The Salacia genus belongs to the family Celastraceae. Several species belonging to this genus have been used in herbal medicine to treat primarily diabetes, obesity, and rheumatism. Salacia contains bioactive compounds that are effective antidiabetic, antiobesity, hepatoprotective, and antioxidant agents of tremendous medicinal value [1]. Salacia species have been widely traded as raw herbal medicines in India, Japan, China, and Korea due to their use in traditional medicine [2]. There are many plants in the family Celastraceae that have medicinal properties. Some plants that are used in herbal medicine in India and other parts of the world are Cassine glauca, Celastrus paniculatus, Euonymus indica, Lophopetalum wallichianum, Gymnosporia montana, Maytenus heyneana, Microtropis wallichiana, and Pleurostylia opposita.
The Western Ghats is one of the world’s major biodiversity hotspots [3], and due to high biodiversity and endemism, areas of South India have attracted attention [4]. With high endemism and species richness in the Western Ghats, taxonomists have had a problem with proper identification, and this problem is profound when the samples are collected in their vegetative state. Eight species of the genus Salacia have been described in the “Flora of Karnataka” by Saldanha and Larsen [5], namely, Salacia beddomei, Salacia chinensis, Salacia fruticosa, Salacia macrosperma, Salacia malabarica, Salacia oblonga, Salacia reticulata, and Salacia talbotii.
The status of conservation for S. beddomei, S. macrosperma, and S. malabarica has been labeled vulnerable, rare, and endangered respectively (http://florakarnataka.ces.iisc.ac.in). The status “Vulnerable” is given to S. oblonga, which is endemic to the Western Ghats of India and Sri Lanka, by the International Union for Nature Conservation. Habitat destruction by human activities through land clearing, for agriculture, for construction of the residence, and expansion of roads and railways, along with indiscriminate collection for its use in various medicinal applications, is the primary cause of the decline of S. oblonga. Species of Salacia are rarely found in deciduous forests. In coastal forests, S. chinensis and S. fruticosa are usually found S. macrosperma and S. oblonga inhabit evergreen forests. Euonymus species such as Euonymus angulatus, Euonymus assamicus, Euonymus paniculatus, Euonymus serratifolius, and Cassine viburnifolia native to India have also been documented as endangered and vulnerable.
Due to the high level of morphological similarities between species, the genus Salacia is often difficult to identify [5]. Dev and Anoop [6], and Devipriya and Devipriya [7], have used internal transcribed spacer (ITS) sequences to carry out phylogenetic studies for eight species and one variety of the genus Salacia collected from Kerala. In addition to ITS sequences, Dev and Anoop [6] have used rbcL, matK, and trnH-psbA sequences in their research.
Using traditional taxonomy, the task of proper identification becomes arduous for the very closely related species. DNA barcoding, irrespective of environmental factors affecting the growth, development, and morphology of an organism, is a complementary method for quick and accurate species identification. Inter-simple sequence repeat (ISSR) is a fast and very sensitive technique in detecting polymorphisms at intra- and interspecific levels of variation using oligonucleotide primers [8]. As the method utilizes longer semi-arbitrary simple sequence repeats primers in high-stringency polymerase chain reaction (PCR) conditions, ISSRs are highly reproducible and polymorphic [9]. The ITS sequence is a popular choice for phylogenetic analyses [10] since it is amplified by a universal set of primers, and also ITS region is a simple sequence which is biparentally inherited, intragenomically uniform, and intergenomically variable with low functional constraints having a length of 500–700 base pairs [11].
The Plant Working Group (2009) of the Consortium for the Barcode of Life (CBOL) proposed multilocus barcoding as an efficient method for barcoding and species discrimination in plants. For plant barcoding, CBOL recommends matK and rbcL regions to be the most effective universal barcodes. In the current study, using ISSR marker, we have assessed the diversity of four Salacia species along with eight genera (C. glauca, C paniculatus, E. indica, L. wallichianum, G. montana, M. heyneana, M. wallichiana, and P. opposita) belonging to the family Celastraceae sampled from Western Ghats of Karnataka and Kerala. Furthermore, three DNA barcodes (matK, rbcL, and ITS) were used to evaluate their ability for species discrimination within Salacia species.
2. MATERIALS AND METHODS
2.1. Collection of Plant Material and DNA Extraction
From different parts of the Western Ghats, leaves of Salacia species and related genera were collected [Table 1]. DNA was extracted by following a modified Stange et al. method [12].
