1. INTRODUCTION
Rapid development and release of untreated wastewater from various metal, electroplating, leather and fertilizer industries, leads to toxicity risk to human survival as well as disrupts ecological balance [1]. In general, chromate-reducing bacteria mostly isolated from chromium-contaminated soil and industrial wastes. Chromium is more toxic in nature and various standard methods have been used to reduce and remove Cr (VI) from the environment. One of the most recent is reduction of Cr (VI) to Cr (III) under aerobic and anaerobic or both conditions by bacteria with less cost and more convenience as trivalent chromium is less toxic than hexavalent chromium and forms insoluble oxides and hydroxides [2,3]. Nowadays, a diversity of bacteria having Cr (VI) reducing properties have been isolated from the environment and some proteins having chromate reducing properties were also puriï¬ed and characterized [4,5] till now. Recent research is focused on some soluble enzymes such as cytoplasmic dimeric flavoproteins have been identiï¬ed as chromate reductases (ChrR), which can fully reduce Cr (VI) – Cr (III) [6,7]. A new approach of chromium reduction can be in situ microbial bioreduction of Cr (VI) to Cr (III) because it may serve as a potential strategy for the detoxification and immobilization of chromate compared with more cost-prohibitive physical and chemical treatment methods [8-10]. Nickel also is the 24th most abundant element and high concentration of this can lead to oxidative stress in cells. Although biosorption, accumulation, precipitation, reduction, and chromate efflux are some of the known mechanisms involved in bioremediation of heavy metals, the best known example of nickel resistance is mediated by efflux pumps such as cnr CBA (cobalt-nickel resistance) from Cupriavidus metallidurans CH34 (formerly Ralstonia metallidurans CH34), NccCBA (Nickel-cobalt-cadmium), NreB (nickel resistance) from Achromobacter xylosoxidans 31A, and CznABC (Cadmium-zinc-nickel) from Helicobacter pylori [11,12]. In future microbes identified in this work can be used for the reduction of chromium and nickel and will be valuable for different bioremediation processes. In this work also the bacterial isolates which were resistant to both chromium and nickel independently was checked for the synergistic effect where it showed resistance to the metals together. This also can be due to some metal resistant genes or proteins [13,14].
2. MATERIALS AND METHODS
2.1. Sample Collection
Soil samples were collected from the East Kolkata Wetland area for three seasons, summer, monsoon, and winter from Brahmi root, chilparajhil area, mathpukurkhal area, metropolitan khal area, Five number jhil area, Seven number jhil area, Natarbheri area, Chingrighata area, Two number jhil area, and Nunebheri area [15,16].
2.2. Soil Sample Preparation
The soil samples were air dried first and then grinding was followed. A mortar or pestle was used for grinding to avoid contamination in the soil sample. After grinding soil samples were sieved using mesh sieve and stored in dry and clean screw cap jars with proper labeling [16,17].
2.3. Soil Parameter Tests
The soil parameter tests include determination of temperature [18], water holding capacity, pH, measurement of electrical conductivity, determination of organic carbon, nitrogen, phosphorous, and presence of heavy metals chromium and nickel [19].
2.4. Isolation of Bacteria from Root Soil
Soil sample collected from Brahmi root was used for the isolation process which falls under Metropolitan area. Samples were diluted in normal saline and inoculated (0.1 ml) on Luria-Bertani (LB) agar plate containing 500–2000 mg/l of potassium dichromate (K2Cr2O7) concentration solutions as Cr(VI) by spread plate method. Plates were incubated at 37°C for 4 days. After 4 days single colonies were selected and preserved in nutrient broth for further studies. Gram staining was performed to check the morphological characteristics of the isolates [20]. For the screening of nickel(II) resistant bacteria, the same chromium-resistant single bacterial colonies were plated on nutrient agar medium with nickel chloride as Ni(II) concentration ranging from 200 to 800 g/L. Again Gram staining was performed to check the morphological characteristics [21].
