomyces clavuligerus and Streptomyces sp. DAUFPE 3060 [7] was optimized by this method, based on the same three nutrients their optimal concentrations were obtained using response surface methodology which we have followed here. The ANOVA results revealed that the first order model for kalafungin production was satisfactory and the following model was established by regression:
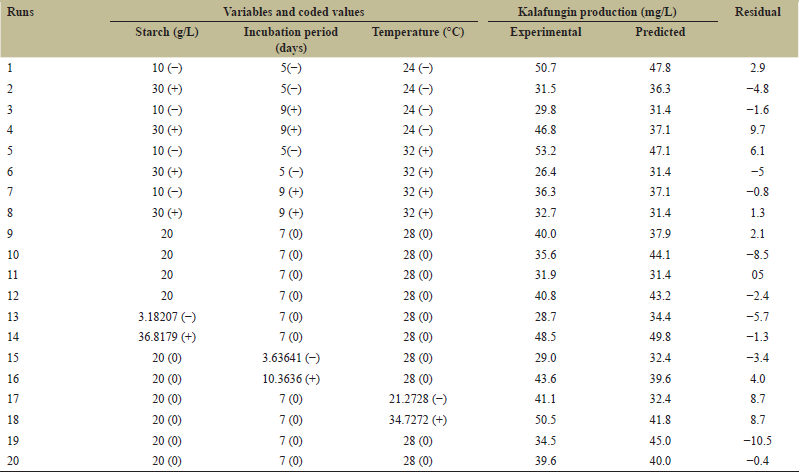 | Table 4: Box–Behnken experimental design and responses for the production of kalafungin by Streptomyces sp. SBRK1. [Click here to view] |
 | Figure 1: The effects of kalafungin production by OFAT method (a) Biomass production and (b) of various selected components in optimized media. [Click here to view] |
Kalafungin (R1) = 48.3232 + 7.11002 * A + 11.8306 * G + 7.23366 * H
Where A is starch, G is incubation period and H is temperature. In this study, the model is significant with the p value 0.024. Values based on ANOVA, the model of regression was found to be significant (P = 0.024), and p-value for starch, incubation period and temperature also found to be significant and shown in Table 5. Based on coefficient estimate of factors, Pareto chart [Figure 2] shows the ranking of chemical or physical factors which influences the growth and kalafungin production. It confirms that the most important factors determining kalafungin production were starch, incubation period, temperature, NaCl, and FeSO4, whereas KNO3, K2HPO4, MgSO4, and pH showed negative effects. From these effects, the high coefficient estimate variables such as starch, incubation period, and temperature were selected to optimize further. The other factors were set as zero level and the negative effects showing sources were excluded for further optimization process.
3.3. Media Optimization for Culture Conditions by Box–Behnken Design
Box–Behnken, a typical Design (BBD) most commonly used RSM analysis, is chosen for optimization process to achieve maximum production of kalafungin. BBD with 20 experiments were performed for three selected factors including starch, incubation period, and temperature at three levels for optimizing kalafungin production in agar plate fermentation. Table 4 shows the design and obtained experimental responses from PBD and the runs produced considerably improved results for kalafungin production. The effects of extraction parameters and production of antimicrobial metabolites from Micromonospora and relative yield was assessed by a three-level three-factor Box–Behnken design and reported [37]. Yang et al. optimized the effective culture conditions of amoxicillin degrading bacteria using RSM [38]. Then, the polynomial model with quadratic process order to describe the correlation between the kalafungin production and the three variable equations as follows:
 | Figure 2: Pareto chart shows the ranking of factors which influences the production of kalafungin, a beta lactamase inhibitor. [Click here to view] |
Kalafungin = 0.0319341 + 0.00155663 * A + −0.00291194 * B + 0.000943945 * C + 0.000175 * AB + −0.0017 * AC + −0.002275 * BC + 0.00162116 * A2 + 0.00441423 * B2 + 0.00386623 * C2
In this test, the obtained model was statistically significant (P <0.0001) based on ANOVA [Table 5].
The lack of fit F-value of this improved model is 0.83 which is not significant when compared to the pure error; hence, it is the required parameter for the improved production of kalafungin. The adjusted R2 value was 0.0659, again this value supports that the experimental data are as predicted positive response. The same enhanced production was also observed and recorded by Chen et al., in the macrolide compound obtained from a marine bacterium [39]. The coefficient estimate was tabulated in Table 6 when all remaining factors are held constant the expected change was observed in factor value of response per unit change. The overall average response for all the runs is the cutoff in an orthogonal design. The variance inflation factor (VIF) is 1; when the factors are orthogonal, the VIFs show multicollinearity when the value is >1, the increase in VIFs is directly proportional to the correlation of factors, the VIFs up to 10 is tolerable. In aid of particular values of the variables along with kalafungin production, that is, response Y a regression model is used to predict future observations.