Table 1: Isolate code and sampling area in the Western Ghats.
| Name | Sample code | Sampling area | Latitude (oN) | Longitude (oE) | Elevation (M) | Reference |
|---|---|---|---|---|---|---|
| Cassine glauca | CG1 | Bisle Ghats, Hassan district | 12° 43’ 51.42’’ | 75° 42’ 41.18’’ | 934 | This study |
| Celastrus paniculatus | CP1 | Hunsur, Mysore district | 12° 7’ 54.41’’ | 76° 10’ 7.30’’ | 781 | |
| CP3 | Mysore Zoo, Mysore district | 12° 18’ 9.95’’ | 76° 39’ 48.98’’ | 748 | ||
| Euonymus indica | EI | Bisle Ghats | 12° 41’ 59.26’’ | 75° 38’ 25.12’’ | 838 | |
| Lophopetalum wallichianum | LW1 | Mysore Zoo, Mysore district | 12° 18’ 9.95’’ | 76° 39’ 48.98’’ | ||
| LW2 | Somwarpet, Kodagu district. | 12° 40’ 28.14’’ | 75° 48’ 7.30’’ | 1106 | ||
| Gymnosporia montana | GC | Bisle Ghats, Hassan district | 12° 42’ 42.70’’ | 75° 41’ 40.62’’ | 803 | |
| Maytenus heyneana | ME | Virajpet, Kodagu district | 12° 15’ 20.28’’ | 75° 45’ 40.82’’ | 907 | |
| Microtropis wallichiana | MS | Sakleshpur, Hassan district | 12° 55’ 7.21’’ | 75° 45’ 4.7952’’ | 914 | |
| Pleurostylia opposita | P | Chamundi Hills, Mysore district | 12° 16’ 21.28’ | 76° 40’ 15.0132’’ | 1024 | |
| Salacia chinensis | SC1, SC2, SC3, SC4, SC5 | Udupi district | 13° 21’ 42.39’’ | 74° 46’ 2.0’’ | 21 | (Bajpe et at. 2018) |
| Hippocrateaceae | SC6, SC7, SC8, SC9, SC10, SC11 | Makuta, Madikeri district | 12° 4’ 29.87’’ | 75° 43’ 31.10’’ | 115 | This study |
| Salacia macrosperma | SM1 | Virajpet, Madikeri district | 12° 11’ 41.94’’ | 75° 48’ 14.40’’ | 907 | (Bajpe et al., 2018) |
| SM2, SM3 | Shanivarsanthe, Kodagu district. | 12° 43’ 35.96’’ | 75° 53’ 8.92’’ | 965 | This study | |
| SM4 | Abbi Falls, Madikeri district | 12° 27’ 29.54’’ | 775° 43’ 10.38’’ | 108 | (Bajpe et al., 2018) | |
| SM5 | Madikeri town, Madikeri district | 12° 25’ 27.91’’ | 75° 44’ 17.46’’ | 1128 | ||
| SM6 | Bisle Ghats, Hassan district | 12° 42’ 33.46’’ | 75° 40’ 38.80’’ | 614 | ||
| SM7 | Kigga, Chikkamagaluru district | 13° 25’ 1.53’’ | 75° 11’ 17.15’’ | 664 | ||
| SM8, SM9 | Somwarpet | 12° 43’ 18.76’’ | 75° 53’ 55.7946’’ | This study | ||
| SM10 | Kigga, Chikkamagaluru District | 13° 25’ 1.53’’ | 75° 11’ 17.15’’ | 664 | (Bajpe et al., 2018) | |
| SM12, SM13, SM14 | Makuta, Madikeri district | 12° 9’ 37.99’’ | 75° 47’ 28.93’’ | 755 | ||
| Salacia fruticosa | SF1, SF2, SF3, SF4, SF5, SF6 | Makuta, Madikeri district | 12° 6’ 26.22’’ | 75° 46’ 47.21’’ | 555 | (Bajpe et al., 2018), This study |
| Salacia oblonga | SO1, SO2, SO3, SO4 | Kigga, Chikkamagaluru district | 13° 27’ 7.60’’ | 75° 11’ 51.60’’ | 657 | (Bajpe et al., 2018), this study |
2.2. PCR Amplification of ISSR Marker
Eleven primers were used in the current study [Table 2]. PCR was performed in a 20 ul volume reaction mixture contained 100 uM of dNTPs; 0.5 unit of Taq DNA polymerase; 10× Taq buffer-A (Merck Biosciences), 5 uM primer; and sterile water. For amplification of ISSR following program was used, initial denaturation at 94°C for 300 s, followed by 45 cycle of 94°C for the 60 s, the primer specific annealing temperature for 60 s, and 72°C for 120 s, and a final extension at 72°C for 300 s in Veriti™ 96-Well Thermal Cycler (Applied Biosystems). The amplified products were separated in 1.8% agarose gel stained with ethidium bromide using a 1× Tris-Borate-EDTA buffer. Using Gel Doc™ XR (Bio-Rad), the band patterns on the gel were photographed. Amplified fragments were score visually for the presence (1) or absence (0) of homologous bands. The polymorphism information content (PIC) value was calculated as suggested by Roldàn-Ruiz and Dendauw [13], and the marker index was calculated as defined by Varshney and Chabane [14]. Genetic relatedness among Salacia species and related genus was studied by Unweighted Pair Group Method with Arithmetic (UPGMA) averages cluster analysis using NTSYSpc software 2.02e [15]. For parametric calculation, software POPGENE 1.31 [16] was used. Principal Coordinates Analysis (PCoA) was done using GenAlEx 6.51b2 [17].