2.5. Biochemical Characterization of Isolated Bacteria
After obtaining pure culture, biochemical tests were performed for the preliminary characterization purposes. These tests were used to identify the isolate according to the Bergey’s manual of systematic bacteriology [21].
2.6. DNA, Protein Extraction, and 16S rRNA Sequencing
The isolated colonies were subjected to Agarose gel electrophoresis and SDS-Page electrophoresis for the extraction of DNA and protein from the samples. For the identification of positive isolates, 16SrRNA sequencing was performed [22].
2.7. Chromium (Vi) Assay
Hexavalent chromium was determined with a spectrophotometer using the S-diphenylcarbazide (DPC) method. The DPC reagent was prepared by adding 24 ml of 85% H3PO4 to 56 ml distilled water. This solution was mixed with 0.076 g DPC previously dissolved in 20 ml of 95% ethanol. The reagent was stored in dark at 4°C. Cr(VI) in the sample was assayed by adding 125 µl of the DPC reagent to 1 ml of chromium samples, mixed gently and kept at room temperature for 20 min. The absorbance of the color produced was measured at 540 nm using a spectrophotometer. Cr(VI) concentration in the sample was calculated from a standard curve using K2Cr2O7 as standard [23].
2.8. Nickel (ii) Assay
The sample broth was transferred to a clean test tube. 10 ml of citrate ammonia solution, 5 ml of iodine solution, and 20 ml of dimethylglyoxime solution were added to nickel ions solutions. The sample was mixed thoroughly with the prepared solutions. Sample mixed with the chemical solution was transferred to the cuvette and measured absorption at the wavelength of 530 nm. The concentrations of Ni ions were observed and calculated from a standard curve [11].
2.9. Reduction of Chromium and Nickel
Cells from overnight grown culture were harvested by centrifugation at 10,000 RPM for 10 min, washed and suspended in sterile phosphate buffer (0.2 M; pH 7.0). Reduction was carried out in sterile medium (20 ml/100 ml flask) containing 20 mg/l Cr(VI) and Ni (II). The flasks were incubated at 30°C under continuous shaking (120 rpm) with different electron donors such as glucose, glycine, peptone, and Na-acetate and the reduction was estimated following usual method [23,24].
2.10. Screening of Synergistic and Antagonistic Effect of Chromium and Nickel Resistant Bacteria
The bacterial isolates were checked for the resistance to both the metals together. Different concentrations of chromium and nickel (500 mg/l–2000 mg/l) were prepared using potassium dichromate and nickel chloride. Then, both the concentrations were plated and cultures were spread onto the plates. The plates were incubated at 37°C for 3–4 days. Then, the colonies were checked for the chromium and nickel assay as mentioned above [25].
2.11. Antimicrobial Activity of Bacillus cereus
2.11.1. Microorganisms
Staphylococcus hominis (MTCC 10220), Staphylococcus cohnii (MTCC 10219), B. cereus (MTCC 430), E. coli (MTCC 443), and Pseudomonas aeruginosa (MTCC 8076) were used for antimicrobial study. All the stock cultures were collected from CSIR – Institute of microbial technology, Chandigarh, India. All of the bacterial strains were grown and maintained on their specific medium. The bacteria were subcultured overnight for further use.
2.11.2. Antimicrobial activity test
The antimicrobial activity of the isolated B. cereus was determined by disk diffusion Technique. Cotton swab was used to inoculate the test tube suspension onto the surface of nutrient agar plate and the plate was allowed to dry. Using a sterilized forcep, sterilized Whatman paper disks were transferred onto the agar surface. Each sterile disk was impregnated with test organism. Amoxicillin was used as control. The experiment was conducted in triplicates. The plates were incubated at 37°C for 24 h. At the end of the period, the inhibition zone against each microorganism by test organism was measured and analyzed [26,27].
2.12. Statistical Analysis
Triplicate measurements were done in all the cases during the observation and assessment of bacterial growth incorporated with different levels of heavy metals. Data were captured into Microsoft Excel Software, version 2010 which was used to calculate means, standard deviations and standard errors [16]. Pearson’s correlation was also performed for the heavy metal accumulation and reductions using IBM SPSS statistics 22 software.