The error term in Box–Behnken design of kalafungin production was verified by linear patterns, the normal plot of residuals is shown in Figure 3a, as the significant diagnostic model, the normal probability plot of the residuals, came up by default. A linear pattern indicated no signs of any mix-up in the analysis. Plot of observed versus predicted response is shown in Figure 3b. In this analysis, the observed range of variables in kalafungin production is directly proportional to predicted values. Figure 3c showed the plot of externally studentized residuals versus predicted values, this is done to rule out constant error. The random scattering of residuals proved the variance of the original observation was constant and found satisfactory. From all these data, we can infer the verifiable model is efficient to detail the production of kalafungin through response surface. The same prediction within the range of selected variable for clavulanic acid production from Streptomyces clavuligerus MTCC 1142 was found to be suitable, it gives valid evidence to our data [40].
 | Table 5: ANOVA of regression model. [Click here to view] |
 | Table 6: Estimated regression coefficients of the second-order polynomial model. [Click here to view] |
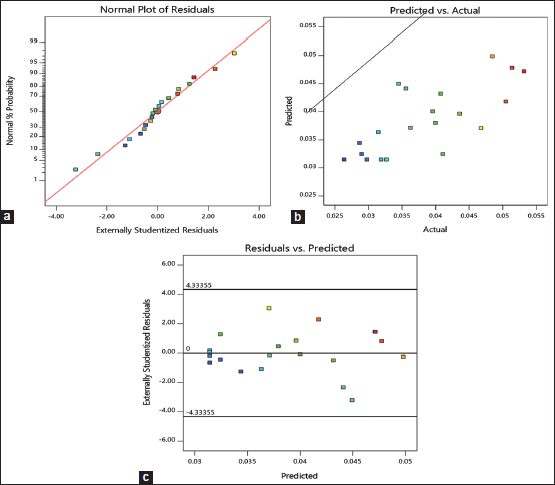 | Figure 3: Residual diagnostics of the quadratic model. (a) Normal probability plot of the externally studentized residuals; (b) the predicted versus observed kalafungin production; (c) plot of internally studentized residuals versus predicted responses. [Click here to view] |
3.4. Response Surface Analysis
In the experimental data to predict the model adequacy and to clarify the signs of any problems, a series of diagnostic plots can be drawn [41]. In the regression equation, the 2D contour plot and the 3D response surface curve are the graphical representation, incorporating two variables which represent the effect on the production of kalafungin while keeping other variables at zero. There is negligible interaction made by the circular plots of response surfaces with the earlier variables. The elliptical or saddle nature of the contour plots confirms the significant interaction between the independent variables. It is clear from Figure 4a-c that there was a significant interaction between starch with incubation period (AB), starch with temperature (AC), and incubation period with temperature (BC). From the three variables starch, incubation period, and temperature, the 3D surface plots will illustrate the pairwise combination, whereas the remaining variable will be held at middle level in the 3D plot which gives the understanding about the interaction effects of these three variables. Increase in incubation period with increase in starch concentration leads to moderate production of kalafungin Figure 4d. Optimum temperature with minimum starch concentration gives more production (4e); the optimum temperature with less days of incubation increases the production, while high temperature affects the metabolite production (4f). The ratio of starch and incubation was not decisive, and it also shows less interaction and had no complementary effects in promoting the production of kalafungin. Moreover, as a typical secondary metabolite, kalafungin mainly accumulated after the rapid growth phase, and starch concentration suitable for growth of Streptomyces sp. SBRK1. The temperature plays an important role in the kalafungin production. This analysis also proved that temperature had significant effect on kalafungin production.
3.5. Experimental Validation
The predicted results obtained from the second order polynomial equation, experimental rechecking was carried out using optimized fermentation conditions with triplicate. Under the optimal conditions [Table 7], the maximum kalafungin production was obtained at 51.4 mg/L and the result was concordant with the predicted value of 50.2 mg/L. Hence, 25.06% increased yield of kalafungin was achieved with 22.621 g/L starch in fermentation media and incubated at 29°C for 8 days which suggests that the RSM model was valid to predict kalafungin production.