Table 2: List of ISSR primers used for the amplification of the 46 samples of Salacia species and closely related members with in Celastraceae family and marker parameters calculated for each ISSR primer used.
| Primer | Primer sequence | Tm | TNB | NPB | PIC | EMR | MI | RP |
|---|---|---|---|---|---|---|---|---|
| ISSR-02 | (CT)8AC | 40 | 8 | 6 | 0.821 | 13.98 | 6.43 | 6.7 |
| ISSR-03 | (CT)8GC | 40 | 9 | 9 | 0.878 | 9.34 | 4.32 | 9.9 |
| ISSR-04 | (CA)6AC | 45 | 13 | 13 | 0.893 | 10.93 | 3.24 | 8.7 |
| ISSR-05 | (CA)6GT | 45 | 9 | 9 | 0.876 | 10.17 | 3.81 | 9.1 |
| ISSR-06 | (CA)6AG | 45 | 11 | 8 | 0.860 | 11.08 | 4.81 | 8.5 |
| ISSR-07 | (CA)6GC | 45 | 9 | 9 | 0.882 | 7.96 | 2.98 | 11.2 |
| ISSR-10 | (GA)6CC | 48 | 10 | 6 | 0.813 | 14.88 | 7.02 | 5.6 |
| ISSR-12 | (CAC)3GC | 37 | 10 | 6 | 0.769 | 16.92 | 6.62 | 5.4 |
| ISSR-13 | (GAG)3GC | 37 | 13 | 10 | 0.891 | 7.62 | 3.19 | 11.8 |
| ISSR-15 | (GTG)3GC | 38 | 7 | 5 | 0.756 | 15.76 | 4.64 | 5.5 |
| ISSR-16 | (GA)9T | 48 | 8 | 5 | 0.791 | 14.23 | 6.68 | 6.6 |
2.3. Barcode Region PCR
For the PCR amplification of the ITS1-5.8s-ITS2 sequence primers, ITS4 and ITS5 were used [18]. For the amplification of matK region primers, 3F_KIM f and 1R_KIM r designed by Ki-Joong Kim (unpublished) were used, and for rbcL region, the primers rbcLa-f and rbcLa-rev were used with PCR protocol as described by the authors.
2.4. Sequence Alignment and Data Analysis
The amplified PCR products were sent to Chromous Biotech, Bengaluru, for sequencing. The sequences were deposited in the NCBI database and the database assigned accession numbers were to the individual samples. The sequences were aligned using the Clustal W option in MEGA 5 software [19]. The sequences were concatenated using the software Sequence Matrix [20]. To estimate the barcoding gap while comparing the distributions of the pairwise intra- and interspecific distance for each barcode candidate with an interval distance of 0.05, software TaxonDNA was used [21], with “pairwise summary function.” Each barcode candidate was measured for the correct identification proportion using TaxonDNA with “best match” (BM) and the “best close match” (BCM) test the species discrimination rates.
3. RESULTS
3.1. Homology Search and Sample Validation
Thirty-two sequences were generated for the ITS region and 18 and 15 sequences were generated for matK and rbcL barcoding regions, respectively. BLASTn was employed for homology search. For ITS sequence, all the samples sequenced in the current study, except S. chinensis samples SC6, SC7, SC8, SC9, SC10, and SC11 showed similarity identity >90% with the Salacia sequence in the database. The sample SC6 to SC10 had a similarity identity ≤85% to Pristimera preussii, and SC11 had a similarity of 85% to Tristemonanthus nigrisilvae, >98% homology with their respective genera. For the matK sequence except for samples SC6 and SC11 which had 99% homology to Apodostigma pallens, all other sequences had 99% homology with Salacia cf. cochinchinensis. For the rbcL sequence, SC10 had 99% homology with Reissantia species. Species P. preussii, T. nigrisilvae, A. pallens, and Reissantia species belong to subfamily Hippocrateoideae which is closely related to subfamily Salacioideae and have most of the morphological characters to be similar. Henceforth, the samples SC6-SC11 were been labeled Hippocrateaceae for the ISSR analysis and these samples were not included in the further analysis.