3. RESULTS AND DISCUSSION
3.1. Soil Parameter Test
The Kolkata Municipal Corporation area generates more than 2500 metric tons of garbage daily, making the soil much more polluted every passing year. This ecosystem having different structural components (water, soil, macrophytes, and plants) acts as ecological features in east Kolkata wetland area. The comparative results obtained from different collected samples show that the temperature is slightly higher in metropolitan khal area, pH and organic carbon content of soil is high in 7 numberjhil area and nunebheriarea. Water holding capacity, Phosphorus, Nitrogen content, and Electrical Conductivity are also higher in nunebheri area, borodhaljhil, chilparajhil, and five numberjhil. Chromium content is higher in chilparajhil area and also Nickel content is high in five numberjhil. pH of the samples collected is in neutral condition, between 7 and 8. On the other hand, nitrogen was found to be similar for each area and the range is between 500 and 800kg/ha. The range of chromium present in all the collected sample is between 2 and 3 mg/kg, whereas nickel content is high in five numberjhil, borodhaljhil, and chilparajhil area, the range is 30–35 mg/kg [Table 1].
The soil sample used for the isolation of heavy metal-resistant bacteria when checked for different parameters have given a satisfactory result indicating a helpful material for plant growth [18,25,28]. The samples collected also consist of a good amount of nutrients such as nitrogen and phosphorus for plant nutrition [29]. However, because of the presence of heavy metals such as chromium and nickel the electrical conductivity value is higher than that of normal [16]. Thus, the microorganisms present in this soil have come up as metal resistant as well as metal reducing agents. The ability of microbial strains to grow in the presence of heavy metals is helpful in many terms including waste water treatment by decomposition of organic matter [30]. Primary study of the collected soil samples for heavy metal resistance ability showed that all samples were positively grown utilizing heavy metals present in their culture media [31-34]. The bacterial isolates were then characterized by morphological, biochemical tests according to Bergey’s manual, DNA and protein extraction, and 16srRNA sequencing [35].
3.2. Isolation of Chromium and Nickel Resistant Bacteria
Chromium and nickel resistance bacteria were isolated using different concentrations of chromium (VI) and nickel (II) by spread plate method. The Brahmi root soil was spread in 500 mg/l–2000 mg/l concentrations and growth was observed till 1500 mg/l whereas no growth was observed in 2000 mg/l [Figure 1a]. The same sample was spread for different concentrations of nickel, from 200 mg/l to 1000 mg/l and growth was observed till 800 mg/l and no growth was observed in 1000 mg/l. Thus, it indicates that bacteria present in this sample can resist chromium and nickel up to 1500 mg/l and 800 mg/l concentrations, respectively [Figure 1b]. From the sample, a total of twenty single colonies were isolated and checked under microscope for morphological and biochemical characterization. The selected potential bacterial isolates resistant to chromium and nickel were subjected to identification by determining its biochemical characteristics as per Bergey’s manual of systemic bacteriology. The isolate was found to be gram positive, catalase positive, rod shaped, spore forming, etc. [Table 2].
Genomic DNA was also extracted from the overnight grown culture of four stressed bacterial sample BRS 5, BRS 11, BRS 17, BRS 19, and one unstressed bacteria B. cereus (MTCC-430). The samples were electrophoresed on 1% agarose gel against 1kb DNA ladder [Figure 2a]. Bacterial samples when exposed to stress condition showed some pigment releasing activity where the color of broth culture changed from yellow to green. Before the molecular weight determination using SDS-PAGE protein concentration of the isolates were checked as per BSA standard curve. As per the result, protein concentration of all the samples increased after the color change [Table 3]. Overnight grown culture of four stressed bacterial sample and one control unstressed sample B. cereus (MTCC-430) electrophoresed using SDS-PAGE electrophoresis for estimation of protein. The protein is separated on the basis of it is molecular weight using protein ladder [Figure 3]. From the biochemical observation, the bacterial isolate was identified as Bacillus sp. Further the result of 16srRNA sequencing was performed which confirms it as B. cereus and the phylogenetic tree is given Figure 2b.