3.6. Quality Checking of Kalafungin by RP-HPLC
Finally, the extracted beta lactamase inhibitor kalafungin from Streptomyces sp. SBRK1 by RSM was assessed spectrophotometrically for its inhibition property and analyzed by RP-HPLC. The mobile phase was used as linear gradient with the following chromatographic parameters: Detection – PDA (200–800 nm); mobile phase solvents A – Water and B – methanol; Gradient: 0–2 min 70% A–30% B, 2–10 min 10% A-90% B, 10–20 min 50% A-50% B, and 20-30 min 70% A-30% B with the flow rate of 1 ml/min. The experimentally optimized kalafungin showed 71% inhibition and purity was detected through HPLC isolation with retention time of 28.515 [Figure 5]. It revealed that the production was regulated under optimized condition.
 | Table 7: Kalafungin production from Streptomyces sp SBRK1 based on the combined effect of variables under optimized and unoptimized condition. [Click here to view] |
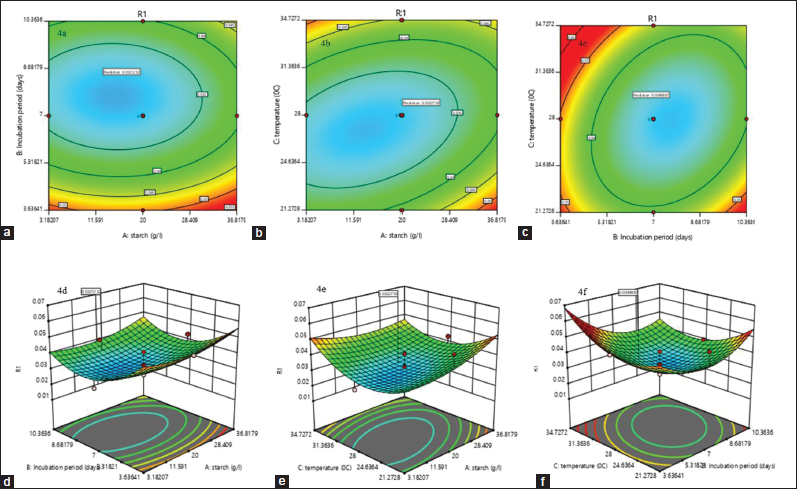 | Figure 4: ContourPlot and response surface plots showing interactive effects of selected variables (a and d) the interaction of starch and incubation period; (b and e) the interaction of starch and temperature; (c and f) the interaction effect of incubation period and temperature; on kalafungin production by Streptomyces sp. SBRK1. [Click here to view] |
 | Figure 5: (a) HPLC profile of kalafungin extracted from Streptomyces sp. SBRK1 under optimized condition. *The peak area of kalafungin with the retention time 28.515 min. (b) HPLC spectrum of pure kalafungin. [Click here to view] |
4. CONCLUSION
RSM designs for optimizing the process parameters have been employed successfully in microbial fermentation for enhanced growth and production of kalafungin. The regular methods used for optimization was inappropriate and failed to consider the interactions between the growth factors. Non-statistical one factor-at-a-time approach gave the betterment of suitable carbon, nitrogen sources, and other physical parameters (pH, temperature, and incubation period) for growth and secondary metabolite production. In this investigation, production of β-lactamase inhibitor kalafungin was enhanced by optimized nutritional and physical growth factors. The maximum kalafungin production of 51.4 mg/L was obtained under optimized conditions than the unoptimized (41.1 mg/L) media with the following optimized factors; starch 22.621 g/L, incubation period of 8 days and temperature at 29°C which is 25.06% more than the unoptimized conditions. The analysis was apparent that PBD and RSM were found more satisfactory for identifying the most significant medium composites with feasible number of experimental trials. Further this study may be useful to develop any large scale fermentation process for the proficient production of targeted metabolite(s) from Streptomyces.
5. AKNOWLEDGMENTS
This work was supported by the Department of Biotechnology, [No.BT/PR8451/MED/29/789/2013] Govt. of India, Drug from Sea, Ministry of Earth Sciences (MoES/09-DS/42015 PC-IV).
6. CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
1. Mohan YS, Sirisha B, Haritha R, Ramana T. Selective screening, isolation and characterization of antimicrobial agents from marine actinomycetes. Int J Pharm Pharm Sci 2013;5:443-9.