3.2. ISSR Analysis
A total of 89 loci were generated from 11 primers. Out of the 89 loci scored, 86 loci (96.63%) were found to be polymorphic. The PIC values of 11 ISSR primers ranged from 0.756 to 0.893. Four primers ISSR-02, ISSR-04, ISSR-05, and ISSR-07 showed 100% polymorphism. High RP scores were observed with the primer ISSR-13 (11.8) and ISSR-07 (11.2), and the low RP scores were seen in primers ISSR-12 (5.4), ISSR-15 (5.5) [Figure 1], and ISSR-10 (5.6) with average RP at 8.09 [Table 2]. Within species, polymorphism of C. glauca was 49.44%, C. paniculatus was 25.84%, Hippocrateaceae was 65.17%, L. wallichianum was 47.19%, S. oblonga was 31.46 %, S. macrosperma was 64.04%, S. chinensis was 28.09%, and S. fruticosa was 48.31%. Overall polymorphism for all the 46 samples was 47.42 ± 7.81%. Na, Ne, I, He, and percentage of polymorphic loci of each population are listed in Table 3.
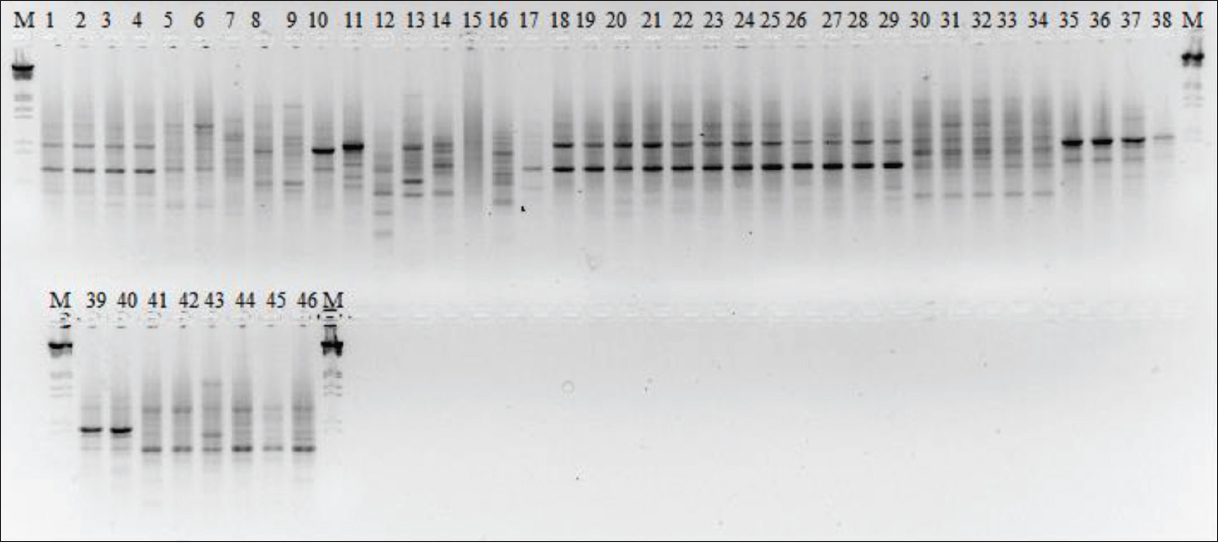 | Figure 1: Inter-simple sequence repeat (ISSR) pattern of 46 samples of Salacia species and closely related members with in Celastraceae family amplified using primer ISSR-15. Lanes 1–4: Salacia oblonga (SO1-SO4), lanes 5–7: Celastrus paniculatus (CP1–CP3), lanes 8 and 9: Cassine glauca (CG1 and CG2), lanes 10 and 11: Lophopetalum wightianum (LW1 and LW2), lane 12: Microtropis stocksii (MS), lane 13: Gymnosporia wallichiana (GC), lane 14: Pleurostylia opposita (p), lane 14: Euonymus indicus (EI), lane 15: Maytenus heyneana (MH). Lanes 16–29: Salacia macrosperma, (SM1–SM14), lanes 30–34: Salacia chinensis (SC1–SC5). Lanes 35–40: Sample belonging to the subfamily Hippocrateoideae (H1–H6), lanes 41–46: Salacia fruticosa (SF1–SF6). M: Molecular weight marker (λ DNA digested with Eco RI and Hind III). [Click here to view] |
Table 3: Genetic variability within the members of Celastraceae family collected from Western Ghats of Karnataka as discerned through ISSR.