 | Figure-1: Metal-resistant bacterial plates (a = Chromium and Nickel plate with 1500 mg/l and 1000 mg/l Cr (VI) and Ni (II) concentration, b = Chromium and Nickel plate with 500 mg/l and 800 mg/l Cr(VI) and Ni(II) concentration). [Click here to view] |
 | Table 1: Different soil parameters tests of collected soil from some regions of East Kolkata Wetland area. [Click here to view] |
 | Table 2: Biochemical characterization of bacterial strain isolated from East Kolkata Wetland area. [Click here to view] |
 | Table 3: Protein concentration of different bacterial isolates using BSA standard curve. [Click here to view] |
Depending on the tests, the isolates were identified as gram positive and the sequencing proved that the strain is B. cereus (BF2) accession number KU955350.1. Bacterial growth was also observed in the presence of heavy metals chromium and nickel using different concentrations that were studied with bacterial growth [24,36,37]. For determination of bacterial genetic information, DNA profiling is important to locate the resistant genes in chromosomes. In our study, the positive strain was used for DNA extraction and observed against 1 kb ladder. The positive isolates were also checked for their protein profiling to understand the molecular weight of the genetic material to confirm the metal resistant genes [38]. According to some reports, ChrA and NerB genes are responsible for chromium and nickel resistance and the molecular weight varies from 43kDa to 66kDa. As per our results, the strains also showed bands between 48 kDa and 65 kDa which confirms the presence of the genes in bacterial sample also the concentration of protein was estimated according to the BSA standard curve [39].
3.3. Accumulation and Reduction of Cr(VI) and Ni(II) in BRS 5, BRS 11, BRS 17, and BRS 19
The current status of the bioremediation of heavy metal is much promising for metal biosorption and detoxification using genetically modified microbes. Biofilm-mediated techniques, microbial gene transfer, and microbial fuel cells-based techniques can be considered as strong approaches in coming years. The peptidoglycan and polysaccharides component presents in the microbial cell wall act as a great binding site for metals help in metal uptake as well as biosorbent [40]. However, some research focuses on the role of bacterium in reducing the heavy metal chromium and nickel from different soil samples and this bacterium also acts as chelating agents. According to some reports, P. aeruginosa and Lactobacillus plantarum MF042018 is also a potent bacterial strain involved in removal of heavy metals. P. aeruginosa was also recorded high removal percentage of different heavy metals at optimum condition for growth such as cadmium, lead, mercury, copper, and zinc other than chromium and nickel [41,42].
Here, in twenty isolated colonies the concentration of nickel is higher than that of chromium [Figure 4]. Influence of different Electron donor (glucose, glycine, Na-acetate, and peptone) on chromium (VI) concentration (20 mg/l) by different time intervals of BRS 5, BRS 11, BRS 17, and BRS 19 bacterial strains [Figure 5a-d]. Influence of different electron donor (glucose, glycine, Na-acetate, and peptone) on nickel (II) concentration (20 mg/l) by different time intervals of BRS 5, BRS 11, BRS 17, and BRS 19 bacterial strains [Figure 6a-d].
Chromium-resistant bacteria that can also reduce the concentrations have been reported earlier in different regions of India and also outside India of different oil contaminated soils but nickel reduction was not so common. The present study clearly indicates the presence of chromium and nickel resistant as well as reducing bacteria in soil of East Kolkata Wetland area. The most potent strain has been identified as B. cereus reduced chromate using different electron sources such as glucose, glycine, Na-acetate, and peptone. As per the result, all the sources have shown a good reduction rate; however, Na-acetate and peptone have shown a remarkable change in reduction of chromium (VI) [43], whereas in nickel glycine along with Na-acetate and peptone has shown a significant reduction rate. The positive isolates when checked with both chromium and nickel concentrations together they have interestingly shown some synergistic and antagonistic effect as well. Previously, the isolates were able to resist chromium and nickel up to 1500 mg/l and 800 mg/l, respectively. However, when the metals were used together the resistant capacity for chromium was reduced to 1000 mg/l whereas the resistant capacity of nickel was increased to 1000 mg/l indicating antagonistic and synergistic effects [44]. For the removal of heavy metals such as chromium and nickel, microbial remediation and reduction is one of the most viable and sustainable methods. For attenuation of excess Cr(VI) accumulation in the environment. Based on the current study, it can be said that for the removal of heavy metal and speciï¬cally Cr(VI) much more work at bigger scale is needed to develop reliable and cost-effective technologies for this problem [45].