2. Narayana KJ, Vijayalakshmi M. Optimization of antimicrobial metabolites production by Streptomyces albidoflavus. Res J Pharmacol 2008;2:4-7.
3. Quinn GA, Banat AM, Abdelhameed AM, Banat IM. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J Med Microbiol 2020;69:1040-8. CrossRef
4. Drawz SM, Papp-Wallace KM, Bonomo RA. New β-lactamase inhibitors: A therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 2014;58:1835-46. CrossRef
5. Nobary SG, Jensen SE. A comparison of the clavam biosynthetic gene clusters in Streptomyces antibioticus Tü1718 and Streptomyces clavuligerus. Can J Microbiol 2012;58:413-25. CrossRef
6. Iniyan AM, Sudarman E, Wink J, Kannan RR, Vincent SG. Ala-geninthiocin, a new broad spectrum thiopeptide antibiotic, produced by a marine Streptomyces sp. ICN19. J Antibiot (Tokyo) 2019;72:99-105. CrossRef
7. de Marques DA, Machado SE, Ebinuma VC, de Duarte CA, Converti A, Porto AL. Production of β-lactamase inhibitors by Streptomyces species. Antibiotics 2018;7:61. CrossRef
8. Mary TR, Kannan RR, Iniyan AM, Ranjith WA, Nandhagopal S, Vishwakarma V, et al. β-lactamase inhibitory potential of kalafungin from marine Streptomyces in Staphylococcus aureus infected zebrafish. Microbiol Res 2020;244:126666. CrossRef
9. Al Farraj DA, Varghese R, Vágvölgyi C, Elshikh MS, Alokda AM, Mahmoud AH. Antibiotics production in optimized culture condition using low cost substrates from Streptomyces sp. AS4 isolated from mangrove soil sediment. J King Saud Univ Sci 2020;32:1528-35. CrossRef
10. Rafieenia R. Effect of nutrients and culture conditions on antibiotic synthesis in Streptomycetes. Asian J Pharm Health Sci 2013;3:3.
11. da Rodrigues KC, de Souza AT, Badino AC, Pedrolli DB, Cerri MO. Screening of medium constituents for clavulanic acid production by Streptomyces clavuligerus. Braz J Microbiol 2018;49:832-9. CrossRef
12. Viana DA, Carneiro-Cunha MN, Araújo JM, Barros-Neto B, Lima-Filho JL, Converti A, et al. Screening of variables influencing the clavulanic acid production by Streptomyces DAUFPE 3060 strain. Appl Biochem Biotechnol 2010;160:1797-807. CrossRef
13. Ser HL, Law JW, Chaiyakunapruk N, Jacob SA, Palanisamy UD, Chan KG, et al. Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: A systematic review. Front Microbiol 2016;7:522. CrossRef
14. Salem-Bekhit MM, Alanazi FK, Alsarra IA. Improvement and enhancement of clavulanic acid production in Streptomyces clavuligerus using vegetable oils. Afr J Biotechnol 2010;9:6806-12.
15. Zhou Y, Han LR, He HW, Sang B, Yu DL, Feng JT, et al. Effects of agitation, aeration and temperature on production of a novel glycoprotein gp-1 by Streptomyces kanasenisi zx01 and scale-up based on volumetric oxygen transfer coefficient. Molecules 2018;23:125. CrossRef
16. Sa-Uth C, Rattanasena P, Chandrapatya A, Bussaman P. Modification of medium composition for enhancing the production of antifungal activity from Xenorhabdus stockiae PB09 by using response surface methodology. Int J Microbiol 2018;2018:3965851. CrossRef
17. Vaidya R, Vyas P, Chhatpar HS. Statistical optimization of medium components for the production of chitinase by Alcaligenes xylosoxydans. Enzyme Microb Technol 2003;33:92-6. CrossRef
18. Xiong ZQ, Tu XR, Tu GQ. Optimization of medium composition for actinomycin X2 production by Streptomyces spp JAU4234 using response surface methodology. J Ind Microbiol Biotechnol 2008;35:729-34. CrossRef
19. Poulikakos P, Falagas ME. Aminoglycoside therapy in infectious diseases. Expert Opin Pharmacother 2013;14:1585-97. CrossRef
20. Latha S, Sivaranjani G, Dhanasekaran D. Response surface methodology: A non-conventional statistical tool to maximize the throughput of Streptomyces species biomass and their bioactive metabolites. Crit Rev Microbiol 2017;43:567-82. CrossRef