| Species | N | Na | Ne | I | He | PPL |
|---|---|---|---|---|---|---|
| Cassine glauca | 2 | 0.899±0.083 | 1.183±0.033 | 0.156±0.028 | 0.107±0.019 | 49.44% |
| Celastrus paniculatus | 3 | 1.225±0.090 | 1.336±0.041 | 0.284±0.032 | 0.193±0.022 | 25.84% |
| Lophopetalum wallichianum | 2 | 1.213±0.088 | 1.334±0.038 | 0.285±0.032 | 0.195±0.022 | 47.19% |
| Salacia oblonga | 4 | 1.124±0.075 | 1.214±0.037 | 0.181±0.029 | 0.123±0.020 | 31.46% |
| Salacia macrosperma | 13 | 1.719±0.071 | 1.573±0.036 | 0.481±0.026 | 0.328±0.018 | 84.27% |
| Salacia fruticosa | 6 | 1.225±0.089 | 1.333±0.042 | 0.162±0.029 | 0.188±0.022 | 48.31% |
| Salacia chinensis 1 | 5 | 1.337±0.098 | 1.200±0.038 | 0.276±0.032 | 0.111±0.020 | 28.09% |
| Hippocrateaceae | 6 | 1.045±0.076 | 1.386±0.039 | 0.343±0.029 | 0.228±0.021 | 65.17% |
| All samples | 46/3.538±0.097 | 0.914±0.024 | 1.197±0.010 | 0.167±0.008 | 0.113±0.005 | 29.21±7.89% |
N: Number of samples, Na: No. of different alleles, Ne: No. of effective alleles, I: Shannon’s Information Index, He: Expected heterozygosity, PPL: Percentage of polymorphic loci
The dendrogram generated from the UPGMA cluster analysis divided 46 samples into two clusters. Cluster 1 contained nine subclusters. Subcluster 1 was further divided into three groups. Group A contained all four samples of S. oblonga and 12 of 13 samples of S. macrosperma. Group B contained five samples of S. chinensis. Group C contained six samples of S. fruticosa.
Subclusters 7 and 9 contained single samples of E. indica and M. wallichiana, respectively. Subclusters 2 and 5 had only samples of C. glauca and C. paniculatus. Subcluster 3 contained P. opposite and M. heyneana. Subcluster 6 contained sample LW2 of L. wallichianum and G. montana. Subcluster 4 contained samples SC10, SC11, and LW1. Subcluster 8 contained samples SC6, SC7, and SC9. Finally, cluster 2 contained sample SC9 and SM1 [Figure 2].
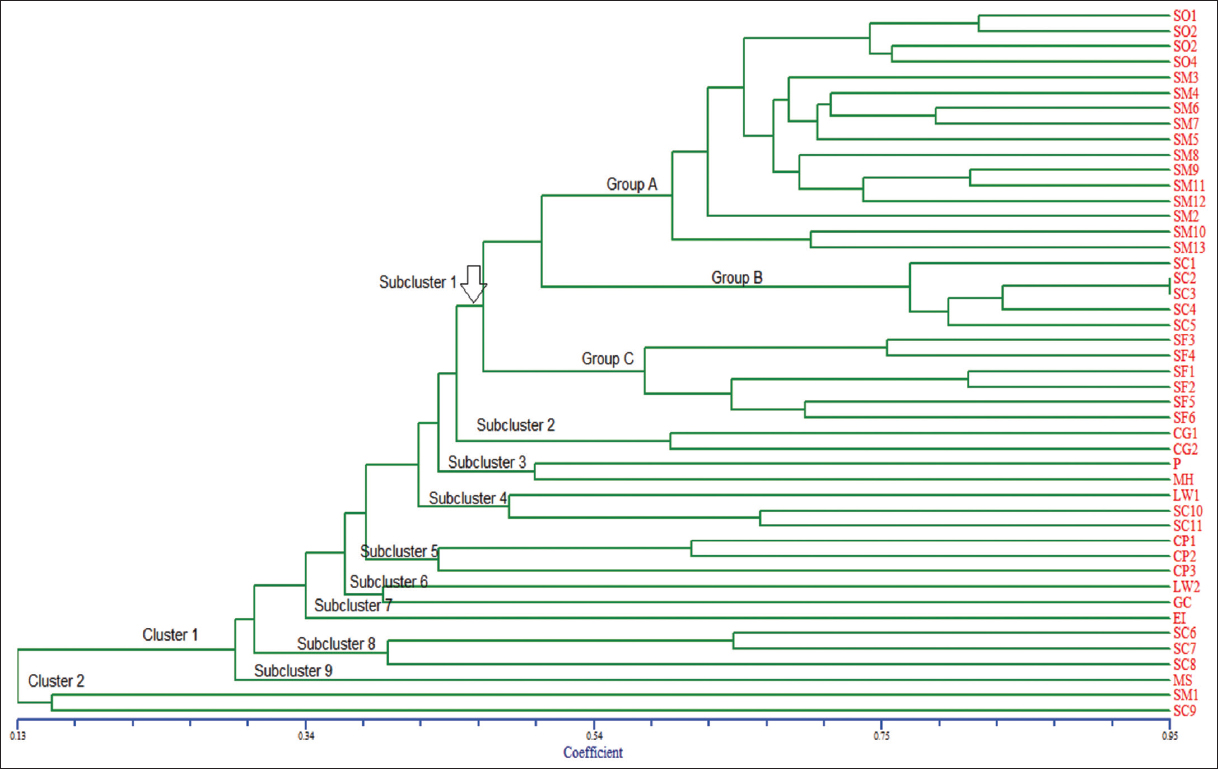 | Figure 2: UPGMA dendrogram of 46 samples of Salacia species and closely related members with in Celastraceae family based on inter-simple sequence repeat marker. [Click here to view] |
The three axes PCoA accounted for a cumulative variation of 46.87% [Figure 3]. The first axis and the second axis accounted for 24.84% and 12.21%, respectively. From the AMOVA data, we found out that 36% of the molecular variance was found among the population while within the population, this value was found to be 64%.