 | Figure 2: Identification of bacterial strain (a) genomic DNA of different bacterial isolates on 1% agarose TAE gel. (b) Phylogenetic tree of the bacterial isolate by 16SrRNA sequencing. [Click here to view] |
3.4. Antimicrobial Activity of Isolates
The antimicrobial activity of isolated BRS 5, BRS 11, BRS 17, and BRS 19 was determined by the disk diffusion method against five different bacterial strains. A standard amoxicillin antibiotic was used in this study as control. None of the samples showed activity against P. aeruginosa MTCC 8076, whereas only BRS 17 showed activity against E. coli MTCC 10220 which indicates it as a potent antimicrobial organism. All four samples showed antimicrobial activity against B. cereus, S. cohnii, and S. hominis [Figure 7].
 | Figure 3: SDS-PAGE (10%) separation of bacterial isolates. [Click here to view] |
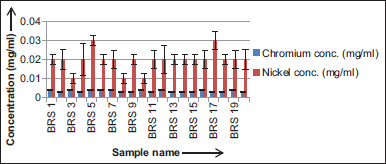 | Figure 4: Cr(VI) and Ni(II) concentration in bacterial isolates – Standard deviation graph of chromium and nickel concentration in bacterial species. [Click here to view] |
The isolates were checked for their antimicrobial activity against different microorganisms collected from CSIR-Institute of microbial technology, Chandigarh, India, where all four samples showed antimicrobial activity against B. cereus, S. cohnii and S. hominis and only BRS 17, which also showed morphological and biochemical characters same as B. cereus, showed activity against E. coli MTCC 10220 indicating it as a potent antimicrobial organism [27,46-48].
3.5. Statistical Analysis using SPSS
The IBM SPSS statistics 22 software was used to understand the following correlation matrix between different electron donors used to accumulate and reduce the metal concentrations of different soil samples using bacterial culture. This test actually is performed to measure the statistical relationship or association between two or more continuous variables as well as the direction of the relationship. As the metals containing different electron sources shown a good result to accumulate and also some synergistic characteristics also came up this correlation was done to check if there is any significant relationship present between them or not. According to correlation matrix, P ≤ 0.05 for the result to become significant and the test is not significant if P ≥ 0.05. As per the table shown below, there is a positive significance present between all the sources used for the accumulation and reduction proving its synergistic effect. Here, two different cultures were used for the analysis of correlation, that is, B. cereus (BRS 11) and also BRS 17, which is morphologically same as BRS 11 and also showed a good reduction and accumulation of metals. As per the following result in BRS 11, all the donors showed significant relationship between each other at 0.01 levels [Table 4], whereas in BRS 17 the relationship was significant at 0.01 and 0.05 levels [Table 5].