21. Singh V, Haque S, Niwas R, Srivastava A, Pasupuleti M, Tripathi CK. Strategies for fermentation medium optimization: An in-depth review. Front Microbiol 2017;7:2087. CrossRef
22. Kavitha G, Kurinjimalar C, Sivakumar K, Kaarthik M, Aravind R, Palani P, et al. Optimization of polyhydroxybutyrate production utilizing waste water as nutrient source by Botryococcus braunii Kütz using response surface methodology. Int J Biol Macromol 2016;93:534-42. CrossRef
23. Iniyan AM, Mary TR, Joseph FJ, Kannan RR, Vincent SG. Cell wall distracting anti-Methicillin-resistant Staphylococcus aureus compound PVI331 from a marine sponge associated Streptomyces. J Appl Biomed 2016;14:273-83. CrossRef
24. Dai X Da, Xiang SH, Li J, Gao Q, Yang KQ. Development of a colorimetric assay for rapid quantitative measurement of clavulanic acid in microbial samples. Sci China Life Sci 2012;55:158-63. CrossRef
25. Azzouz Z, Bettache A, Djinni I, Boucherba N, Benallaoua S. Biotechnological production and statistical optimization of fungal xylanase by bioconversion of the lignocellulosic biomass residues in solid-state fermentation. Biomass Convers Biorefinery 2020;2020:96. CrossRef
26. Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika 1946;33:305-25. CrossRef
27. Box GE, Behnken DW. Some new three level designs for the study of quantitative variables. Technometrics 1960;2:455-75. CrossRef
28. Saurav K, Kannabiran K. Diversity and Optimization of Process Parameters for the Growth of Streptomyces VITSVK9 spp ?solated From Bay of Bengal, India. J Nat Environ Sci 2010;1:56-65.
29. Kiranmayi MU, Sudhakar P, Sreenivasulu K, Vijayalakshmi M. Optimization of culturing conditions for improved production of bioactive metabolites by Pseudonocardia sp. VUK-10. Mycobiology 2011;39:174-81. CrossRef
30. Muhammad SA, Ahmad S, Hameed A. Antibiotic production by thermophilic Bacillus specie SAT-4. Pak J Pharm Sci 2009;22:339-45.
31. Sujatha P, Raju KV, Ramana T. Studies on a new marine streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant Staphylococcus aureus. Microbiol Res 2005;160:119-26. CrossRef
32. Abdelghani T. Production of antibacterial and antifungal metabolites by (S. albovinaceus) strain no.10/2 and media optimization. Am Int J Biol 2017;5:1-24. CrossRef
33. Jain P, Pundir RK. Effect of fermentation medium, pH and temperature variations on antibacterial soil fungal metabolite production. J Agric Technol 2011;7:247-69.
34. Augustine SK, Bhavsar SP, Kapadnis BP. A non-polyene antifungal antibiotic from Streptomyces albidoflavus PU 23. J Biosci 2005;30:201-11. CrossRef
35. Stanbury PF, Whitaker A, Hall SJ. Principles of Fermentation Technology. Netherlands: Elsevier; 2013.
36. Raja A, Prabakarana P. Actinomycetes and drug-an overview. Am J Drug Discov Dev 2011;1:75-84. CrossRef
37. Wang L, Zhang M, Li Y, Cui Y, Zhang Y, Wang Z, et al. Application of response surface methodology to optimize the production of antimicrobial metabolites by Micromonospora Y15. Biotechnol Biotechnol Equip 2017;31:1016-25. CrossRef
38. Yang X, Guo P, Li M, Li H, Hu Z, Liu X, et al. Optimization of culture conditions for amoxicillin degrading bacteria screened from pig manure. Int J Environ Res Public Health 2020;17:1973. CrossRef
39. Chen H, Wu M, Chen Z, Wang M, Lin J, Yang L. Enhancing production of a 24-membered ring macrolide compound by a marine bacterium using response surface methodology. J Zhejiang Univ Sci B 2013;14:346-54. CrossRef
40. Vastrad BM, Neelagund SE. Statistical based media optimization and production of clavulanic acid by solid state fermentation in jackfruit seed powder as novel substrate using Streptomyces clavuligerus Mtcc 1142. Biosci Eng 2014;1:59-77. CrossRef
41. Fang XL, Han LR, Cao XQ, Zhu MX, Zhang X, Wang YH. Statistical optimization of process variables for antibiotic activity of Xenorhabdus bovienii. PLoS One 2012;7:1-11. CrossRef