 | Figure 3: Inter-simple sequence repeat data principal coordinates analysis (PCoA) of 46 samples of Salacia species and closely related members with in Celastraceae family. PCoA accounting for 46.87% of cumulative variation, with the first, second, and third component accounting for 24.84%, 12.21%, and 9.82%, respectively. [Click here to view] |
3.3. Intra- and Interspecific Diversity and Barcoding Gap
The aligned sequence length of ITS was 870 bp, matK was 848 bp, and rbcL was 700 bp. Among the single barcode, ITS had the most variable sites of 588 bp and parsimony-informative characters of 265 bp [Table 4]. The pairwise intraspecific and interspecific distances in the seven barcode combinations ranged from 0.0% to a maximum of 58% and 0% to 63%, respectively. The mean intraspecific and interspecific distances were the lowest for rbcL and highest for ITS [Table 5]. Among seven barcodes, none had visible barcoding gaps. All barcodes had overlaps between their intra- and interspecific distances without distinct barcoding gaps [Figure 4].
Table 4: Evaluation of three DNA barcodes and combinations of the barcodes.
| Barcode | Number of samples | No. of species | Length of aligned sequence (bp) | No. of parsimony informative sites/variable sites | Ability to discriminate (tree-based method-NJ) |
|---|---|---|---|---|---|
| ITS | 99 | 11 | 870 | 240/380 | 67.13% |
| matK | 94 | 9 | 848 | 37/56 | 8.59% |
| rbcL | 91 | 9 | 700 | 14/37 | 10.39% |
| ITS+matK | 99 | 12 | 1718 | 275/419 | 36.60% |
| ITS+rbcL | 99 | 12 | 1570 | 253/407 | 41.98% |
| matK+rbcL | 72 | 9 | 1545 | 15/30 | 5.09% |
| ITS+matK+rbcl | 99 | 12 | 2418 | 288/446 | 31.30% |
Table 5: Summary of the pairwise intraspecific and interspecific distances in the barcode loci of Salacia species of Western Ghats (Karnataka and Kerala).
| Barcode locus | Intraspecific distance | Mean | Interspecific distance | Mean | Threshold |
|---|---|---|---|---|---|
| Maximum | Maximum | ||||
| ITS | 59% | 1.64 | 63% | 5.42 | 33.08% |
| matK | 4% | 0.11 | 6% | 0.56 | 0.61% |
| rbcL | 2.5% | 0.07 | 3.5% | 0.38 | 1.71% |
| ITS+matK | 58.0% | 0.72 | 63% | 3.34 | 24.52% |
| ITS+rbcL | 59% | 0.88 | 63% | 3.59 | 25.32% |
| matK+rbcL | 4% | 0.11 | 6% | 0.53 | 1.16% |
| ITS+matK+rbcL | 58% | 0.58 | 63% | 2.93 | 23.65% |
Minimum interspecific distance is 0% for all the barcodes.
Threshold as computed for the pairwise summary
 | Figure 4: (a-g) Relative distribution of intraspecific and interspecific Kimura 2-parameter distance for three DNA barcodes and its combination in genus Salacia of Western Ghats of Karnataka and Kerala. [Click here to view] |
3.4. Species Discrimination
Analysis using TaxonDNA’s BM and BCM and neighbor-joining (NJ) TBM to evaluate the discriminatory power of the three barcodes in Salacia species provided varied results. In both methods, the highest species discrimination success was obtained by ITS (BC and BCM – 59.59% and TBM – 67%) among the three single barcodes. The other two plastid DNA barcodes, that is, matK and rbcL had 41.48% (BC and BCM) and 8.59% (TBM), 30.76% (BC and BCM), and 10.39% (TBM) species identification rate, respectively [Tables 4 and 6]. When barcodes were combined, the highest discrimination rate of 41.98% was observed in ITS + rbcL (TBM) and 49.55% in ITS + matK (BC and BCM) [Tables 4 and 6]. The bootstrap support values of single barcodes were higher than the barcode combinations. Both the BM and BCM methods provided the same species discrimination success [Table 2].