 | Figure 5: Reduction of Cr(VI) by different electron donor sources. (a) Reduction of Cr (VI) influence by glucose source as electron donor. (b) Reduction of Cr (VI) influence by Glycine source as electron donor. (c) Reduction of Cr (VI) influence by Na-acetate source as electron donor. (d) Reduction of Cr (VI) influence by peptone source as electron donor. [Click here to view] |
 | Table 4: Pearson’s correlation matrix of BRS 11 (Bacillus cereus) culture by IBM SPSS 22. [Click here to view] |
 | Figure 6: Reduction of Ni (II) by different electron donor sources. (a) Reduction of Ni (II) influence by glucose source as electron donor. (b) Reduction of Ni (II) influence by Glycine source as electron donor. (c) Reduction of Ni (II) influence by Na-acetate source as electron donor. (d) Reduction of Ni (II) influence by Peptone source as electron donor. [Click here to view] |
 | Table 5: Pearson’s correlation matrix of BRS 17 culture by IBM SPSS 22. [Click here to view] |
 | Figure 7: Antimicrobial activity of different bacterial isolates against some specific Gram (+ve) and Gram (−ve) bacteria such as Pseudomonas aeruginosa (MTCC 8076), Staphylococcus cohnii (MTCC 10219), Staphylococcus hominis (MTCC 10220), E. coli (MTCC 443) and Bacillus cereus (MTCC 430). [Click here to view] |
4. CONCLUSION
The ability of microbial stains to grow in the presence of heavy metals would be helpful in the contaminated soil treatment. The isolated strain B. cereus is characterized with remarkable tolerance against heavy metals chromium and nickel and when both were mixed together it showed some synergistic as well as antagonistic effects. It can be used as potential agents for the development of a soil inoculant applicable in bio augmentation, biosorption, accumulation, and bioremediation of heavy metals in polluted sites.
5. AUTHORSHIP AND ACKNOWLEDGEMENTS
S.D.; performed the experiments, analysis of data for the work and co-wrote the paper. K.F.; interpretation and analysis of the data and co-wrote the paper. S.C.; supervised the research.
6. ACKNOWLEDGMENT
The authors acknowledge Dr. Avijit Sengupta, director and Dr.Swati Chakraborty assistant professor of Guru Nanak Institute of Pharmaceutical Science and Technology who gave me this opportunity to do this wonderful research work.
7. FUNDING SOURCE
The author acknowledge the financial support from the Department of Science and Technology, Government of West Bengal vide sanction Number 35 (Sanc)/ST/P/S&T/2G-5/2015.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
Not applicable.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Beszedits S. Chromium removal from industrial wastewaters. In: Chromium in the Natural and Human Environments. Vol. 13. New York: Wiley; 1989. p. 232-63. CrossRef
2. Sheppard SC. Chromium in natural and human environments (Advances in environmental sciences and technology, volume 20). J Environ Q 1989;18:132-3. CrossRef
3. Srinath T, Khare S, Ramteke PW. Isolation of hexavalent chromium-reducing facultative anaerobes from tannery effluent. J Gen Appl Microbiol 2001;47:307-12. CrossRef
4. Kabir MM, Fakhruddin AN, Chowdhury MA, Pramanik MK, Fardous Z. Isolation and characterization of chromium (VI)-reducing bacteria from tannery effluents and solid wastes. World J Microbiol Biotechnol 2018;34:1-17. CrossRef
5. Ackerley DF, Gonzalez CF, Park CH, Blake R, Keyhan M, Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl Environ Microbiol 2004;70:873-82. CrossRef
6. Ansari MI, Grohmann E, Malik A. Conjugative plasmids in multi-resistant bacterial isolates from Indian soil. J Appl Microbiol 2008;104:1774-81. CrossRef
7. Bhattacharya A, Gupta A. Evaluation of Acinetobacter sp. B9 for Cr(VI) resistance and detoxification with potential application in bioremediation of heavy-metals-rich industrial wastewater. Environ Sci Pollut Res 2013;20:6628-38. CrossRef
8. Campos VL, Moraga R, Yanez J, Zaror CA, Mondaca MA. Chromate reduction in Serratiam arcescens isolated from tannery effluent. Bull Environ Contam Toxicol 2005;75:400-6. CrossRef
9. Cheung KH, Gu JD. Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: A review. Int Biodeteriorior Biodegradat 2007;59:8-15. CrossRef
10. Essahale A, Malki M, Marin I, Moumni M. Hexavalent chromium reduction and accumulation by Acinetobacter AB1 isolated from Fez tanneries in Morocco. Indian J Microbiol 2012;52:48-53. CrossRef
11. Alboghobeish H, Tahmourespour A, Doudi M. The study of nickel resistant bacteria (NiRB) isolated from wastewaters polluted with different industrial sources. J Environ Health Sci Eng 2014;12:1-7. CrossRef
12. Chauhan M, Solanki M. Isolation and molecular characterization of three Bacillus strains for their tolerance against various heavy metals. J Microbiol Biotechnol Food Sci 2017;6:1065-9. CrossRef
13. Fulladosa E, Desjardin V, Murat JC, Gourdon R, Villaescusa I. Cr(VI) reduction into Cr(III) as a mechanism to explain the low sensitivity of Vibrio fischeri bioassay to detect chromium pollution. Chemosphere 2006;65:644-50. CrossRef
14. Parisa K, Mehran H, Arezoo T. Multimetal resistance study of bacteria highly resistant to Mercury isolated from dental clinic effluent. Afr J Microbiol Res 2011;5:831-7. CrossRef
15. Dutta S, Chakraborty S. The effect of pollution on hydrological parameters analysis of East Kolkata Wetland area. Int J Dev Res 2017;7:15452-8.
16. Firdous K, Chakraborty S. Correlation of different soil parameters and evaluation of its effect in a ramsar site East Kolkata Wetland. Int J Sci Res Sci Technol 2018;4:966-76.
17. Singh KS, Athokpam H. Physico-chemical characterization of farmland soil in some villages of chandel hill district, Manipur (India). Int J Curr Microbiol Appl Sci 2018;7:417-25. CrossRef
18. Viana RM, Ferraz JBS, Neves AF, Vieira G, Pereira BFF. Soil quality indicators for different restoration stages on Amazon rainforest. Soil Tillage Res 2014;140:1-7. CrossRef
19. Dregne HE, Willis WO. American Society of Agronomy, Crop Science Society of America. Vol. 148. Madison, Wisconsin, USA: Soil Science Society of America; 1985. p. 590-1.
20. Ilias M, Rafiqullah IM, Debnath BC, Mannan KS, MozammelHoq M. Isolation and characterization of chromium(VI) reducing bacteria from tannery effluents. Indian J Microbiol 2011;51:76-81. CrossRef
21. Ge S, Zhou M, Dong Z, Lu Y, Ge S. Distinct and effective biotransformation of hexavalent chromium by a novel isolate under aerobic growth followed by facultative anaerobic incubation. Appl Microbiol Biotechnol 2013;97:2131-7. CrossRef
22. Bru D, Martin-Laurent F, Philippot L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S r RNA gene as an example. Appl Environ Microbiol 2008;74:1660-3. CrossRef
23. Hora A, Shetty KV. Inhibitory and stimulating effect of single and multi-metal ions on hexavalent chromium reduction by Acinetobacter sp. Cr-B2. World J Microbiol Biotechnol 2014;30:3211-9. CrossRef
24. Garg SK, Tripathi M, Srinath T. Strategies for chromium bioremediation of tannery effluent. Rev Environ Contam Toxicol 2012;217:75-140. CrossRef
25. Ibrahim AS, El-Tayeb MA, Elbadawi YB, Al-Salamah AA. Isolation and characterization of novel potent Cr(VI) reducing alkaliphilic Amphibacillus sp. KSUCr3 from hypersaline soda lakes. Electron J Biotechnol 2011;14:1-15. CrossRef
26. Senthilkumar P, Sudha S. Antioxidant and antibacterial properties of methanolic extract of green seaweed Chaetomorpha linum from gulf of Mannar: Southeast coast of India. Jundishapur J Microbiol 2012;5:411-5. CrossRef
27. Imada C, Koseki N, Kamata M, Kobayashi T, Hamada-Sato N. Isolation and characterization of antibacterial substances produced by marine actinomycetes in the presence of seawater. Actinomycetologica 2007;21:27-31. CrossRef
28. Parvathy CA. Study on the ichthyofaunal diversity of kole wetlands of puzhakkalkole with emphasis on its water quality parameters, Kerala. Int J Res Appl Sci Eng Technol 2018;6:612-21. CrossRef
29. Klimek B. Effect of long-term zinc pollution on soil microbial community resistance to repeated contamination. Bull Environ Contam Toxicol 2012;88:617-22. CrossRef
30. Kavita B, Keharia H. Reduction of hexavalent chromium by Ochrobactrum intermedium BCR400 isolated from a chromiumcontaminated soil. 3 Biotech 2012;2:79-87. CrossRef
31. Narasimhulu K, Sreenivasa P, Vinod A. Isolation and identification of bacterial strains and study of their resistance to heavy metals and antibiotics. J Microb Biochem Technol 2010;2:74-6. CrossRef
32. Nandi S, Srivastava RC, Agarwal KM. Lead and Cadmium accumulation in fresh water fishes Labeorohita and Catlacatla. J Environ Res Dev 2012;6:748-53.