Table 6: Identification success based on the “best match” and “best close match” function of the program TaxonDNA.
| Barcode | Best match | Best close match | T | Cl | |||||
|---|---|---|---|---|---|---|---|---|---|
| C | A | I | C | A | I | No match | |||
| ITS | 59 (59.59%) | 32 (32.32%) | 8 (8.08%) | 58 (58.58%) | 32 (32.32%) | 8 (8.08%) | 1 (1.01%) | 3 | 4 |
| matK | 39 (41.48%) | 53 (56.38%) | 2 (2.12%) | 39 (41.48%) | 53 (56.38%) | 2 (2.12%) | 0 (0.0%) | 0.61 | 2 |
| rbcL | 28 (30.76%) | 57 (62.63%) | 6 (6.59%) | 28 (30.76%) | 57 (62.63%) | 6 (6.59%) | 0 (0.0%) | 1.71 | 1 |
| ITS+matK | 56 (49.55%) | 45 (39.82%) | 12 (10.61%) | 56 (49.55%) | 45 (39.82%) | 11 (9.73%) | 1 (0.88%) | 3 | 3 |
| ITS+rbcL | 49 (42.6%) | 58 (50.43%) | 8 (6.95%) | 48 (41.73%) | 58 (50.43%) | 8 (6.95%) | 1 (0.86%) | 3 | 2 |
| matK+rbcL | 34 (32.69%) | 64 (61.53%) | 6 (5.76%) | 34 (32.69%) | 64 (61.53%) | 5 (4.8%) | 1 (0.96%) | 1.16 | 3 |
| ITS+matK+rbcl | 42 (35.59%) | 64 (54.23%) | 12 (10.16%) | 42 (35.59%) | 64 (54.23%) | 11 (9.32%) | 1 (0.84%) | 3 | 2 |
4. DISCUSSION
ISSR motifs GA, CA, CT, GTG, CAC, and GAG were used in the present study, of which CA and GAG motifs produced maximum scorable loci. The overall mean percentage of polymorphism of only Salacia species (28 samples) was 48.03 ± 12.86%. This could be due to the addition of seven more samples in the current study. Similarly, higher polymorphism for medicinal plants sampled at a particular geographical region within India is observed in Oroxylum indicum (49.61%) [13]. ISSR has proved to be useful in species discrimination as it can detect very low variation with closely related species [9,14].
From our sample survey, we found out that the identification of Salacia species is indeed difficult as S. oblonga and S. macrosperma had very similar morphological characters. Salacia species can be easily misidentified and incorrect species can be used in the medicinal preparations, especially when two or more species are present in the area of sampling. Have used the random amplified polymorphic DNA marker of identifying genetic similarity in seven Salacia species samples from Wayanad, Kerala.
From Nei genetic distance and identity, S. oblonga is closely associated with S. macrosperma than S. chinensis and S. fruticosa. Within Salacia species, S. macrosperma has the highest percentage of polymorphic of 84.27% followed by S. fruticosa (48.31%) and S. oblonga. Since some of the species of Salacia are endangered, the effort to conserve germplasm by micropropagation of S. chinensis [15], S. reticulata [16], and S. oblonga [17] has been carried out. Molecular analysis suggests that S. chinensis has the lowest variation within the species, making it a suitable candidate for large-scale micropropagation, conservation, and alternative for other endangered species of Salacia. The use of S. reticulata in traditional medicine in Sri Lanka has created pressure for the supply of this plant from the wild. Therefore, the use of S. chinensis as a substituent has been suggested by Keeragalaarachchi and Dharmadasa [18]. Salacia species considered in the current study have been known to contain catechins, quinones, friedo-oleanones, quinone methide, and triterpenoids in them, which may be responsible for their pharmacological and medicinal properties.
Dev and Anoop [6] analyzing Salacia sampled from Kerala by NJ tree-based method observed that ITS2 and trnH-psbA region had 100% and 40% efficiency in species discrimination, while matK and rbcL failed in species discrimination. However, our present study using TaxonDNA software indicated that matK and rbcL barcodes are also useful in the correct identification of Salacia species sampled from Western Ghats of Karnataka and Kerala.
CBOL has recommended a combination of matK+rbcL as a universal barcode for plants. However, in the present study, the single barcode had higher discriminatory success than the combination of two or three barcodes. A combination of matK+rbcL had the lower species resolution of 32.69% in “BM” and lowest in the NJ tree-based method (5.09%) among all the combinations. This may be due to the low substitution rates of these coding genes. A very low species delineation was also observed in genus Nepenthes concerning rbcL+matK barcodes [19].
5. CONCLUSION
The ISSR DNA markers and barcodes (ITS, matK, and rbcL) are useful tools for the molecular validation of taxonomically described plant organisms. No single DNA marker is sufficient for the proper authentication of Salacia species and its associated species. It is essential to identify the most effective marker and its efficiency of species discrimination, for proper molecular identification. This study shows that Salacia ISSR marker is useful for the assessment of genetic diversity and that the ITS sequence can also be used for identification in a satisfactory manner.
6. ACKNOWLEDGMENT
The authors express their sincere thanks to Head of Department, Botany and Biotechnology, Dr. B. A Kumara Hegde and Dr. B. Yashovarma, Secretary, Sri Dharmasthala Manjunatheshwara Educational Society, Ujire, for encouragement and facilities.
7. CONFLICTS OF INTEREST
All the authors declare that there are no conflicts of interest regarding the publication of this paper.
8. FINANCIAL SUPPORT
None.
9. AUTHORS’ CONTRIBUTIONS STATEMENT
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
REFERENCES
1. Chawla A, Singh S, Sharma AK. Salacia oblonga wall: A review on its pharmacognostic, phytochemical and pharmacological aspects. Int J Res Pharm Biomed Sci 2013;4:1215-28.
2. Singh BA, Duggal S, Singh A. Salacia spp: Hypoglycemic principles and possible role in diabetes management. Integr Med 2010;9:40-3.
3. Myers N. Threatened biotas: "Hot spots" in tropical forests. Environmentalist 1988;8:187-208. [CrossRef]
4. Prasad V, Farooqui A, Tripathi SM, Garg R, Thakur B. Evidence of late palaeocene-early eocene equatorial rain forest refugia in Southern Western Ghats, India. J Biosci 2009;34:777-97. [CrossRef]
5. Udayan P, Yohannan R, Devipriya M, Devipriya V, Pradeep A. A new species of Salacia (Hippocrateaceae) from south India. Edinb J Bot 2012;69:255-8. [CrossRef]
6. Dev SA, Anoop B, Anoja K, Udayan P, Muralidharan E. Species discrimination through DNA barcoding in the genus Salacia of the Western Ghats in India. Nord J Bot 2015;33:722-8. [CrossRef]
7. Devipriya M, Devipriya V, Udayan P, Yohannan R. Molecular systematics of Indian Salacia based on ITS sequences of nrDNA. Int J Adv Res 2015;3:1513-21.
8. Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994;20:176-83. [CrossRef]
9. Reddy MP, Sarla N, Siddiq E. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002;128:9-17. [CrossRef]
10. Kress WJ, Erickson DL, Swenson NG, Thompson J, Uriarte M, Zimmerman JK. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS One 2010;5:e15409. [CrossRef]
11. Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 2003;29:417-34. [CrossRef]
12. Ginwal H, Mittal N. An efficient genomic DNA isolation protocol for RAPD and SSR analysis in Acorus calamus L. Indian J Biotechnol 2010;9:213-6.
13. Jayaram K, Prasad M. Genetic diversity in Oroxylum indicum (L.) Vent. (Bignoniaceae), a vulnerable medicinal plant by random amplified polymorphic DNA marker. Afr J Biotechnol 2008;7:253-62.
14. Fernández M, Figueiras A, Benito C. The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor Appl Genet 2002;104:845-51. [CrossRef]
15. Chavan JJ, Ghadage DM, Bhoite AS, Umdale SD. Micropropagation, molecular profiling and RP-HPLC determination of mangiferin across various regeneration stages of Saptarangi (Salacia chinensis L.). Ind Crop Prod 2015;76:1123-32. [CrossRef]
16. Dhanasri G, Reddy MS, Naresh B, Cherku D. Micropropagation of Salacia reticulata-an endangered medicinal plant. Plant Tissue Cult Biotechnol 2013;23:221-9. [CrossRef]
17. Deepak K, Suneetha G, Surekha C. A simple and effective method for vegetative propagation of an endangered medicinal plant Salacia oblonga Wall. J Nat Med 2016;70:115-9. [CrossRef]
18. Keeragalaarachchi K, Dharmadasa R, Wijesekara R, Kudavidanage EP. Natural antidiabetic potential of Salacia chinensis L. (Celastraceae) based on morphological, phytochemical, physico-chemical and bioactivity: A promising alternative for Salacia reticulata Thw. World J Agric Res 2016;4:49-55.
19. Gogoi B, Bhau BS. DNA barcoding of the genus Nepenthes (Pitcher plant): A preliminary assessment towards its identification. BMC Plant Biol 2018;18:153. https://doi.org/10.1186/s12870-018-1375-5