33. Nandi S, Srivastava RC, Agarwal KM. Bioaccumulation of heavy metal in Brassica oleracea effects on human health. Int J Med Sci 2012;5:21-3.
34. Nandi S, Srivastava RC, Agarwal KM. Heavy metal concentration in surface water of east Kolkata Wetland. Indian J Public Health Res Dev 2013;4:227. CrossRef
35. Liu YG, Xu WH, Zeng GM, Li X, Gao H. Cr(VI) reduction by Bacillus sp. isolated from chromium landfill. Proc Biochem 2006;41:1981-6. CrossRef
36. Gokarn K, Pal R. Activity of siderophores against drug resistant gram-positive and gram-negative bacteria. Infect Drug Resist 2018;11:61-75. CrossRef
37. Roane TM. Lead resistance in two bacterial isolates from heavy metal contaminated soil. Microb Ecol 1999;37:218-24. CrossRef
38. Masood F, Malik A. Hexavalent chromium reduction by Bacillus sp. strain FM1 isolated from heavy-metal contaminated soil. Bull Environ Contam Toxicol 2011;86:114-9. CrossRef
39. Cervantes CM, Hadjeb N, Newman L, Price CA. ChrA is a carotenoid-binding protein in chromoplasts of Capsicum annuum. Plant Physiol 1990;92:1241-3. CrossRef
40. Igiri BE, Okoduwa SR, Idoko GO, Akabuogu EP, Adeyi AO, Ejiogu IK. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery waste water: A review. J Toxicol 2018;2018:16. CrossRef
41. Abdelbary S, Elgamal MS, Farrag A. Trends in Heavy Metals Tolerance and Uptake by Pseudomonas aeruginosa. London: IntechOpen; 2019. p. 85875. CrossRef
42. Ameen FA, Hamdan AM, El-Naggar MY. Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci Rep Nat Res 2020;10:314. CrossRef
43. Dorian A, Hernandez B, Burgos LIR, Paramo ED, Linares LF. Zinc and lead biosorption by Delftia tsuruhatensis: A bacterial strain resistant to metals isolated from mine tailings. J Water Resour Prot 2012;4:207-16. CrossRef
44. Aboudrar W, Schwartz C, Morel JL, Boularbah A. Effect of nickel-resistant rhizosphere bacteria on the uptake of nickel by the hyperaccumulator Noccaea caerulescens under controlled conditions. J Soils Sediment 2013;13:501-7. CrossRef
45. Bhattacharya A, Gupta A, Kaur A, Malik D. Alleviation of hexavalent chromium by using microorganisms: Insight into the strategie sand complications. Water Sci Technol 2019;79:411-24. CrossRef
46. Ashwini U, Asha S. In vitro antibacterial activity of Acalypha indica Linn leaves extract against gram negative bacteria. Int J Res Ayurveda Pharm 2017;8:195-8. CrossRef
47. Yilmaz M, Soran H, Beyatli Y. Antimicrobial activities of some Bacillus spp. strains isolated from the soil. Microbiol Res 2006;161:127-31. CrossRef
48. Bizani D, Brandelli A. Characterization of a bacteriocin produced by a newly isolated Bacillus sp. Strain 8 A. J Appl Microbiol 2002;93:512-9. CrossRef